Abstract
The pregnancy-associated glycoproteins (PAGs) represent a complex group of putative aspartic peptidases expressed exclusively in the placentas of species in the Artiodactyla order. The ruminant PAGs segregate into two classes -the ‘ancient’ and ‘modern’ PAGs. Some of the modern PAGs possess alterations in the catalytic center that are predicted to preclude their ability to act as peptidases. The ancient ruminant PAGs in contrast are thought to be peptidases, although, no proteolytic activity has been described for these members. The goal of this present study was to investigate (1) if the ancient bovine PAGs (PAGs-2 and -12) have proteolytic activity, and (2) if there are any differences in activity between these two closely related members. Recombinant bovine PAGs-2 and -12 were expressed in a baculovirus expression system and the purified proteins were analyzed for proteolytic activity against a synthetic fluorescent cathepsin D/E substrate. Both proteins exhibited proteolytic activity with acidic pH optima. The kcat/KM for bovine PAG-2 was 2.7×105 M−1s−1 and for boPAG-12 it was 6.8×104 M−1s−1. The enzymes were inhibited by pepstatin A with a Ki of 0.56 and 7.5 nM for boPAG-2 and boPAG-12, respectively. This is the first report describing proteolytic activity in PAGs from ruminant ungulates.
Keywords: aspartic peptidases, cattle, placenta, pregnancy, trophoblast
Introduction
Aspartic peptidases (AP) are a class of proteolytic enzymes that typically require acidic conditions for optimal activity (Davies, 1990). Consequently, most mammalian AP tend to be found in low pH environments such as those present in gastric secretions (e.g. pepsin A, pepsin C, chymosin), lysosomes (e.g. cathepsin D) and other intracellular compartments (e.g. cathepsin E) within the cell (Davies, 1990; Dunn, 2002). One notable exception to this trend is renin, which is operational under physiological pH conditions (Cody, 1994).
The mammalian APs are bi-lobed proteins with both the N-terminal and C-terminal lobes being roughly symmetrical to one another (Tang et al., 1973). The lobes form a binding cleft that can accommodate a peptide substrate of seven to eight amino acids. The catalytic aspartic acid residues Asp32 and Asp215 are located at the middle of the cleft (the numbering represents that for porcine pepsin). Aspartic peptidases are produced as zymogens/pro-enzymes (Tang et al., 1973). Functional activation of the zymogens involves proteolytic processing of the amino-terminal propeptide.
One hallmark of the mammalian APs is the presence of highly conserved residues that flank the catalytic aspartic acids: hydrophobic-F/I/L-D-T-G-S in the N-terminal domain, and hydrophobic-D-T/S-GS/T in the C-terminal domain (Cooper et al., 1990). The carboxyl groups of these two aspartic acids, Asp32 and A215 interact with one another and also with a solvent water molecule by a network of hydrogen bonds. The proteolytic mechanism involves activation of the water molecule positioned between these aspartates to act as a nucleophile on the carbonyl carbon of the scissile peptide bond (Davies, 1990; Szecsi, 1992; Dunn, 2002). The current model suggests that Asp215 acts as a general base to remove a proton from the water molecule while Asp32 donates a proton to the carbonyl oxygen of the scissile bond. In the resulting tetrahedral intermediate, Asp215 is hydrogen bonded to the attacking oxygen atom (originally part of the water molecule), while the hydrogen remaining on that oxygen is hydrogen bonded to an oxygen on Asp32 (Davies, 1990; Szecsi, 1992; Dunn, 2002). The transfer of a proton from Asp215 to the nitrogen of the scissile peptide bond completes its hydrolysis.
The pregnancy-associated glycoproteins (PAGs) are a recently discovered family of genes that belong to the AP gene family (Xie et al., 1997; Green et al., 1998; Hughes et al., 2003; Telugu et al., 2009). They are expressed exclusively by trophoblasts -the outer cells of the placenta that are in direct contact with maternal tissues. The PAG gene family is found only in species within the Artiodactyla order (Green et al., 1998; Garbayo et al., 2000; Green et al., 2000; Hughes et al., 2003).
In ruminant ungulates, the PAG gene family is particularly large and complex. Dozens of distinct cDNAs, and numerous variants, have been cloned from cattle, sheep, goat and deer placentae (Szafranska et al., 1995; Xie et al., 1997; Garbayo et al., 1998; Garbayo et al., 2000; Green et al., 2000; Brandt et al., 2007; Telugu et al., 2009). The PAG gene family in ruminants is comprised of two evolutionarily distinct groups (Green et al., 2000; Hughes et al., 2000; Hughes et al., 2003). One grouping, the ‘modern PAGs’, is transcribed exclusively in specialized, moderately invasive trophoblasts known as ‘binucleate’ cells (BNC) (Green et al., 2000; Hughes et al., 2003; Wooding et al., 2005). The other grouping, known as the ‘ancient PAGs’, is transcribed in all trophoblast cell types (Green et al., 2000; Wooding et al., 2005).
The ancient PAGs are packaged in vesicles within both mononucleate and binucleate trophoblasts and, upon secretion, they accumulate at the microvillar junction of the maternal-fetal interface (Wooding et al., 2005). Coincident with differences in spatial expression, there are variations in temporal expression patterns as well. For example, some PAGs are expressed relatively early in gestation, while other PAGs do not appear until later in pregnancy (Green et al., 2000; Patel et al., 2004). Notably, there are also obvious differences in their levels of expression. For example, bovine (bo) PAG-2 is the most abundant transcript among all the PAGs identified to date. A closely related family member, boPAG-12, is substantially less prevalent in the placenta (Telugu et al., 2009).
