Abstract
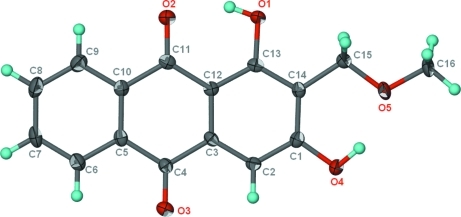
The title compound, C16H12O5, common name: lucidin ω-methyl ether, exists as a planar molecule (r.m.s. deviation = 0.04 Å). Within the molecule, the 1-hydroxy group forms a hydrogen bond to the adjacent carbonyl O atom, and the 3-hydroxy group forms a hydrogen bond to the adjacent methoxy O atom. The methoxy O atom is disordered over two positions of equal occupancy.
Related literature
The title compound has been isolated from several plants: Rubia tinctorum L. (Boldizsar et al., 2004 ▶), taurina subsp. caucasica (Ozgen et al., 2006 ▶), Prismatomeris fragrans (Kanokmedhakul et al., 2005 ▶), Crucianella maritima L. (El-Lakany et al., 2004 ▶), Rubia wallichiana Decne (Wu et al., 2003 ▶), Morinda elliptica (Ali et al., 2000 ▶; Ismail et al., 1997 ▶; Ismail et al., 2002 ▶), Ophiorrhiza pumila (Kitajima et al., 1998 ▶), Morinda officinalis How. (Yoshikawa et al., 1995 ▶), Galiumspurium var. echinospermon (Koyama et al., 1993 ▶), Damnacanthus indicus (Koyama et al., 1992 ▶), Rubia cordifolia L. (Vidal-Tessier et al., 1987 ▶), Faramea cyanea (Ferrari et al., 1985 ▶), Morinda parvifolia (Chang & Lee, 1984 ▶) and Galium album (Kupier & Labadie, 1984 ▶).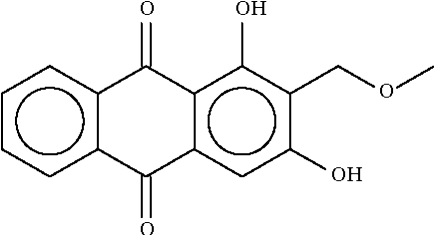
Experimental
Crystal data
C16H12O5
M r = 284.26
Monoclinic,
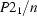
a = 4.6725 (1) Å
b = 39.685 (1) Å
c = 6.9869 (2) Å
β = 107.654 (2)°
V = 1234.55 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.12 mm−1
T = 100 K
0.30 × 0.07 × 0.02 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
10046 measured reflections
2825 independent reflections
1888 reflections with I > 2σ(I)
R int = 0.041
Refinement
R[F 2 > 2σ(F 2)] = 0.054
wR(F 2) = 0.156
S = 1.01
2825 reflections
201 parameters
4 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.43 e Å−3
Δρmin = −0.46 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809017607/tk2447sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809017607/tk2447Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1o⋯O2 | 0.85 (1) | 1.79 (2) | 2.557 (2) | 150 (3) |
| O4—H4o⋯O5 | 0.84 (1) | 1.77 (2) | 2.546 (7) | 152 (4) |
| O4—H4o⋯O5′ | 0.84 (1) | 1.77 (2) | 2.539 (7) | 152 (4) |
Acknowledgments
We thank Universiti Teknologi MARA and the University of Malaya for supporting this study.
supplementary crystallographic information
Experimental
About 1 kg of the root of Rennelia elliptica Korth., which was collected from the Kuala Keniam National Park, Malaysia, was extracted with dichloromethane. The solvent was removed to give a crude material (approx. 10 g) that was fractionated on a chromatography column (60 x 5 cm) packed with silica. The silica had been previously immersed in 4% oxalic acid and then activated by heating to 363 K. The fractions were eluted with hexane–dichloromethane and dichloromethane–methanol in increasing polarity. The fraction eluted with hexane–dichloromethane (2:8 v/v) was purified by thin layer chromatography (2 mm). The product was recrystallized from dichloromethane to furnish yellow crystals. The formulation was established by 1H- and 13C-NMR spectroscopy.
