Abstract
The title compound, C11H19NO6, is an important intermediate for the synthesis of cephalosporin derivatives. The N atom is in a planar configuration. In the crystal, molecules are linked into zigzag layers parallel to (100) by O—H⋯O hydrogen bonds.
Related literature
The condensation of the title compound with cephalosporin may improve the pharmacokinetics, see: Sakagami et al. (1990 ▶, 1991 ▶); Uhrich & Frechet (1992 ▶).
Experimental
Crystal data
C11H19NO6
M r = 261.27
Orthorhombic,

a = 10.632 (2) Å
b = 14.559 (3) Å
c = 18.257 (4) Å
V = 2826.1 (11) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 292 K
0.60 × 0.50 × 0.44 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: none
2979 measured reflections
2601 independent reflections
1050 reflections with I > 2σ(I)
R int = 0.008
3 standard reflections every 200 reflections intensity decay: 1.3%
Refinement
R[F 2 > 2σ(F 2)] = 0.054
wR(F 2) = 0.171
S = 1.09
2601 reflections
175 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.22 e Å−3
Δρmin = −0.25 e Å−3
Data collection: DIFRAC (Gabe & White, 1993 ▶); cell refinement: DIFRAC; data reduction: NRCVAX (Gabe et al., 1989 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809018911/ci2799sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809018911/ci2799Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3O⋯O4i | 0.98 (5) | 1.68 (5) | 2.653 (4) | 174 (4) |
| O5—H5O⋯O2ii | 0.94 (5) | 1.70 (5) | 2.628 (3) | 168 (4) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported by the National 973 Project under grant No. 2004CB518800.
supplementary crystallographic information
Comment
The title compound is an important intermediate for the synthesis of a new type of cephalosporin. The condensation of the title compound with cephalosporin may improve the pharmacokinetics of the cephalosporin (Sakagami et al., 1990). It has two carboxylic acid functionalities that are available for the condensation with the amino group of cephalosporin, while the protected amine can be easily activated by deprotection, so that it can be condensed with the carboxyl of cephalosporin. The condensation with cephalosporin may increase the drug concentration, control the release of drug and reduce the drug toxicity (Uhrich & Frechet, 1992; Sakagami et al., 1991).
The N atom has a trigonal planar configuration, with sum of bond angles around N1 being 359.8 °. The molecules are linked into zigzag layers parallel to the (100) by O—H···O hydrogen bonds.
Experimental
Dimethyl 3,3'-azanediyldipropanoate (5.67g, 30 mol) was treated with NaOH solution (4.0g NaOH in 20 ml H2O) and stirred at room temperature for 2 h. Then a solution of (Boc)2O (7.0g, 32mmol) (Boc is tert-butoxycarbonyl) in tertiary butyl alcohol (10 ml) was added dropwise at 283 K. The contents were stirred for 30 min at room temperature. The reaction mixture was washed with n-pentane (10 ml × 3) and the aqueous layer was adjusted to a pH of 1.0 with hydrochloric acid and extracted with ethyl acetate. The organic layer was dried (MgSO4) and evaporated in vacuo and recrystallized in cyclohexane-ethyl acetate to get colourless crystals.
Refinement
Hydroxyl H atoms were located in a difference map and refined freely. The remaining H atoms were positioned geometrically (C-H = 0.96–0.97 Å) and refined using a riding model, with Uiso(H) = 1.2–1.5Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level.
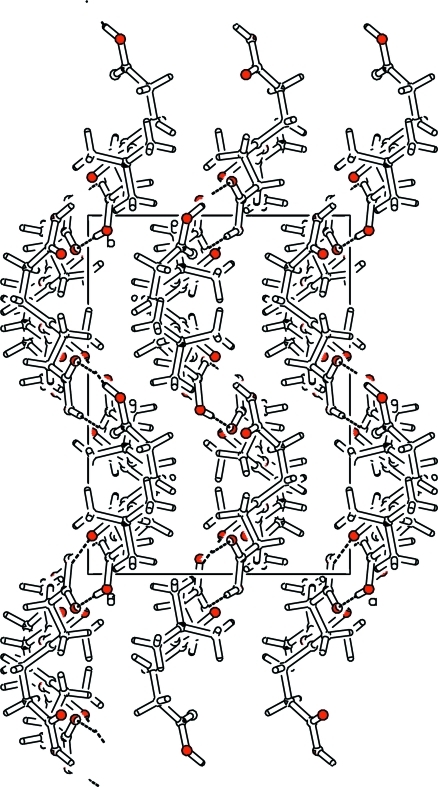
Fig. 2.
A packing diagram of the title compound. Hydrogen bonds are shown as dashed lines.