Interestingly, many of the modern PAGs possess atypical residues in amino acid positions known to be involved in catalysis or in substrate-binding (Guruprasad et al., 1996; Green et al., 2000). Since PAGs are closely related to pepsin, molecular models (based on porcine pepsin and bovine chymosin crystal structures) revealed that some of the alterations within the catalytic center are likely to render some modern PAGs incapable of acting as proteolytic enzymes (Xie et al., 1995; Guruprasad et al., 1996; Green et al., 2000). On the other hand, most of the ancient PAGs have retained the characteristics of typical APs and are known or predicted to possess proteolytic activity (Green et al., 1998; Wooding et al., 2005; Telugu and Green, 2008). In those PAGs suspected to be peptidases, there are differences in residues known to contribute to catalytic activity and substrate specificity, suggesting that different members of the ancient PAG grouping probably possess distinct substrate preferences and activities (Xie et al., 1991; Guruprasad et al., 1996; Xie et al., 1997).
In the experiments described in this report, we sought to determine if some ancient PAGs in cattle are capable of proteolytic activity. Two paralogous ancient PAG members were chosen as the focus of the analysis – these were boPAG-2 and boPAG-12. Both of these proteins accumulate at the placenta-maternal interface (Wooding et al., 2005 and unpublished data). However, they also exhibit distinct temporal patterns of expression throughout pregnancy and they differ substantially in their relative level of transcript abundance in the placenta. Bovine PAG-2 is the most abundantly transcribed PAG gene in cattle, whereas boPAG-12 mRNA is much less abundant – differing by as much as two to three orders of magnitude at any given time-point during pregnancy (Telugu et al., 2009).
Results
Expression and purification of recombinant boPAG-2 and -12
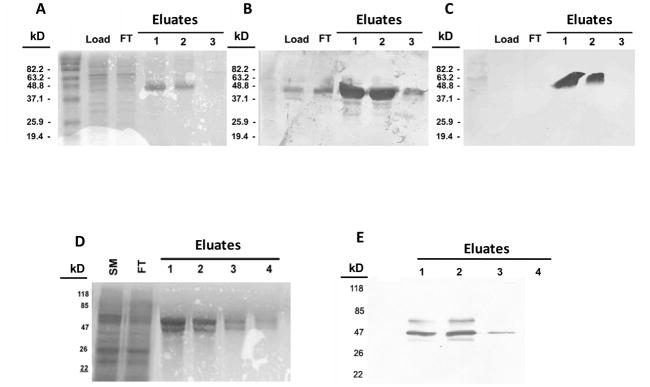
Recombinant bovine PAGs-2 and -12 were expressed in a Baculovirus insect cell expression system as fusion proteins with an N-terminal FLAG peptide engineered in-frame with the pro-peptide. The FLAG peptide allowed for affinity purification of fusion proteins by using an anti-FLAG M2 antibody matrix (Figure 1). Typical yields for the FLAG-zymogens were approximately 75 μg of purified protein per one confluent (80%) 75 cm2 culture dish. The expressed and purified proteins had the molecular weight expected of the full length fusion proteins(approximately 45,000 MW). Accompanying the regular pro-form of boPag-12, however, was some limited glycosylation, which manifested as a somewhat higher molecular weight fraction (Figure 1). This probably resulted from it being cloned into the pACMP2 transfer vector that facilitates limited cytoplasmic glycosylation (Pharmingen’s Baculovirus expression manual). Both PAGs as well as the higher molecular fraction of boPAG-12 were identified in Western blots by using an anti-FLAG antibody(Figure 1). The presence of boPAG-2 was further confirmed with a polyclonal antibody raised against boPAG-2 (Wooding et al., 2005).
Figure 1. SDS-PAGE gels of expressed recombinant bovine PAGs-2 and 12 stained either with Coomassie blue or by Western blotting.
Panel (A) shows Coomassie blue staining of fractions from a preparation of recombinant boPAG-2. Lane-1 contains the total proteins from the lysed insect cell pellet. Lane-2 contains the flow-through of lysate from the anti-FLAG column. Lanes 3–5 contain elution fractions of recombinant boPAG-2 in order of emergence from the column. Panels (B) and (C) show Western blot images of boPAG-2-containing fractions transferred from identically loaded gels and immuno-blotted with anti-PAG-2 polyclonal and anti-FLAG monoclonal antibodies, respectively. (D) Coomassie blue staining of fractions from a preparation of recombinant boPAG-12. Lane-1 contains total proteins from the insect cell lysate. Lane-2 contains the flow-through from the anti-FLAG column. Lanes 3–5 contain fractions in order of their elution from the column. (E) Western blot image of boPAG-12-containing fractions immunoblotted and detected with anti-FLAG monoclonal antibody.
Dependence of activities on pH
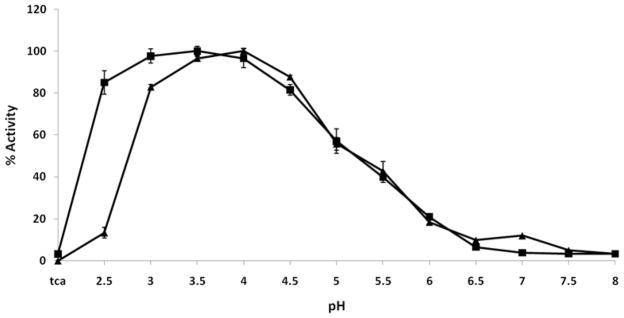
The presence of proteolytic activity and the preferred pH optima for each recombinant PAG zymogen was tested against a commercial cathepsin D/E FRET substrate. The preferred pH for maximal activity for boPAG-2 was found to be around pH 4.0, with high activity from pH 3.0 to 4.5 (Figure 2). Bovine PAG-12 had an optimal activity at pH 3.5, with high activity over a broader range of pH 2.5 to 4.5 (Figure 2). Some differences were notable. BoPAG-12 had high activity at pH 2.5 while boPAG-2 had relatively little activity at this pH. The optimal activity of boPAG-2 and 12 under acidic conditions is consistent with the pH preferences of most other aspartic peptidases (Davies, 1990; Kay and Dunn, 1992; Szecsi, 1992; Dunn, 2002).
