Abstract
The enantiomerically pure title compound, C11H15NO4S, contains a pyramidal N atom with an S—N bond length of 1.6262 (8) Å. In the crystal, molecules are linked to form chains parallel to the a axis by the hydrogen bond from NH to the carbonyl oxygen. C—H⋯O interactions are also present.
Related literature
For the applications of esters in the food and cosmetics industries, see: Soni et al. (2002 ▶). For their use as intermediates in the synthesis of heterocyclic compounds, see: Akhtar et al. (2007 ▶); Chen et al. (2007 ▶); Kucukguzel et al. (2007 ▶). For their use in the pharmaceutical industry, see: Iqbal & Chaudhry (2008 ▶). For the pharmacological activity of sulfonamides, see:Akhtar et al. (2008 ▶); Zareef et al. (2007 ▶). For a description of the Cambridge Structural Database, see: Allen (2002 ▶).
Experimental
Crystal data
C11H15NO4S
M r = 257.30
Orthorhombic,
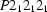
a = 7.1948 (2) Å
b = 11.2552 (3) Å
c = 15.3311 (4) Å
V = 1241.50 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.26 mm−1
T = 100 K
0.35 × 0.30 × 0.20 mm
Data collection
Oxford Diffraction Xcalibur E diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2008 ▶) T min = 0.972, T max = 1.000 (expected range = 0.922–0.949)
55416 measured reflections
4284 independent reflections
4060 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.022
wR(F 2) = 0.063
S = 1.05
4284 reflections
161 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.37 e Å−3
Δρmin = −0.31 e Å−3
Absolute structure: Flack (1983 ▶), 1818 Friedel pairs
Flack parameter: 0.01 (4)
Data collection: CrysAlis CCD (Oxford Diffraction, 2008 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2008 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP (Siemens, 1994 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809017371/hg2507sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809017371/hg2507Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N—H01⋯O1i | 0.865 (16) | 2.057 (16) | 2.9097 (10) | 168.6 (14) |
| C10—H10⋯O1i | 0.95 | 2.62 | 3.3696 (11) | 136 |
| C9—H9⋯O2ii | 0.95 | 2.63 | 3.4065 (11) | 140 |
| C7—H7⋯O3iii | 0.95 | 2.67 | 3.6167 (11) | 172 |
| C4—H4C⋯O4iv | 0.98 | 2.49 | 3.3453 (13) | 145 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Acknowledgments
The authors are grateful to the Higher Education Commission of Pakistan for financial support through project Nos. 20–674/R & D/06/1764 under the National Research Program for Universities.
supplementary crystallographic information
Comment
Because of their versatility and harmlessness, esters have found many applications in the food and cosmetics industries (Soni et al., 2002). They have been reported as emulsifiers, dispersants, or thickeners. In addition to their use as intermediates in the synthesis of a large number of heterocyclic compounds (Chen et al., 2007; Kucukguzel et al., 2007; Akhtar et al., 2007), esters have also been used in the pharmaceutical industry as nervous system depressants, intestinal antiseptics and antibacterials (Iqbal et al., 2008). Sulfonamides, on the other hand, constitute an important class of drugs with several types of pharmacological activities (Akhtar et al., 2008; Zareef et al., 2007). The title compound (I) was synthesized in our laboratory as an intermediate for onward conversion to 1,3,4-oxadiazole derivatives, with a view to explore their anti-HIV and anti-HCV activities.
The molecule of (I) is shown in Fig. 1. Bond lengths and angles may be regarded as normal. In particular, the N—S bond length is 1.6262 (8) Å; a search of the Cambridge Database (Allen, 2002; Version 1.11) revealed 817 examples of the fragment Ph—SO2—NH—C(sp3) (including substituted Ph rings) with a mean bond length of 1.614 Å. The nitrogen atom displays a pyramidal geometry, with N lying 0.293 (7) Å out of the plane of S, C2 and H01. The database hits showed a highly skewed distribution for this displacement, with many values of exactly zero; this is presumably attributable to fixing the hydrogen assuming planar geometry (AFIX 43 in the SHELX system), whereby the assumption of planarity is clearly not justified. Neglecting the zero values, the average deviation for 462 values is 0.24 Å. The sulfonyl group is oriented such that the bond S—O4 is approximately coplanar with the aromatic ring (torsion angle O4—S—C5—C6 - 6.70 (9)°).