Refinement
Hydrogen atoms were placed at calculated positions (C–H 0.95–0.99 Å) and were treated as riding on their parent carbon atoms, with U(H) set to 1.2–1.5 times Ueq(C). The hydroxy H-atoms were located in a difference Fourier map, and were refined with a distance restraint of O–H 0.84±0.01 Å; their temperature factors were refined.
The methoxy oxygen atom is disordered over two positions, but the occupancy could not be refined. The disorder was assumed to be 50:50. The C–O/C–O' bonds to the aryl group were restrained to within 0.01 Å of each other, as were those to the alkyl group. The anisotropic displacement factors of the primed atom were restrained to those of the umprimed one.
Figures
Fig. 1.
Thermal ellipsoid plot (Barbour, 2001) of the molecule of C16H12O5 at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius. The disorder is not shown
Crystal data
| C16H12O5 | F(000) = 592 |
| Mr = 284.26 | Dx = 1.529 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1810 reflections |
| a = 4.6725 (1) Å | θ = 3.1–27.9° |
| b = 39.685 (1) Å | µ = 0.12 mm−1 |
| c = 6.9869 (2) Å | T = 100 K |
| β = 107.654 (2)° | Plate, yellow |
| V = 1234.55 (6) Å3 | 0.30 × 0.07 × 0.02 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 1888 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.041 |
| graphite | θmax = 27.5°, θmin = 1.0° |
| ω scans | h = −5→6 |
| 10046 measured reflections | k = −51→51 |
| 2825 independent reflections | l = −9→8 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.054 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.156 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0691P)2 + 1.3159P] where P = (Fo2 + 2Fc2)/3 |
| 2825 reflections | (Δ/σ)max = 0.001 |
| 201 parameters | Δρmax = 0.43 e Å−3 |
| 4 restraints | Δρmin = −0.46 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.5327 (4) | 0.35893 (4) | 0.3196 (2) | 0.0173 (4) | |
| H1o | 0.476 (7) | 0.3392 (4) | 0.338 (5) | 0.039 (9)* | |
| O2 | 0.2892 (4) | 0.31121 (4) | 0.4610 (3) | 0.0200 (4) | |
| O3 | 0.0049 (4) | 0.39767 (4) | 0.9709 (2) | 0.0189 (4) | |
| O4 | 0.5244 (4) | 0.46858 (4) | 0.5956 (3) | 0.0202 (4) | |
| H4o | 0.601 (8) | 0.4724 (9) | 0.503 (4) | 0.051 (11)* | |
| O5 | 0.777 (3) | 0.45857 (15) | 0.3246 (15) | 0.027 (2) | 0.50 |
| O5' | 0.694 (3) | 0.46028 (15) | 0.2862 (15) | 0.027 (2) | 0.50 |
| C1 | 0.4569 (5) | 0.43535 (6) | 0.5813 (3) | 0.0144 (5) | |
| C2 | 0.3075 (5) | 0.42345 (6) | 0.7145 (3) | 0.0137 (5) | |
| H2 | 0.2585 | 0.4386 | 0.8052 | 0.016* | |
| C3 | 0.2318 (5) | 0.38992 (6) | 0.7142 (3) | 0.0129 (5) | |