Figure 2. The relative activity of boPAG-2 (triangles) and -12 (squares) zymogens as a function of pH.
Each activity point is normalized by that protease’s maximum activity (relative fluorescence units) at its pH optimum. The error bars represent standard deviation in results obtained from duplicate reads from two separate experiments.
Confirmation that observed activity was not from co-purifying insect cell peptidases
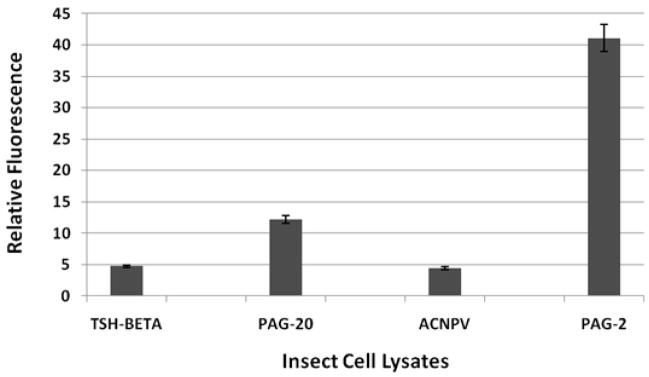
An immunoprecipitation experiment was performed against several insect cell lysates (with the anti-FLAG antibody) followed by peptidase assays on the immunoprecipitated material. The lysates tested represented insect cells infected with wildtype virus and insect cells infected with baculovirus driving expression of recombinant equine TSH-beta (eTSHβ), bovine PAG-2 and boPAG-20 (a modern PAG). The activity obtained from the boPAG-2 lysate was substantially greater than was observed in the two control lysates [eTSHβ and cells infected with wild-type baculovirus (ACNPV)]. Bovine PAG-20 also exhibited some activity, albeit much less than boPAG-2 (Figure 3).
Figure 3. Activity measured after an immuno-precipitation assay performed on insect cell lysates.
Anti-FLAG antibody was used as a capturing antibody and protein G matrix was used as a solid support. Immunoprecipitated proteins were reacted with the cathepsin D/E FRET substrate for one hour in pH 3.5 buffer. Liberated product was measured on a fluorescent plate reader.
pH dependencies of activities of zymogen and activated forms
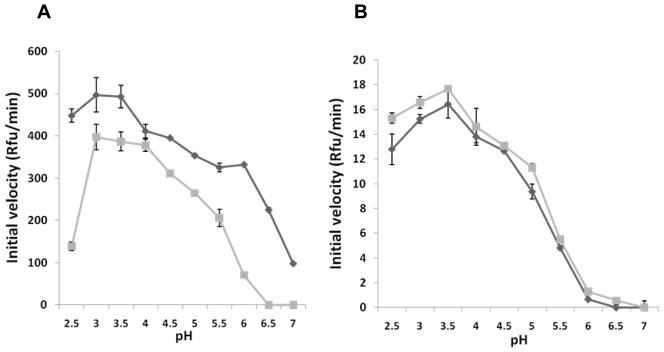
One hallmark of many APs is their ability to remove or displace their propeptides when placed in an acidic environment (Hazel et al., 1992; Koelsch et al., 1994; Richter et al., 1998; Wittlin et al., 1999; Barrett et al., 2004). When boPAG-2 zymogen was incubated in activation buffer (0.25 M glycine-HCl, pH 3.5) for 1 min at RT, the ability of boPAG-2 to exhibit activity at higher pH became evident. When the pre-incubated protein was compared in the assay alongside boPAG-2, which had not been subjected to a pre-incubation step, there was a moderate increase in activity in the pre-incubated form. The difference in activity was most pronounced at pH 2.5 and between pH 5.5 and 6.5 (Figure 4a). The greatest initial rate activity was recorded within the pH optima of 3.5 to 4.0.
Figure 4. Influence of pHon PAG-2 and PAG-12 activity.
The pH-dependence of the activity of recombinant boPAG-2 (A) and boPAG-12 (B) before activation (squares) and following activation (triangles). The initial velocities (in relative fluorescent units/min) of each PAG against the fluorescent FRET-cathepsin D/E substrate at each pH were estimated from kinetic reads for 10 min.
Similar measurements were made on boPAG-12. In contrast to boPAG-2, exposing the zymogen of boPAG-12 to pH 3.5 for 1 min did not result in an increased activity. However, exposing the protein to pH 3.0 for 1 min did produce a marginal increase in activity in the range from pH 2.5 to 5.0 (Figure 4b). Also unlike the situation that was observed with PAG-2, there was no marked improvement in activity between the pre-incubated protein and the intact zymogen between pH 5.5 and 6.5. These experiments were replicated multiple times and the observations were consistent among the experiments.
Comparison of steady-state kinetics of recombinant boPAG-2 and -12
The Michaelis-Menten kinetic parameters kcat, Km and kcat/Km were determined from progress curves for the full-length PAG-FLAG fusions (Figure 5; Table 1). The Ki for pepstatin A was also determined (Table 1). These data revealed differences in catalytic turnover rate and efficiency toward the cathepsin D/E substrate between boPAG-2 and -12. Under identical reaction conditions, the catalytic efficiency of boPAG-2 (2.72 × 105 M−1s−1) was found to be approximately 4-fold greater than boPAG-12 (6.86 × 104 M−1s−1). The kcat for boPAG-2 (0.96 ± 0.08 s−1) was approximately 3.3 fold higher than boPAG-12 (0.29 ± 0.03 s−1). The Km values of 3.53 μM and 4.2 μM for boPAG-2 and -12, respectively, being within 20% of each other indicated that the two enzymes have similar affinity for the substrate. The Ki for pepstatin A was lower by more than an order of magnitude in boPAG-2 (0.56 nM) compared to boPAG-12 (7.5 nM) (Table 1).