The molecules are linked by classical hydrogen bonds from the NH group to the carbonyl oxygen (and not, as might have been expected, to a sulfonyl oxygen); the packing diagram (Fig. 2) shows chains of molecules parallel to the a axis. The four "weak" hydrogen bonds crosslink these chains to a three-dimensional pattern.
Experimental
Alanine (0.02 mol) was dissolved in an aqueous solution of potassium carbonate (0.06 mol, 50 ml) and a solution of 4-methylbenzenesulfonyl chloride (0.027 mol) in toluene (30 ml) was added. On completion of the reaction, the organic layer was separated and the aqueous layer acidified using dilute hydrochloric acid. The 2-(4-methylphenylsulfonylamino)propanoic acid thus obtained was filtered off and recrystallized from aqueous ethanol.
2-(4-Methylphenylsulfonylamino)propanoic acid (0.02 mol) was dissolved in methanol (30 ml), sulfuric acid (0.5 ml) was added and the mixture subjected to reflux while the reaction was monitored by TLC at regular intervals. After completion of the reaction, the reaction mixture was concentrated on the rotary evaporator to remove excess methanol. The product thus obtained was poured into water, neutralized with sodium bicarbonate and extracted with ethyl acetate (3 × 50 ml). The combined organic extracts were dried over anhydrous sodium sulfate and the solvent evaporated on a rotary evaporator. The product was recrystallized from acetone/water.
Refinement
The NH hydrogen was refined freely. Methyl H atoms were located in difference syntheses, idealized to C—H 0.98 Å and H—C—H 109.5°, and refined as rigid groups allowed to rotate but not tip. Other H atoms were placed in calculated positions and refined using a riding model with C—H 0.95 Å for aromatic H and 1.00 Å for methine CH. Hydrogen U values were fixed at 1.5 × U(eq) of the parent atom for methyl H and 1.2 × U(eq) of the parent atom for other H. The compound is enantiomerically pure and its absolute configuration (S at C2) was confirmed by the Flack (1983) parameter.
Figures
Fig. 1.
The molecule of the title compound. Ellipsoids correspond to 50% probability levels.
Fig. 2.
Packing diagram of the title compound, showing classical hydrogen bonds (dashed lines).
Crystal data
| C11H15NO4S | Dx = 1.377 Mg m−3 |
| Mr = 257.30 | Melting point = 363–365 K |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 40101 reflections |
| a = 7.1948 (2) Å | θ = 2.2–32.6° |
| b = 11.2552 (3) Å | µ = 0.26 mm−1 |
| c = 15.3311 (4) Å | T = 100 K |
| V = 1241.50 (6) Å3 | Irregular block, colourless |
| Z = 4 | 0.35 × 0.30 × 0.20 mm |
| F(000) = 544 |
Data collection
| Oxford Diffraction Xcalibur E diffractometer | 4284 independent reflections |
| Radiation source: fine-focus sealed tube | 4060 reflections with I > 2σ(I) |
| graphite | Rint = 0.028 |