| C4 | 0.0737 (5) | 0.37804 (6) | 0.8575 (3) | 0.0135 (5) | |
| C5 | 0.0051 (5) | 0.34142 (6) | 0.8590 (3) | 0.0139 (5) | |
| C6 | −0.1291 (5) | 0.32923 (6) | 0.9985 (4) | 0.0183 (5) | |
| H6 | −0.1774 | 0.3443 | 1.0900 | 0.022* | |
| C7 | −0.1917 (6) | 0.29523 (6) | 1.0034 (4) | 0.0217 (5) | |
| H7 | −0.2798 | 0.2869 | 1.1001 | 0.026* | |
| C8 | −0.1268 (6) | 0.27307 (6) | 0.8677 (4) | 0.0229 (6) | |
| H8 | −0.1728 | 0.2498 | 0.8707 | 0.028* | |
| C9 | 0.0054 (6) | 0.28509 (6) | 0.7283 (4) | 0.0203 (5) | |
| H9 | 0.0490 | 0.2700 | 0.6352 | 0.024* | |
| C10 | 0.0745 (5) | 0.31922 (6) | 0.7240 (3) | 0.0146 (5) | |
| C11 | 0.2280 (5) | 0.33153 (6) | 0.5794 (3) | 0.0141 (5) | |
| C12 | 0.3044 (5) | 0.36698 (6) | 0.5816 (3) | 0.0132 (5) | |
| C13 | 0.4545 (5) | 0.37961 (6) | 0.4486 (3) | 0.0129 (5) | |
| C14 | 0.5296 (5) | 0.41375 (6) | 0.4458 (3) | 0.0131 (5) | |
| C15 | 0.6804 (5) | 0.42442 (6) | 0.2919 (3) | 0.0153 (5) | |
| H15A | 0.8555 | 0.4097 | 0.3022 | 0.018* | 0.50 |
| H15B | 0.5378 | 0.4220 | 0.1551 | 0.018* | 0.50 |
| H15C | 0.8858 | 0.4149 | 0.3277 | 0.018* | 0.50 |
| H15D | 0.5654 | 0.4157 | 0.1578 | 0.018* | 0.50 |
| C16 | 0.8677 (6) | 0.47240 (6) | 0.1631 (4) | 0.0221 (6) | |
| H16A | 0.9308 | 0.4958 | 0.1938 | 0.033* | 0.50 |
| H16B | 0.6989 | 0.4716 | 0.0390 | 0.033* | 0.50 |
| H16C | 1.0357 | 0.4592 | 0.1459 | 0.033* | 0.50 |
| H16D | 0.9262 | 0.4958 | 0.1984 | 0.033* | 0.50 |
| H16E | 0.7474 | 0.4711 | 0.0215 | 0.033* | 0.50 |
| H16F | 1.0482 | 0.4585 | 0.1853 | 0.033* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0221 (9) | 0.0145 (9) | 0.0194 (9) | −0.0005 (7) | 0.0124 (7) | −0.0022 (7) |
| O2 | 0.0264 (9) | 0.0162 (8) | 0.0209 (9) | −0.0008 (7) | 0.0122 (8) | −0.0020 (7) |
| O3 | 0.0220 (9) | 0.0190 (9) | 0.0184 (9) | 0.0000 (7) | 0.0102 (7) | −0.0021 (7) |
| O4 | 0.0303 (10) | 0.0136 (8) | 0.0205 (9) | −0.0041 (7) | 0.0134 (8) | −0.0010 (7) |
| O5 | 0.036 (6) | 0.0142 (10) | 0.043 (3) | 0.0027 (18) | 0.033 (4) | 0.0050 (14) |
| O5' | 0.036 (6) | 0.0142 (10) | 0.043 (3) | 0.0027 (18) | 0.033 (4) | 0.0050 (14) |
| C1 | 0.0138 (11) | 0.0130 (11) | 0.0156 (11) | −0.0001 (9) | 0.0032 (9) | 0.0012 (8) |
| C2 | 0.0135 (11) | 0.0143 (11) | 0.0138 (11) | 0.0003 (8) | 0.0050 (9) | −0.0016 (8) |
| C3 | 0.0099 (11) | 0.0171 (12) | 0.0121 (11) | 0.0005 (9) | 0.0038 (9) | 0.0006 (8) |
| C4 | 0.0122 (11) | 0.0151 (11) | 0.0131 (11) | 0.0003 (9) | 0.0040 (9) | 0.0008 (9) |
| C5 | 0.0103 (11) | 0.0164 (11) | 0.0147 (11) | −0.0009 (8) | 0.0032 (9) | 0.0023 (9) |