Crystal data
| C11H19NO6 | F(000) = 1120 |
| Mr = 261.27 | Dx = 1.228 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 20 reflections |
| a = 10.632 (2) Å | θ = 5.7–6.8° |
| b = 14.559 (3) Å | µ = 0.10 mm−1 |
| c = 18.257 (4) Å | T = 292 K |
| V = 2826.1 (11) Å3 | Block, colourless |
| Z = 8 | 0.60 × 0.50 × 0.44 mm |
Data collection
| Enraf–Nonius CAD-4 diffractometer | Rint = 0.008 |
| Radiation source: fine-focus sealed tube | θmax = 25.5°, θmin = 2.2° |
| graphite | h = −1→12 |
| ω/2–θ scans | k = −3→17 |
| 2979 measured reflections | l = −10→22 |
| 2601 independent reflections | 3 standard reflections every 200 reflections |
| 1050 reflections with I > 2σ(I) | intensity decay: 1.3% |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.054 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.171 | w = 1/[σ2(Fo2) + (0.0701P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.001 |
| 2601 reflections | Δρmax = 0.22 e Å−3 |
| 175 parameters | Δρmin = −0.25 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0127 (17) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.1690 (2) | 0.18631 (13) | 0.36711 (10) | 0.0709 (7) | |
| O2 | 0.0153 (3) | 0.10681 (16) | 0.31021 (11) | 0.0841 (8) | |
| O3 | 0.0720 (3) | −0.04162 (19) | 0.08342 (15) | 0.0995 (10) | |
| H3O | 0.031 (4) | −0.058 (3) | 0.037 (3) | 0.129 (16)* | |
| O4 | 0.0484 (3) | 0.09467 (16) | 0.03663 (14) | 0.1078 (11) | |
| O5 | 0.1228 (3) | 0.49187 (18) | 0.26626 (13) | 0.0833 (8) | |
| H5O | 0.066 (5) | 0.526 (3) | 0.238 (2) | 0.137 (18)* | |
| O6 | 0.1031 (3) | 0.39281 (16) | 0.17583 (15) | 0.1142 (11) | |
| N1 | 0.1433 (3) | 0.20065 (16) | 0.24663 (13) | 0.0669 (8) | |
| C1 | 0.2392 (4) | 0.1974 (2) | 0.48679 (17) | 0.0898 (12) | |
| H1A | 0.3223 | 0.1849 | 0.4687 | 0.135* | |
| H1B | 0.2328 | 0.1772 | 0.5367 | 0.135* | |
| H1C | 0.2233 | 0.2622 | 0.4843 | 0.135* | |
| C2 | 0.1697 (4) | 0.0451 (2) | 0.4391 (2) | 0.0981 (14) | |
| H2A | 0.1055 | 0.0146 | 0.4112 | 0.147* | |
| H2B | 0.1700 | 0.0217 | 0.4882 | 0.147* | |
| H2C | 0.2501 | 0.0342 | 0.4169 | 0.147* | |
| C3 | 0.0128 (4) | 0.1701 (3) | 0.4649 (2) | 0.1067 (14) | |
| H3A | −0.0026 | 0.2343 | 0.4571 | 0.160* | |
| H3B | 0.0038 | 0.1560 | 0.5160 | 0.160* | |
| H3C | −0.0467 | 0.1347 | 0.4371 | 0.160* | |
| C4 | 0.1436 (4) | 0.1469 (2) | 0.44047 (16) | 0.0686 (10) | |
| C5 | 0.1046 (4) | 0.1604 (2) | 0.30812 (18) | 0.0623 (9) | |
| C6 | 0.0904 (4) | 0.1711 (2) | 0.17744 (16) | 0.0688 (10) | |
| H6A | 0.1042 | 0.2185 | 0.1409 | 0.083* | |
| H6B | 0.0003 | 0.1629 | 0.1829 | 0.083* | |
| C7 | 0.1484 (3) | 0.0821 (2) | 0.15127 (17) | 0.0747 (11) | |
| H7A | 0.2365 | 0.0922 | 0.1400 | 0.090* | |
| H7B | 0.1437 | 0.0368 | 0.1902 | 0.090* | |
| C8 | 0.0838 (4) | 0.0459 (3) | 0.08533 (19) | 0.0719 (10) | |
| C9 | 0.2554 (4) | 0.2605 (2) | 0.24626 (18) | 0.0762 (10) | |