Figure 5. Determination of kinetic parameters for PAG-2 and PAG-12.
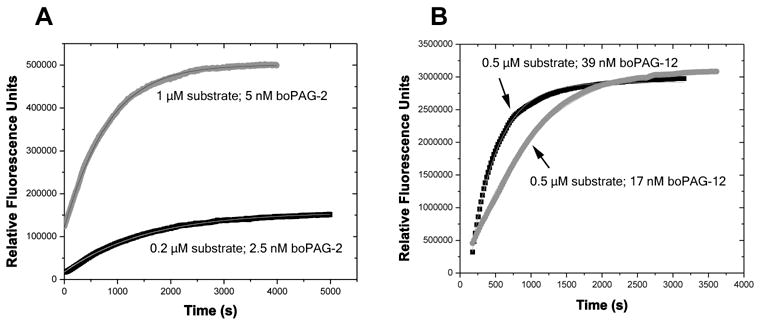
Progress curves global fitted with kcat and Km for digestion of cathepsin D/E substrate by recombinant boPAG-2 (A) and boPAG-12 (B) at pH 4.0 and 37°C. In (A), the FRET substrate and boPAG-2 concentrations for the upper and lower series are 1 μM substrate and 5 nM boPAG-2 enzyme and 0.2 μM substrate and 2.5 nM boPAG-2 enzyme, respectively. In (B), the substrate and boPAG-12 concentrations are 0.5 μM substrate and 39 nM boPAG-12 enzyme and 0.5 μM substrate and 17 nM boPAG-12 enzyme. The lines representing each progess curve are indicated in the panels.
Table 1.
Steady-state enzyme kinetic parameters for bovine PAG-2 and PAG-12.
| KM (μM) | kcat (s−1) | kcat/KM (M−1s−1) | Pepstatin Ki(nM) | |
|---|---|---|---|---|
| BoPAG-2 | 3.53±0.03 | 0.96±0.08 | 272,168±277 | 0.56 |
| BoPAG-12 | 4.2±0.3 | 0.29±0.03 | 68,638±415 | 7.5 |
Substrate preferences of boPAG-2 and -12 in comparison to porcine gastric pepsin and bovine spleen cathepsin D
To gain insight into the primary peptide specificity for boPAG-2 and -12, both full-length forms were investigated for their activity against commercially available FRET-25X peptide libraries alongside two canonical mammalian APs, porcine pepsin (from gastric mucosa) and bovine cathepsin D (from spleen). While porcine pepsin and cathepsin D displayed good activity against the primary substrate libraries, boPAG-2 and -12 failed to produce reasonable velocities (Table 2). Of the two boPAGs, boPAG-12 exhibited a detectable, albeit minor, release of product. In contrast, boPAG-2 failed to exhibit detectable activity, even with relatively high concentrations of the enzymes (100 nM of active-site titrated enzyme) (Table 2).
Table 2.
Initial velocities (RFU/min) obtained from the proteolysis of 25-X FRET substrate libraries.
| Ala | Arg | Asn | Asp | Gln | Glu | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Tyr | Trp | Val | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pepsin | 489 | 871 | 431 | 347 | 480 | 397 | 268 | 469 | 2063 | 2168 | 0 | 1860 | 335 | 450 | 341 | 717 | 272 | 449 | 1578 |
| Cathepsin D | 197 | 293 | 395 | 321 | 369 | 303 | 156 | 0 | 120 | 307 | 0 | 235 | 0 | 528 | 250 | 492 | 0 | 176 | 0 |
| Pag-2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pag-12 | 3.6 | 6.5 | 2.4 | 2.1 | 38.9 | 22 | 5.2 | 2.4 | 3.7 | 4.3 | 5.5 | 4.2 | 19.4 | 5.2 | 1.8 | 2.4 | 14.2 | 4.8 | 5.6 |
Homology-based modeling of the structures of boPAGs -2 and -12
The homology models for boPAGs-2 and -12 were created by using crystal coordinates of human pepsin bound to pepstatin A (1psn.pdb). SWISS-MODEL predicted this as one of the best templates for generating the model. Previously reported homology modeling of a modern PAG, boPAG-1, identified key residues predicted to contribute to the substrate binding pockets (Guruprasad et al., 1996).
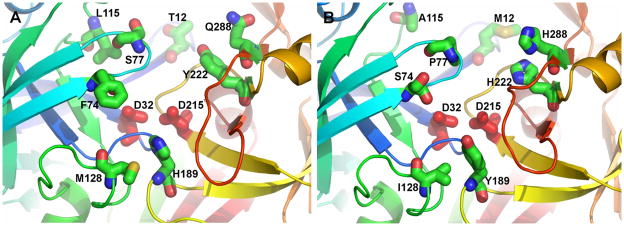
Based on this exercise, it was determined that eight sequence positions may distinguish the active sites of pepsin, boPAG-2, boPAG-12, and homologues (Figure 6). Their differing side chains may tune their respective substrate specificities. The subsites proceed in the views of Figure 6 from S4 in the upper background towards S3′ (primed) in the lower foreground. The subsites were numbered according to the structure of the complex of pepsin with pepstatin (Fujinaga et al., 1995). Either Thr12 and Gln288 or Met12 and His288 appear at S4 in the respective homology models of boPAG-2 and -12 (toward the back at top in Figure 6). Leu115 or Ala115 contribute to both S3 and S1 in boPAG-2 and -12, respectively. Either Ser77 or Pro77 of the flap of these two proteases contribute to both S2 and S1. Either Tyr222 or His222 form the other wall of S2 on the opposite side of the cleft (Figure 6). Aromatic groups also differentiate S1′ with either His189 of boPAG -2 or Tyr189 of boPAG-12. Long aliphatic chains of either Met 128 or beta-branched Ile128 appear at S2′ in the two respective enzymes. For the flap’s contribution to S3′, the bulky hydrophobic Phe74 of boPAG -2 is a notable contrast to the small polar Ser74 of boPAG-12.