| C6 | 0.0180 (12) | 0.0196 (12) | 0.0189 (12) | 0.0006 (10) | 0.0082 (10) | 0.0024 (9) |
| C7 | 0.0217 (13) | 0.0231 (13) | 0.0230 (13) | −0.0038 (10) | 0.0110 (10) | 0.0064 (10) |
| C8 | 0.0245 (13) | 0.0147 (12) | 0.0308 (14) | −0.0030 (10) | 0.0102 (11) | 0.0037 (10) |
| C9 | 0.0217 (13) | 0.0151 (12) | 0.0248 (13) | −0.0005 (9) | 0.0083 (11) | −0.0011 (10) |
| C10 | 0.0135 (11) | 0.0144 (11) | 0.0154 (12) | 0.0015 (9) | 0.0038 (9) | 0.0023 (9) |
| C11 | 0.0116 (11) | 0.0166 (11) | 0.0137 (11) | 0.0035 (9) | 0.0031 (9) | 0.0007 (9) |
| C12 | 0.0128 (11) | 0.0136 (11) | 0.0124 (11) | 0.0008 (8) | 0.0027 (9) | 0.0002 (8) |
| C13 | 0.0098 (11) | 0.0163 (11) | 0.0114 (11) | 0.0022 (8) | 0.0015 (8) | −0.0008 (8) |
| C14 | 0.0104 (11) | 0.0147 (11) | 0.0143 (11) | 0.0003 (8) | 0.0036 (9) | 0.0025 (9) |
| C15 | 0.0168 (12) | 0.0146 (11) | 0.0157 (11) | −0.0002 (9) | 0.0068 (9) | 0.0001 (9) |
| C16 | 0.0273 (14) | 0.0184 (12) | 0.0257 (14) | −0.0043 (10) | 0.0156 (11) | 0.0049 (10) |
Geometric parameters (Å, °)
| O1—C13 | 1.349 (3) | C7—C8 | 1.392 (4) |
| O1—H1o | 0.848 (10) | C7—H7 | 0.9500 |
| O2—C11 | 1.249 (3) | C8—C9 | 1.387 (3) |
| O3—C4 | 1.222 (3) | C8—H8 | 0.9500 |
| O4—C1 | 1.353 (3) | C9—C10 | 1.395 (3) |
| O4—H4o | 0.842 (10) | C9—H9 | 0.9500 |
| O5—C15 | 1.425 (6) | C10—C11 | 1.488 (3) |
| O5—C16 | 1.431 (6) | C11—C12 | 1.450 (3) |
| O5'—C15 | 1.426 (6) | C12—C13 | 1.415 (3) |
| O5'—C16 | 1.432 (6) | C13—C14 | 1.401 (3) |
| C1—C14 | 1.393 (3) | C14—C15 | 1.514 (3) |
| C1—C2 | 1.404 (3) | C15—H15A | 0.9900 |
| C2—C3 | 1.377 (3) | C15—H15B | 0.9900 |
| C2—H2 | 0.9500 | C15—H15C | 0.9900 |
| C3—C12 | 1.412 (3) | C15—H15D | 0.9900 |
| C3—C4 | 1.490 (3) | C16—H16A | 0.9800 |
| C4—C5 | 1.489 (3) | C16—H16B | 0.9800 |
| C5—C6 | 1.396 (3) | C16—H16C | 0.9800 |
| C5—C10 | 1.399 (3) | C16—H16D | 0.9800 |
| C6—C7 | 1.383 (3) | C16—H16E | 0.9800 |
| C6—H6 | 0.9500 | C16—H16F | 0.9800 |
| C13—O1—H1o | 107 (2) | C3—C12—C13 | 117.9 (2) |
| C1—O4—H4o | 105 (2) | C3—C12—C11 | 121.7 (2) |
| C15—O5—C16 | 113.1 (5) | C13—C12—C11 | 120.4 (2) |
| C15—O5'—C16 | 113.0 (5) | O1—C13—C14 | 117.32 (19) |
| O4—C1—C14 | 123.4 (2) | O1—C13—C12 | 120.8 (2) |
| O4—C1—C2 | 115.5 (2) | C14—C13—C12 | 121.9 (2) |
| C14—C1—C2 | 121.1 (2) | C1—C14—C13 | 118.1 (2) |
| C3—C2—C1 | 120.2 (2) | C1—C14—C15 | 124.9 (2) |
| C3—C2—H2 | 119.9 | C13—C14—C15 | 116.92 (19) |
| C1—C2—H2 | 119.9 | O5—C15—C14 | 110.1 (3) |
| C2—C3—C12 | 120.8 (2) | O5'—C15—C14 | 109.5 (3) |
| C2—C3—C4 | 119.0 (2) | O5—C15—H15A | 109.6 |
| C12—C3—C4 | 120.3 (2) | O5'—C15—H15A | 123.2 |