| Detector resolution: 16.1419 pixels mm-1 | θmax = 32.0°, θmin = 2.2° |
| ω–scan | h = −10→10 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2008) | k = −16→16 |
| Tmin = 0.972, Tmax = 1.000 | l = −22→22 |
| 55416 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.022 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.063 | w = 1/[σ2(Fo2) + (0.0451P)2 + 0.0628P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 4284 reflections | Δρmax = 0.37 e Å−3 |
| 161 parameters | Δρmin = −0.31 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1818 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.01 (4) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S | 0.77287 (3) | 0.499255 (18) | 0.576519 (12) | 0.01308 (5) | |
| O1 | 0.26383 (10) | 0.24453 (5) | 0.58832 (5) | 0.01963 (14) | |
| O2 | 0.29792 (10) | 0.39044 (5) | 0.68639 (4) | 0.01622 (12) | |
| O3 | 0.95092 (9) | 0.44220 (6) | 0.58061 (5) | 0.02115 (14) | |
| O4 | 0.70626 (11) | 0.56695 (6) | 0.64934 (4) | 0.02111 (14) | |
| C1 | 0.31824 (12) | 0.34256 (7) | 0.60812 (5) | 0.01252 (14) | |
| C2 | 0.42632 (12) | 0.42436 (7) | 0.54745 (5) | 0.01322 (14) | |
| H2 | 0.4070 | 0.5085 | 0.5662 | 0.016* | |
| C3 | 0.36297 (14) | 0.41083 (10) | 0.45331 (6) | 0.0239 (2) | |
| H3A | 0.4314 | 0.4668 | 0.4164 | 0.036* | |
| H3B | 0.2295 | 0.4275 | 0.4493 | 0.036* | |
| H3C | 0.3872 | 0.3295 | 0.4336 | 0.036* | |
| C4 | 0.19070 (16) | 0.32121 (10) | 0.74852 (7) | 0.0258 (2) | |
| H4A | 0.0614 | 0.3160 | 0.7288 | 0.039* | |
| H4B | 0.1950 | 0.3598 | 0.8058 | 0.039* | |
| H4C | 0.2433 | 0.2411 | 0.7529 | 0.039* | |
| C5 | 0.77229 (12) | 0.59314 (7) | 0.48448 (5) | 0.01268 (14) | |
| C6 | 0.69566 (12) | 0.70659 (7) | 0.48923 (5) | 0.01463 (14) | |
| H6 | 0.6427 | 0.7348 | 0.5421 | 0.018* | |
| C7 | 0.69813 (12) | 0.77808 (7) | 0.41487 (6) | 0.01543 (15) | |
| H7 | 0.6459 | 0.8555 | 0.4174 | 0.019* | |
| C8 | 0.77573 (13) | 0.73814 (7) | 0.33690 (5) | 0.01487 (15) | |
| C9 | 0.84746 (13) | 0.62251 (8) | 0.33331 (6) | 0.01649 (16) | |
| H9 | 0.8974 | 0.5933 | 0.2801 | 0.020* | |
| C10 | 0.84653 (12) | 0.55006 (7) | 0.40655 (6) | 0.01525 (15) | |
| H10 | 0.8959 | 0.4719 | 0.4037 | 0.018* | |
| C11 | 0.78446 (17) | 0.81761 (9) | 0.25811 (6) | 0.02262 (18) | |
| H11A | 0.6738 | 0.8683 | 0.2564 | 0.034* | |
| H11B | 0.7893 | 0.7689 | 0.2052 | 0.034* | |
| H11C | 0.8960 | 0.8674 | 0.2614 | 0.034* | |
| N | 0.62422 (10) | 0.39329 (6) | 0.55827 (5) | 0.01262 (13) | |
| H01 | 0.661 (2) | 0.3437 (14) | 0.5189 (10) | 0.032 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S | 0.01306 (9) | 0.01406 (8) | 0.01212 (8) | −0.00207 (7) | −0.00239 (6) | 0.00121 (7) |
| O1 | 0.0243 (3) | 0.0136 (3) | 0.0210 (3) | −0.0038 (2) | −0.0008 (3) | −0.0026 (2) |
| O2 | 0.0175 (3) | 0.0183 (3) | 0.0129 (3) | −0.0021 (2) | 0.0007 (2) | −0.0013 (2) |