Figure 6. Homology models to predict substrate binding site differences between bovine PAG-2 and PAG-12.
Residues that distinguish the active sites of boPAG-2 (A) and boPAG-12 (B) and pepsin (not shown). The homology models were constructed by using the crystal structure of mature human pepsin bound to pepstatin (PDB code 1PSN) as template by using the SWISS-MODEL server (http://swissmodel.expasy.org//SWISS-MODEL.html) (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006). The backbone ribbon progresses through the colors of the rainbow from blue at the N-terminus to red at the C-terminus. The conserved aspartate side chains critical for peptidase activity are red. Side chains of the active site cleft differing between boPAG-2 and -12 are plotted with standard atom colors of blue for nitrogen, red for oxygen, and green for carbon. The “flap” is light blue.
Discussion
Pregnancy-associated glycoproteins are related to APs and are abundantly expressed products of the placenta of ruminants and other species within the Artiodactyla order. Although PAGs have been studied extensively, their role(s) within the ungulate placenta remains largely obscure. Despite being structurally related to APs, some of the modern PAGs have accumulated mutations within key residues in and around the catalytic site, including the critical catalytic aspartates (Xie et al., 1991; Xie et al., 1997; Green et al., 1998). Those modern PAGs carrying such mutations are likely to be incapable of acting as peptidases. The ancient PAGs of cattle, sheep and goats, in contrast, have all the definitive characteristics of typical APs and were predicted to have proteolytic activity (Green et al., 1998; Wooding et al., 2005). Consequently, the goal of this project was to address two important questions, (1) do ancient bovine PAGs have proteolytic activity and, if so, (2) how do closely related family members compare to one another in regard to kinetic parameters? To address these fundamental questions, boPAGs-2 and -12 were chosen as candidates for these experiments. BoPAG-2 was an obvious choice since it is the most abundantly transcribed member of the bovine PAG grouping (Telugu et al., 2009). It is also among the first PAGs to have been identified and characterized in regard to its expression by trophoblasts and in regard to its abundant localization at the placenta-uterine interface (Xie et al., 1994; Wooding et al., 2005). BoPAG-12 is closely related to boPAG-2, sharing 89% identity at the nucleotide level and 83% identity in amino acid composition (Green et al., 2000; Hughes et al., 2000). However, its level of expression is orders of magnitude below that for boPAG-2, despite that it is also localized to the placenta-uterine interface (Telugu et al., 2009).
A baculovirus insect cell expression system was chosen for expression of soluble recombinant full-length PAGs. This system is amenable to expression of acid peptidases because the cytoplasm in these cells is not strongly reducing and is maintained close to a pH of 7.0 (Medina et al., 1995). Such conditions were predicted to help prevent the activation and subsequent loss of acid peptidases by autocatalysis. The inclusion of a synthetic coding sequence for the FLAG peptide permitted the isolation of relatively homogenous preparations of purified proteins by using standard anti-FLAG affinity purification protocols.
The purified full-length protein preparations were tested for optimal activity against a synthetic FRET substrate MOCAC-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(Dnp)-D-Arg-NH2. This peptide was derived from a consensus substrate sequence for APs [Lys-Pro-Ile-Gln-Phe*Nph-Arg-Leu(Nph: nitro-phenylalanine)] described elsewhere (Beyer et al., 2005). The fluorescent substrate was initially designed and tested for utility against other well-characterized APs, Cathepsin D and E (Yasuda et al., 1999). Both boPAG-2 and -12 exhibited activity toward this substrate (Table 1).
The optimal activity for PAG-2 and -12 was around pH 4 and 3.5 respectively (Figure 2). Such acidic pH optima are typical for most APs. Of the two candidate PAGs, boPAG-12 had greater activity at pH 2.5 compared to boPAG-2. When comparing their kinetic parameters from progress curves, it was apparent that boPAG-2 is almost four-fold more active than boPAG-12 at their pH optima, despite that they are closely related and have similar affinity for the cathepsin D/E substrate. It was determined that boPAG-2 had a catalytic turnover rate more than 3 times greater than boPAG-12 under identical experimental conditions. In addition, the affinity of boPAG-2 for pepstatin A was also more than an order of magnitude greater than boPAG-12. Finally, boPAG-2 and boPAG-12 differed from one another in regard to the increase in activity observed upon a short pre-incubation at low pH prior to performing kinetic measurements (Figure 4). In most aspartic peptidases, acidification of the protein permits an intra-or intermolecular cleavage of the propeptide (Hazel et al., 1992; Koelsch et al., 1994; Richter et al., 1998; Wittlin et al., 1999; Barrett et al., 2004). The loss of the propeptide is thought to permit more ready access of substrate to the active site and also produces an increased mobility by SDS-PAGE due to the remove of the N-terminal peptide (40–50 amino acids, depending on the protein). In the pre-incubation experiments, boPAG-2 exhibited characteristics similar to other aspartic peptidases in that an increase in activity was observed after low pH exposure. Bovine PAG-12, in contrast, exhibited little increase in activity. After extensive incubation periods at low pH it was found that boPAG-12 exhibited no shifts in mobility (i.e. no cleavage of the propeptide) by SDS-PAGE (data not shown). Bovine PAG-2 did appear to process itself after incubation periods, but the processing did not involve the loss of the N-terminus and, instead, seemed to consist of non-specific cleavage during the incubation period (data not shown). One interpretation of these observations are that boPAG-2 and boPAG-12, while peptidases, are not able to remove their own propeptides and may require pro-protein converting peptidases to perform this function.