| O3—C4—C5 | 121.2 (2) | C14—C15—H15A | 109.6 |
| O3—C4—C3 | 121.1 (2) | O5—C15—H15B | 109.6 |
| C5—C4—C3 | 117.73 (19) | C14—C15—H15B | 109.6 |
| C6—C5—C10 | 119.8 (2) | H15A—C15—H15B | 108.2 |
| C6—C5—C4 | 119.1 (2) | O5'—C15—H15C | 109.8 |
| C10—C5—C4 | 121.0 (2) | C14—C15—H15C | 109.8 |
| C7—C6—C5 | 119.9 (2) | O5'—C15—H15D | 109.8 |
| C7—C6—H6 | 120.0 | C14—C15—H15D | 109.8 |
| C5—C6—H6 | 120.0 | H15C—C15—H15D | 108.2 |
| C6—C7—C8 | 120.5 (2) | O5—C16—H16A | 109.5 |
| C6—C7—H7 | 119.7 | O5—C16—H16B | 109.5 |
| C8—C7—H7 | 119.7 | H16A—C16—H16B | 109.5 |
| C9—C8—C7 | 119.8 (2) | O5—C16—H16C | 109.5 |
| C9—C8—H8 | 120.1 | H16A—C16—H16C | 109.5 |
| C7—C8—H8 | 120.1 | H16B—C16—H16C | 109.5 |
| C8—C9—C10 | 120.3 (2) | O5—C16—H16D | 107.0 |
| C8—C9—H9 | 119.9 | O5'—C16—H16D | 109.5 |
| C10—C9—H9 | 119.9 | H16B—C16—H16D | 110.0 |
| C9—C10—C5 | 119.7 (2) | H16C—C16—H16D | 111.3 |
| C9—C10—C11 | 119.7 (2) | O5'—C16—H16E | 109.5 |
| C5—C10—C11 | 120.6 (2) | H16D—C16—H16E | 109.5 |
| O2—C11—C12 | 121.9 (2) | O5'—C16—H16F | 109.5 |
| O2—C11—C10 | 119.5 (2) | H16D—C16—H16F | 109.5 |
| C12—C11—C10 | 118.62 (19) | H16E—C16—H16F | 109.5 |
| O4—C1—C2—C3 | −179.2 (2) | C2—C3—C12—C11 | 179.7 (2) |
| C14—C1—C2—C3 | 0.5 (3) | C4—C3—C12—C11 | 0.2 (3) |
| C1—C2—C3—C12 | 0.4 (3) | O2—C11—C12—C3 | 179.3 (2) |
| C1—C2—C3—C4 | 179.9 (2) | C10—C11—C12—C3 | −1.2 (3) |
| C2—C3—C4—O3 | 1.7 (3) | O2—C11—C12—C13 | −0.5 (3) |
| C12—C3—C4—O3 | −178.8 (2) | C10—C11—C12—C13 | 179.0 (2) |
| C2—C3—C4—C5 | −177.7 (2) | C3—C12—C13—O1 | 179.4 (2) |
| C12—C3—C4—C5 | 1.8 (3) | C11—C12—C13—O1 | −0.9 (3) |
| O3—C4—C5—C6 | −2.7 (3) | C3—C12—C13—C14 | −0.3 (3) |
| C3—C4—C5—C6 | 176.8 (2) | C11—C12—C13—C14 | 179.5 (2) |
| O3—C4—C5—C10 | 177.7 (2) | O4—C1—C14—C13 | 178.4 (2) |
| C3—C4—C5—C10 | −2.9 (3) | C2—C1—C14—C13 | −1.2 (3) |
| C10—C5—C6—C7 | 0.3 (3) | O4—C1—C14—C15 | −2.6 (4) |
| C4—C5—C6—C7 | −179.4 (2) | C2—C1—C14—C15 | 177.8 (2) |
| C5—C6—C7—C8 | −1.1 (4) | O1—C13—C14—C1 | −178.5 (2) |
| C6—C7—C8—C9 | 0.8 (4) | C12—C13—C14—C1 | 1.1 (3) |
| C7—C8—C9—C10 | 0.3 (4) | O1—C13—C14—C15 | 2.4 (3) |
| C8—C9—C10—C5 | −1.1 (4) | C12—C13—C14—C15 | −178.0 (2) |
| C8—C9—C10—C11 | 177.4 (2) | C16—O5—C15—O5' | −77.9 (18) |
| C6—C5—C10—C9 | 0.8 (3) | C16—O5—C15—C14 | −168.5 (6) |
| C4—C5—C10—C9 | −179.5 (2) | C16—O5'—C15—O5 | 77.2 (17) |
| C6—C5—C10—C11 | −177.7 (2) | C16—O5'—C15—C14 | 172.5 (6) |
| C4—C5—C10—C11 | 2.0 (3) | C1—C14—C15—O5 | 8.4 (6) |
| C9—C10—C11—O2 | 1.1 (3) | C13—C14—C15—O5 | −172.6 (6) |
| C5—C10—C11—O2 | 179.6 (2) | C1—C14—C15—O5' | −8.8 (6) |