| H9A | 0.2844 | 0.2675 | 0.1962 | 0.091* | |
| H9B | 0.3220 | 0.2308 | 0.2738 | 0.091* | |
| C10 | 0.2318 (4) | 0.3548 (2) | 0.27853 (18) | 0.0762 (11) | |
| H10A | 0.1970 | 0.3475 | 0.3273 | 0.091* | |
| H10B | 0.3116 | 0.3865 | 0.2833 | 0.091* | |
| C11 | 0.1451 (4) | 0.4125 (2) | 0.2344 (2) | 0.0732 (11) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0897 (19) | 0.0642 (13) | 0.0588 (12) | −0.0127 (13) | −0.0146 (12) | −0.0004 (10) |
| O2 | 0.097 (2) | 0.0806 (16) | 0.0750 (16) | −0.0276 (16) | −0.0109 (14) | −0.0061 (12) |
| O3 | 0.152 (3) | 0.0724 (19) | 0.0737 (17) | 0.0156 (18) | −0.0244 (17) | −0.0096 (14) |
| O4 | 0.164 (3) | 0.0759 (17) | 0.0831 (17) | 0.0102 (17) | −0.0439 (18) | −0.0010 (15) |
| O5 | 0.094 (2) | 0.0750 (17) | 0.0803 (16) | 0.0181 (15) | −0.0104 (14) | 0.0071 (14) |
| O6 | 0.153 (3) | 0.0836 (18) | 0.106 (2) | 0.0274 (18) | −0.053 (2) | −0.0026 (16) |
| N1 | 0.078 (2) | 0.0626 (15) | 0.0595 (16) | −0.0032 (16) | −0.0056 (15) | 0.0027 (14) |
| C1 | 0.094 (3) | 0.103 (3) | 0.073 (2) | 0.004 (2) | −0.017 (2) | −0.012 (2) |
| C2 | 0.132 (4) | 0.072 (3) | 0.090 (3) | 0.002 (3) | −0.012 (3) | 0.015 (2) |
| C3 | 0.091 (4) | 0.132 (3) | 0.097 (3) | 0.009 (3) | 0.013 (3) | −0.017 (3) |
| C4 | 0.080 (3) | 0.069 (2) | 0.0572 (18) | 0.002 (2) | −0.0009 (18) | −0.0036 (17) |
| C5 | 0.066 (3) | 0.054 (2) | 0.067 (2) | −0.0066 (19) | −0.0100 (19) | −0.0035 (17) |
| C6 | 0.080 (3) | 0.065 (2) | 0.060 (2) | 0.011 (2) | −0.0078 (17) | −0.0011 (16) |
| C7 | 0.073 (3) | 0.082 (2) | 0.069 (2) | 0.014 (2) | −0.0123 (18) | −0.0100 (18) |
| C8 | 0.081 (3) | 0.071 (3) | 0.064 (2) | 0.025 (2) | −0.0029 (19) | −0.009 (2) |
| C9 | 0.069 (3) | 0.075 (2) | 0.084 (2) | 0.005 (2) | −0.0019 (19) | 0.014 (2) |
| C10 | 0.078 (3) | 0.064 (2) | 0.087 (2) | −0.010 (2) | −0.023 (2) | 0.0155 (17) |
| C11 | 0.087 (3) | 0.062 (2) | 0.071 (2) | −0.006 (2) | −0.011 (2) | 0.0116 (19) |
Geometric parameters (Å, °)
| O1—C5 | 1.331 (4) | C2—H2B | 0.96 |
| O1—C4 | 1.482 (3) | C2—H2C | 0.96 |
| O2—C5 | 1.229 (4) | C3—C4 | 1.499 (5) |
| O3—C8 | 1.281 (4) | C3—H3A | 0.96 |
| O3—H3O | 0.98 (5) | C3—H3B | 0.96 |
| O4—C8 | 1.199 (4) | C3—H3C | 0.96 |
| O5—C11 | 1.315 (4) | C6—C7 | 1.513 (4) |
| O5—H5O | 0.94 (5) | C6—H6A | 0.97 |
| O6—C11 | 1.193 (4) | C6—H6B | 0.97 |
| N1—C5 | 1.331 (4) | C7—C8 | 1.482 (5) |
| N1—C6 | 1.448 (4) | C7—H7A | 0.97 |
| N1—C9 | 1.477 (4) | C7—H7B | 0.97 |
| C1—C4 | 1.514 (5) | C9—C10 | 1.515 (4) |
| C1—H1A | 0.96 | C9—H9A | 0.97 |
| C1—H1B | 0.96 | C9—H9B | 0.97 |
| C1—H1C | 0.96 | C10—C11 | 1.486 (5) |
| C2—C4 | 1.508 (4) | C10—H10A | 0.97 |
| C2—H2A | 0.96 | C10—H10B | 0.97 |
| C5—O1—C4 | 121.8 (3) | O1—C5—N1 | 113.5 (3) |
| C8—O3—H3O | 108 (2) | N1—C6—C7 | 111.8 (3) |
| C11—O5—H5O | 110 (3) | N1—C6—H6A | 109.3 |
| C5—N1—C6 | 119.0 (3) | C7—C6—H6A | 109.3 |