The experiments performed with the synthetic 25-x FRET substrate libraries suggested that boPAG-2 and -12 may be relatively selective in regard to the length or composition of the substrate required for detectable activity. Both porcine pepsin and bovine cathepsin D exhibited activity toward these libraries in a manner consistent with what is already known about their substrate specificities (Abad-Zapatero et al., 1990; Scarborough and Dunn, 1994; Barrett et al., 2004). However, both boPAG-2 and -12, when used at the same molar concentration, failed to produce significant product liberation from these libraries(Table 2). This lack of activity may be reminiscent of studies performed with furin, which is fastidious when it comes to substrate sequences required for optimal activity. In those experiments, furin failed to show activity against these same substrate libraries (information provided by Peptides International). The lack of activity against these libraries could be explained if the library failed to provide peptide sequences of the appropriate length or composition for optimal activity with both PAG-2 and 12. However, it is worth mentioning that PAG-12 did produce at least some activity when compared to PAG-2, which suggests that PAG-12 might be more flexible in regard to its substrate requirements.
The fact that bovine PAG-2 and -12 have distinct pH profiles and inhibitor affinities, suggested that there are likely to be differences in residues that are known to interact with the substrate to potentially stabilize it in the substrate binding cleft. In fact, such differences were revealed from comparative modeling of these proteins based on the crystal coordinates of human pepsin (Figure 6). For example Thr12 in boPAG-2 that contributes to the S4 pocket, is substituted by Met12 in boPAG-12. Such a change may alter the preference for a hydrophobic amino acid in the substrate occupying this sub pocket. Similarly, Leu115 contributes to the S3 and S1 sites in boPAG-2, while the somewhat smaller Ala115 shares the comparable position in boPAG-12. Another major change was that of Ser77 in boPAG-2 by Pro77 in boPAG-12, this position is of crucial importance since, it forms part of the hairpin ‘flap’ structure. The flap is a conserved fold in the mature molecule that arches over the substrate binding cleft and interacts and stabilizes the substrate within the substrate binding cleft (Hartsuck et al., 1992; Guruprasad et al., 1996; Richter et al., 1998; Dunn, 2002). It makes contact with numerous residues in the substrate. Those residues at position 77 contribute to the S2 and S1 pockets. The replacement of Ser77 in boPAG-2 with Pro77 in boPAG-12 marks another deviation between the two PAGs that might alter the dynamics of substrate stabilization within the substrate binding cleft. Another change within the flap was the substitution of the bulky hydrophobic aromatic amino acid Phe74 in boPAG-2 with the small polar Ser74. These residues contribute to the S3′ pocket. Collectively, these differences, as well as those described in the Results section, suggest the following speculations on potential specificity differences between boPAG-2 and -12: BoPAG-12 might be able to accommodate a bulkier side chain at P3 and/or P1 of putative substrates since the Leu115 of boPAG-2 fills these pockets more than does the Ala115 of boPAG-12. BoPAG-12 would be predicted to accommodate a bulkier or more polar side chain at P3′ since it presents Ser74 (instead of Phe74) at S3′.
Although the two PAGs investigated in these experiments are active peptidases, there was little indication that they act as general peptidases, like pepsin. These data suggested that they may have somewhat refined roles in cleaving protein substrates, either within the trophoblast itself or at the fetal-maternal interface. The PAGs are secreted glycoproteins, hence they are exposed to acidic environments (potentially as low as pH 5.2) within the secretory pathway (Paroutis et al., 2004). The PAGs might have a major role within the secretory pathway, where they might be activated by an unidentified peptidase or by auto-activation (Davies, 1990; Dunn, 2002). These activated PAGs might then participate in proteolytic processing of other proteins within the secretory vesicles before they exit the cell. Alternatively, they might be diverted towards the endo-lysosomal pathway where they could function by digestion of endocytosed agonist-bound receptors or proteins taken up from uterine secretions (uterotroph). Since both the identified PAGs seem to be somewhat selective in regard to substrate preference, it may be safe to hypothesize that the ancient PAGs do not function as general degradative enzymes like cathepsin D and E, which are lysosomal and endosomal peptidases, respectively (Godbold et al., 1998; Ishidoh and Kominami, 2002). Bovine PAG-2/-12 are constitutively secreted and accumulate at the maternal-fetal interface (Wooding et al., 2005). Therefore, they might play roles as ‘sheddases’ to activate latent growth factors or as peptidases involved in inactivating growth factor binding proteins or other proteins within the pericellular microenvironment (Munger et al., 1998; Rifkin et al., 1999). Some data do exist to indicate that this environment may be slightly acidic (pH 6.5) (Punturieri et al., 2000). In either situation, such proteolytic conversion of trophic factors may have an important role in promoting trophoblast/placentomal growth and differentiation in the ruminant placenta (Ko et al., 1991; Mathialagan and Roberts, 1994; Grundker and Kirchner, 1996; Yelich et al., 1997; Tanaka et al., 1998; Rifkin et al., 1999; Spencer and Bazer, 2004). Future efforts will be directed toward exploring these varied possibilities.