| C9—C10—C11—C12 | −178.4 (2) | C13—C14—C15—O5' | 170.2 (6) |
| C5—C10—C11—C12 | 0.1 (3) | C15—O5—C16—O5' | 77.8 (18) |
| C2—C3—C12—C13 | −0.5 (3) | C15—O5'—C16—O5 | −77.3 (17) |
| C4—C3—C12—C13 | 179.98 (19) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1o···O2 | 0.85 (1) | 1.79 (2) | 2.557 (2) | 150 (3) |
| O4—H4o···O5 | 0.84 (1) | 1.77 (2) | 2.546 (7) | 152 (4) |
| O4—H4o···O5' | 0.84 (1) | 1.77 (2) | 2.539 (7) | 152 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2447).
References
- Ali, A. M., Ismail, N. H., Mackeen, M. M., Yazan, L. S., Mohamed, S. M., Ho, A. S. H. & Lajis, N. H. (2000). Pharm. Biol.38, 298–301. [DOI] [PubMed]
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Boldizsar, I., Laszlo-Bencsik, A., Szucs, Z. & Danos, B. (2004). Acta Pharm. Hung.74, 142–148. [PubMed]
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chang, P. & Lee, K. S. (1984). Phytochemistry, 23, 1733–1736.
- El-Lakany, A. M., Aboul-Ela, M. A., Abdel-Kader, M. S., Badr, J. M., Sabri, N. N. & Goher, Y. (2004). Nat. Prod. Sci.10, 63–68.
- Ferrari, F., Delle Monache, G. & Alves de Lima, R. (1985). Phytochemistry, 24, 2753–2755.
- Ismail, N. H., Ali, A. M., Aimi, N., Kitajima, M., Takayama, H. & Lajis, N. H. (1997). Phytochemistry, 45, 1723–1725.
- Ismail, N. H., Mohamad, H., Mohidin, A. & Lajis, N. H. (2002). Nat. Prod. Sci.8, 48–51.
- Kanokmedhakul, K., Kanokmedhakul, S. & Phatchana, R. (2005). J. Ethnopharmacol.100, 284–288. [DOI] [PubMed]
- Kitajima, M., Fischer, U., Nakamura, M., Ohsawa, M., Ueno, M., Takayama, H. & Unger, M. (1998). Phytochemistry, 48, 107–111.
- Koyama, J., Ogura, T. & Tagahara, K. (1993). Phytochemistry, 33, 1540–1542.
- Koyama, J., Okatani, T., Tagahara, K., Kouno, I. & Irie, H. (1992). Phytochemistry, 31, 709–710.
- Kupier, J. & Labadie, R. P. (1984). Planta Med.42, 390–399. [DOI] [PubMed]
- Ozgen, U., Kazaz, C., Secen, H. & Coskun, M. (2006). Turk. J. Chem.30, 15–20.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Vidal-Tessier, A. M., Delaveau, P. & Champion, B. (1987). Ann. Pharm. Fr.45, 261–267. [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
- Wu, T.-S., Lin, D.-M., Shi, L.-S., Damu, A. G., Kuo, P.-C. & Kuo, Y.-H. (2003). Chem. Pharm. Bull.51, 948–950. [DOI] [PubMed]
- Yoshikawa, M., Yamaguchi, S., Nishisaka, H., Yamahara, J. & Murakami, N. (1995). Chem. Pharm. Bull.43, 1462–1465. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809017607/tk2447sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809017607/tk2447Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report