| C5—N1—C9 | 120.9 (3) | N1—C6—H6B | 109.3 |
| C6—N1—C9 | 119.0 (3) | C7—C6—H6B | 109.3 |
| C4—C1—H1A | 109.5 | H6A—C6—H6B | 107.9 |
| C4—C1—H1B | 109.5 | C8—C7—C6 | 111.8 (3) |
| H1A—C1—H1B | 109.5 | C8—C7—H7A | 109.2 |
| C4—C1—H1C | 109.5 | C6—C7—H7A | 109.2 |
| H1A—C1—H1C | 109.5 | C8—C7—H7B | 109.2 |
| H1B—C1—H1C | 109.5 | C6—C7—H7B | 109.2 |
| C4—C2—H2A | 109.5 | H7A—C7—H7B | 107.9 |
| C4—C2—H2B | 109.5 | O4—C8—O3 | 122.6 (3) |
| H2A—C2—H2B | 109.5 | O4—C8—C7 | 122.5 (4) |
| C4—C2—H2C | 109.5 | O3—C8—C7 | 114.9 (3) |
| H2A—C2—H2C | 109.5 | N1—C9—C10 | 113.5 (3) |
| H2B—C2—H2C | 109.5 | N1—C9—H9A | 108.9 |
| C4—C3—H3A | 109.5 | C10—C9—H9A | 108.9 |
| C4—C3—H3B | 109.5 | N1—C9—H9B | 108.9 |
| H3A—C3—H3B | 109.5 | C10—C9—H9B | 108.9 |
| C4—C3—H3C | 109.5 | H9A—C9—H9B | 107.7 |
| H3A—C3—H3C | 109.5 | C11—C10—C9 | 113.9 (3) |
| H3B—C3—H3C | 109.5 | C11—C10—H10A | 108.8 |
| O1—C4—C3 | 110.5 (3) | C9—C10—H10A | 108.8 |
| O1—C4—C2 | 109.4 (3) | C11—C10—H10B | 108.8 |
| C3—C4—C2 | 113.4 (3) | C9—C10—H10B | 108.8 |
| O1—C4—C1 | 101.2 (3) | H10A—C10—H10B | 107.7 |
| C3—C4—C1 | 110.4 (3) | O6—C11—O5 | 122.7 (3) |
| C2—C4—C1 | 111.3 (3) | O6—C11—C10 | 125.6 (3) |
| O2—C5—O1 | 123.5 (3) | O5—C11—C10 | 111.6 (3) |
| O2—C5—N1 | 123.0 (3) | ||
| C5—O1—C4—C3 | −63.2 (4) | C9—N1—C6—C7 | 89.6 (3) |
| C5—O1—C4—C2 | 62.3 (4) | N1—C6—C7—C8 | 173.1 (3) |
| C5—O1—C4—C1 | 179.9 (3) | C6—C7—C8—O4 | 39.8 (5) |
| C4—O1—C5—O2 | 4.4 (5) | C6—C7—C8—O3 | −142.1 (3) |
| C4—O1—C5—N1 | −177.7 (3) | C5—N1—C9—C10 | −76.4 (4) |
| C6—N1—C5—O2 | −8.8 (5) | C6—N1—C9—C10 | 115.9 (3) |
| C9—N1—C5—O2 | −176.5 (3) | N1—C9—C10—C11 | −67.1 (4) |
| C6—N1—C5—O1 | 173.3 (3) | C9—C10—C11—O6 | −5.7 (6) |
| C9—N1—C5—O1 | 5.6 (4) | C9—C10—C11—O5 | 177.0 (3) |
| C5—N1—C6—C7 | −78.3 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3O···O4i | 0.98 (5) | 1.68 (5) | 2.653 (4) | 174 (4) |
| O5—H5O···O2ii | 0.94 (5) | 1.70 (5) | 2.628 (3) | 168 (4) |
Symmetry codes: (i) −x, −y, −z; (ii) −x, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI2799).
References
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Gabe, E. J., Le Page, Y., Charland, J.-P., Lee, F. L. & White, P. S. (1989). J. Appl. Cryst.22, 384–387.
- Gabe, E. J. & White, P. S. (1993). DIFRAC American Crystallographic Association Meeting, Pittsburgh, Abstract PA 104.
- Sakagami, K., Atsumi, K. & Tamura, A. (1990). J. Antibiot.8, 1047–1050. [DOI] [PubMed]
- Sakagami, K., Atsumi, K. & Yamamoto, Y. (1991). Chem. Pharm. Bull.39, 2433–2436. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Uhrich, K. E. & Frechet, J. M. J. (1992). J. Chem. Soc. Perkin Trans. 1, pp. 1623–1630.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809018911/ci2799sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809018911/ci2799Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report