Materials and methods
Cloning and expression of recombinant bovine PAGs-2 and 12
The BD Baculogold™ Baculovirus insect cell expression system (BD Biosciences Pharmingen, San Diego, CA) was used to express the recombinant proteins. BoPAG-2 was cloned into the pVL92 transfer vector by using the following oligonucleotides, sense: 5′ GAC TGA GCGGCCGCATGGATTACAAGGACGATGACGATAAGATAGTCATTTTGCCTCTA 3′ and antisense: 5′ GTCAGTC AGAGTCAGAGTCATGACTAGAGTCTAGATGACTATTACACTGCCGGAGCCAG 3′. Bovine PAG-12 was cloned into the pACMP3 transfer vector with the following oligonucleotides, sense: 5′ GACTCTAGAATGGATTACAAGGACGATGACGATAAG ATAGTCATTTTGCCTCTA 3′ and antisense: 5′GATCTATGATCTCAGTACT GCGGCCGCTCACTATTACACCTGTGCCAGGCCAAT 3′. The recombinant proteins were expressed as fusion proteins with a FLAG-tag in the N-terminus of the protein. A sequence encoding FLAG peptide (DYKDDDDK), shown as regular bold in the sense oligonucleotide, was engineered into the sequence so that this peptide would be incorporated into the N-terminus of the proforms of both PAG-2 and -12. Sequences encoding restriction enzymes (bold italicized) Not-1 and Bgl-2 (New England Biolabs, MA USA) were also engineered into the sense and antisense oligonucleotides to permit directional cloning into the corresponding transfer plasmids. Once the integrity and frame of the sequences in the transfer vectors was verified by sequencing, the vectors were transfected into Sf-9 cells along with BD Baculogold linearized Baculovirus DNA by using the BD Baculogold transfection kit according to the manufacturer’s recommendations. Following transfection, the viruses were extracted, amplified and used to infect Sf-9 cells to generate recombinant proteins as described elsewhere (O’Reilly et al., 1992). Infected cells were harvested, chilled on ice, centrifuged at 600 g for 5 min at 4 °C followed by two wash cycles under similar conditions with cold 1x PBS (2.68 mM KCl, 1.47 mM KH2PO4, 136.89 mM NaCl and 8.10 mM Na2HPO4, pH 7.2). The final cell pellet was stored at −80 °C until use.
Purification of recombinant boPAG-2 and 12
For purification of the recombinant protein, the corresponding frozen pellets were lysed on ice with I-Per insect cell protein extraction reagent (Pierce, IL, USA). A standard cocktail of protease inhibitors, which included 0.4 mM Pefabloc SC-AEBSF (Roche Applied Science), 5 μg/mL aprotinin, 10 μM E-64, 1 mM EDTA along with 1 mM DTT, (Sigma, MO, USA) was supplemented to the lysis buffer just before use. Following mixing and incubation with lysis buffer for at least 15 min on ice, the lysate was cleared by centrifugation at 15,000 × g for 30 min and dialyzed overnight in a 30 kD MWCO dialysis tubing in a buffer containing 20 mM Tris-HCl, pH 7.4, 250 mM NaCl at 4 °C. All purification procedures were performed in a refrigerated room at 4–6 °C. The dialysed lysate was fractionated on a Sephadex-200 size exclusion column (1.5 cm × 106 cm) equilibrated in 20 mM Tris-HCl, pH 7.4, 150 mM NaCl. All fractions that were determined to have the FLAG peptide present (by dot-blot with an anti-FLAG M2 antibody) were pooled and subsequently affinity purified by using anti-FLAG M2 agarose (Sigma, MO, USA). For affinity chromatography, the matrix was equilibrated with TBS buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.4), following which, the FLAG-containing protein samples were loaded twice onto the column by gravitational flow at approximately 0.2 mL/min. The column was then subjected to subsequent washes with 20 column volumes of wash buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.4), 20 column volumes of high salt buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.4) and finally 20 column volumes of high salt buffer supplemented with 0.1% Tween. The anti-FLAG M2 agarose was re-equilibrated with 10 column volumes of wash buffer to remove residual detergent, followed by 10 column volumes of pre-elution buffer (10 mM phosphate buffer, pH 7.2). The column was then eluted with 5 column volumes of 50 mM phosphate buffer, 2 M MgCl2, pH 7.2. The protein sample was desalted by dialysis in 20 mM Tris-HCl, 250 mM NaCl, pH 8.0 and concentrated by using an Amicon Ultra-15 with Ultra cell-30 membrane (Millipore, MA, USA). The concentrated protein samples were protected with the addition of the inhibitor cocktail and glycerol to 10% (v/v) at 4°C. In most cases, the protein samples were immediately used in the assays. For long term storage, the protein sample was stored in 50% glycerol at −80 °C.