| O3 | 0.0131 (3) | 0.0248 (3) | 0.0256 (3) | −0.0009 (2) | −0.0058 (3) | 0.0072 (3) |
| O4 | 0.0310 (4) | 0.0199 (3) | 0.0125 (3) | −0.0053 (3) | 0.0004 (3) | −0.0036 (2) |
| C1 | 0.0109 (3) | 0.0130 (3) | 0.0137 (3) | 0.0008 (3) | −0.0016 (3) | 0.0000 (3) |
| C2 | 0.0113 (3) | 0.0137 (3) | 0.0146 (3) | 0.0008 (3) | −0.0005 (3) | 0.0020 (3) |
| C3 | 0.0167 (4) | 0.0388 (5) | 0.0163 (4) | −0.0023 (4) | −0.0046 (3) | 0.0078 (4) |
| C4 | 0.0269 (5) | 0.0319 (5) | 0.0187 (4) | −0.0047 (4) | 0.0050 (4) | 0.0060 (4) |
| C5 | 0.0123 (3) | 0.0128 (3) | 0.0129 (3) | −0.0013 (3) | −0.0004 (3) | 0.0003 (2) |
| C6 | 0.0141 (3) | 0.0140 (3) | 0.0158 (3) | 0.0005 (3) | 0.0016 (3) | −0.0013 (3) |
| C7 | 0.0145 (3) | 0.0128 (3) | 0.0190 (4) | 0.0006 (3) | −0.0003 (3) | 0.0004 (3) |
| C8 | 0.0150 (3) | 0.0153 (3) | 0.0143 (3) | −0.0032 (3) | −0.0031 (3) | 0.0019 (3) |
| C9 | 0.0194 (4) | 0.0164 (4) | 0.0137 (4) | −0.0014 (3) | 0.0022 (3) | −0.0011 (3) |
| C10 | 0.0167 (4) | 0.0129 (3) | 0.0162 (4) | 0.0009 (3) | 0.0014 (3) | −0.0012 (3) |
| C11 | 0.0299 (5) | 0.0199 (4) | 0.0180 (4) | −0.0044 (4) | −0.0027 (4) | 0.0050 (3) |
| N | 0.0112 (3) | 0.0109 (3) | 0.0157 (3) | 0.0003 (2) | −0.0002 (2) | −0.0002 (2) |
Geometric parameters (Å, °)
| S—O4 | 1.4341 (7) | C9—C10 | 1.3878 (12) |
| S—O3 | 1.4344 (7) | C2—H2 | 1.0000 |
| S—N | 1.6262 (8) | C3—H3A | 0.9800 |
| S—C5 | 1.7628 (8) | C3—H3B | 0.9800 |
| O1—C1 | 1.2094 (10) | C3—H3C | 0.9800 |
| O2—C1 | 1.3235 (10) | C4—H4A | 0.9800 |
| O2—C4 | 1.4525 (11) | C4—H4B | 0.9800 |
| C1—C2 | 1.5223 (11) | C4—H4C | 0.9800 |
| C2—N | 1.4755 (11) | C6—H6 | 0.9500 |
| C2—C3 | 1.5213 (13) | C7—H7 | 0.9500 |
| C5—C6 | 1.3929 (11) | C9—H9 | 0.9500 |
| C5—C10 | 1.3956 (11) | C10—H10 | 0.9500 |
| C6—C7 | 1.3955 (11) | C11—H11A | 0.9800 |
| C7—C8 | 1.3938 (12) | C11—H11B | 0.9800 |
| C8—C9 | 1.4012 (12) | C11—H11C | 0.9800 |
| C8—C11 | 1.5043 (12) | N—H01 | 0.865 (16) |
| O4—S—O3 | 120.15 (5) | C2—C3—H3B | 109.5 |
| O4—S—N | 107.69 (4) | H3A—C3—H3B | 109.5 |
| O3—S—N | 105.46 (4) | C2—C3—H3C | 109.5 |
| O4—S—C5 | 107.69 (4) | H3A—C3—H3C | 109.5 |
| O3—S—C5 | 107.79 (4) | H3B—C3—H3C | 109.5 |
| N—S—C5 | 107.48 (4) | O2—C4—H4A | 109.5 |
| C1—O2—C4 | 115.78 (7) | O2—C4—H4B | 109.5 |
| O1—C1—O2 | 124.28 (8) | H4A—C4—H4B | 109.5 |
| O1—C1—C2 | 124.32 (8) | O2—C4—H4C | 109.5 |
| O2—C1—C2 | 111.35 (7) | H4A—C4—H4C | 109.5 |
| N—C2—C3 | 111.84 (7) | H4B—C4—H4C | 109.5 |
| N—C2—C1 | 106.32 (7) | C5—C6—H6 | 120.6 |
| C3—C2—C1 | 111.48 (7) | C7—C6—H6 | 120.6 |
| C6—C5—C10 | 120.98 (7) | C8—C7—H7 | 119.3 |
| C6—C5—S | 120.57 (6) | C6—C7—H7 | 119.3 |
| C10—C5—S | 118.43 (6) | C10—C9—H9 | 119.6 |
| C5—C6—C7 | 118.74 (7) | C8—C9—H9 | 119.6 |
| C8—C7—C6 | 121.32 (7) | C9—C10—H10 | 120.3 |
| C7—C8—C9 | 118.74 (7) | C5—C10—H10 | 120.3 |
| C7—C8—C11 | 120.90 (8) | C8—C11—H11A | 109.5 |
| C9—C8—C11 | 120.36 (8) | C8—C11—H11B | 109.5 |
| C10—C9—C8 | 120.81 (8) | H11A—C11—H11B | 109.5 |
| C9—C10—C5 | 119.37 (8) | C8—C11—H11C | 109.5 |
| C2—N—S | 118.70 (6) | H11A—C11—H11C | 109.5 |
| N—C2—H2 | 109.0 | H11B—C11—H11C | 109.5 |
| C3—C2—H2 | 109.0 | C2—N—H01 | 111.8 (10) |
| C1—C2—H2 | 109.0 | S—N—H01 | 113.1 (10) |
| C2—C3—H3A | 109.5 | ||
| C4—O2—C1—O1 | −4.64 (13) | C5—C6—C7—C8 | −0.17 (13) |
| C4—O2—C1—C2 | 177.89 (8) | C6—C7—C8—C9 | 1.82 (13) |
| O1—C1—C2—N | −87.54 (10) | C6—C7—C8—C11 | −177.58 (9) |
| O2—C1—C2—N | 89.93 (8) | C7—C8—C9—C10 | −1.88 (13) |
| O1—C1—C2—C3 | 34.58 (12) | C11—C8—C9—C10 | 177.52 (9) |
| O2—C1—C2—C3 | −147.95 (8) | C8—C9—C10—C5 | 0.29 (13) |
| O4—S—C5—C6 | −6.70 (9) | C6—C5—C10—C9 | 1.42 (13) |
| O3—S—C5—C6 | −137.70 (7) | S—C5—C10—C9 | −179.99 (7) |
| N—S—C5—C6 | 109.07 (7) | C3—C2—N—S | 108.25 (8) |
| O4—S—C5—C10 | 174.70 (7) | C1—C2—N—S | −129.86 (6) |
| O3—S—C5—C10 | 43.70 (8) | O4—S—N—C2 | 52.67 (7) |
| N—S—C5—C10 | −69.53 (8) | O3—S—N—C2 | −177.90 (6) |
| C10—C5—C6—C7 | −1.48 (13) | C5—S—N—C2 | −63.10 (7) |
| S—C5—C6—C7 | 179.97 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N—H01···O1i | 0.865 (16) | 2.057 (16) | 2.9097 (10) | 168.6 (14) |
| C10—H10···O1i | 0.95 | 2.62 | 3.3696 (11) | 136 |
| C9—H9···O2ii | 0.95 | 2.63 | 3.4065 (11) | 140 |
| C7—H7···O3iii | 0.95 | 2.67 | 3.6167 (11) | 172 |
| C4—H4C···O4iv | 0.98 | 2.49 | 3.3453 (13) | 145 |
Symmetry codes: (i) x+1/2, −y+1/2, −z+1; (ii) −x+3/2, −y+1, z−1/2; (iii) x−1/2, −y+3/2, −z+1; (iv) −x+1, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2507).
References
- Akhtar, T., Hameed, S., Al-Masoudi, N. A. & Khan, K. M. (2007). Heteroatom. Chem.18, 316–322.
- Akhtar, T., Hameed, S., Al-Masoudi, N. A., Loddo, R. & La Colla, P. (2008). Acta Pharm.58, 135–149. [DOI] [PubMed]
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Chen, C. J., Song, B.-A., Yang, S. & Xu, G.-F. (2007). Bioorg. Med. Chem.15, 3981–3989. [DOI] [PubMed]
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Iqbal, M. J. & Chaudhry, M. A. (2008). J. Mol. Liq.143, 75–80.
- Kucukguzel, S. G., Kucukguzel, I. & Tatar, E. (2007). Eur. J. Med. Chem.42, 893–901. [DOI] [PubMed]
- Oxford Diffraction (2008). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd, Abingdon, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siemens (1994). XP Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
- Soni, M. G., Taylor, S. L., Greenberg, N. A. & Burdock, G. A. (2002). Food Chem. Toxicol.40, 1335–1373. [DOI] [PubMed]
- Zareef, M., Iqbal, R., Al-Masoudi, N. A., Zaidi, J. H., Arfan, M. & Shahzad, S. A. (2007). Phosphorus Sulfur Silicon Relat. Elem.182, 281–298.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809017371/hg2507sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809017371/hg2507Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