Western blot analysis
For the Western blot experiments the recombinant proteins were electrophoretically transferred onto Immobilon PVDF-membrane (Millipore, MA, USA). The membranes were washed once with an excess of 1x TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween, pH 7.5) and blocked in blocking buffer that consisted of 3% bovine serum albumin and 3% non fat dry milk (Sigma, St. Louis, MO, USA) in 1x TBST. The blots were subsequently incubated with either a 1:1000 dilution of monoclonal anti-FLAG antibody (Sigma) or 1:2000 polyclonal anti-boPAG-2 anti-serum (to further confirm the presence of boPAG-2) in blocking buffer. The blots were then washed and incubated with 1:2000 dilution of anti-mouse (for anti-flag) or anti-rabbit IgG (for boPAG-2 antiserum) conjugated to alkaline phosphatase for 45 min (Promega, WI, USA). The blots were finally washed and stained with a mixture of NBT (Nitro-Blue Tetrazolium chloride) and BCIP (5-bromo-4-chloro-3′-indolyphosphate p-toluidine Salt) according to the manufacturer’s instructions (Promega, WI, USA). The production and characterization of the boPAG-2 antiserum has been described previously (Wooding et al., 2005)
Determining optimal pH for activity studies
To estimate the optimal pH for each PAG, the recombinant PAGs were incubated in various buffers. All the pH activity experiments were conducted at 35 °C. A synthetic FRET (fluorescence resonance energy transfer) cathepsin D/E substrate, MOCAC-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(Dnp)-D-Arg-NH2 [MOCAc: (7-methoxycoumarin-4-yl) acetyl; Dnp: 2,4-dinitrophenyl] (Peptides International, KY, USA) was used to investigate the activity of bovine PAG-2 and -12. The substrate was dissolved in 10% DMSO to a final concentration of 200 μM, aliquoted into 10 μL samples, and stored in the dark at −80 °C until use. Equal amounts of protein sample(approximately 500 ng in 20 μL) were incubated with 20 μM of substrate. For determination of optimal pH, the following buffers were used: 0.1 M glycine-HCl buffer for pH 2.5, 3.0 and 3.5; 0.1 M sodium citrate-citric acid buffer for pH 4 and 4.5; 0.1 M sodium acetate-acetic acid for 5 and 5.5; 0.1 M Bis-tris-HCl buffer for pH 6 and 6.5; 0.1 M HEPES for pH 7; and 0.1 M Tris-HCl for pH 7.5 and 8.0 buffers. Each reaction solution also contained NaCl at a final concentration of 0.1 M. The final volume of the reaction was maintained at 100 μL. The reaction was performed for 20 min and was terminated by addition of 900 μL of 5% trichloroacetic acid (TCA), as described previously (Yasuda et al., 1999). The resultant mixture was further diluted to 2 mL with 5% TCA and the resultant fluorescence in the mixture was read in a PC1™ Photon counting spectofluorimeter at 328 nm (excitation) and 393 nm (emission) wavelengths as described previously (Yasuda et al., 1999). All the experiments were set up in duplicate. The results from duplicate reads and from two successive experiments were used to compile the data.
Estimation of contamination by insect cell-derived acid peptidases
A parallel purification experiment was performed to confirm that the activity being measured was due to recombinant PAGs and not from residual insect cell-or baculovirus-derived peptidases that might have been retained after isolation of the PAGs. The lysates used for this experiment were obtained from infected insect cells expressing boPAG-2, boPAG-20 and, as a negative control, equine-thyroid stimulating protein-beta (eTSHβ). The boPAG-20 and eTSHβ were cloned and engineered to contain a FLAG peptide in a similar approach to that used to express boPAG-2 and boPAG-12. An additional control for this experiment consisted of lysed insect cells that had been infected with wild-type baculovirus (ACNPV). One mL of each lysate was pre-cleared by incubating with 100 μL of protein G (50%) bead slurry (BioRad, USA) on a rocker shaker at 4°C for half hour. The protein G beads were removed by centrifugation at 14,000G at 4°C for 10 min. Equal amounts (500μg) of insect cell lysates were mixed with 5 μg of anti-FLAG antibody and rotated at 4°C for 2 h. Protein G agarose matrix was added at 100 μL (50% slurry) and the resulting mixture was incubated overnight. The matrix was isolated by centrifugation and washed with 20 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.1% Tween buffer three times. After the final wash, the matrix was incubated with 0.1 M glycine-HCl buffer pH 3.5 containing 0.1M NaCl and 20 μM fluorescent substrate (described above) in a final volume of 100 μL at 40°C for 1 h. The resulting solution was centrifuged at 14,000 G for 10 min to precipitate the protein G matrix and 50 μL of supernatant was mixed with equal amounts of 10% TCA and the resultant fluorescence was measured by using a Fluroscan plate reader (Ascent, USA).
Establishing pH profiles for pro-and activated forms of recombinant boPAG-2 and boPAG-12
A feature of APs is the ability to auto-activate. In most cases, this phenomenon involves removal of the propeptide by either an intra-or intermolecular mechanism (Hazel et al., 1992; Koelsch et al., 1994; Richter et al., 1998; Wittlin et al., 1999; Barrett et al., 2004). After defining activation procedures for boPAG-2 (data not shown), an optimal scheme was identified that involved adding one-third volume of 0.25 M glycine-HCl, pH 3.5 (the activation buffer) to the sample of boPAG-2 (stored in 20 mM Tris-HCl, pH 8.0, 250 mM NaCl buffer), mixing and incubating for 1 min at RT. A similar activation procedure for boPAG-12 entailed adding one-third volume of 0.25 M glycine-HCl, pH 3.0 to the sample of boPAG-12, mixing and incubating for 1 min at RT. To estimate the profile of the ‘activated’ protein as a function of pH and to compare it to the activity of the unactivated protein, approximately 620 ng of boPAG-2 or 540 ng of boPAG-12 total protein (either the unactivated or activated form) was incubated with 20 μM of substrate in buffers of varying pH that were described above. The samples were placed in a 96-well Costar black microtitre plate (Corning, USA) and subsequently incubated at 37°C for 10 min in a Synergy-HT Fluorescent plate reader (Biotek, USA). Following the addition of substrate, the kinetic reads were obtained for the first 10 min of the reaction and the calculated initial rates were displayed as relative fluorescent units (rfu)/min. The experiment was conducted with samples in triplicate to estimate the experimental noise. The same experiment was replicated multiple times and data from a representative experiment was shown.
Determination of steady-state enzyme kinetics of boPAG-2 and -12
The steady-state enzyme kinetic parameters for boPAG-2 and -12 were estimated by fitting to progress curves as described elsewhere (Palmier and Van Doren, 2007). The concentration of total protein in the purified samples was estimated by BCA protein assay (Pierce, Rockford, IL). Accurate estimates of specific activity of PAG preparations relied upon accurate active enzyme concentrations measured by active site titrations with the tight-binding inhibitor of aspartic peptidases known as pepstatin A, fitted as described (Knight, 1995; Copeland, 2000). The Ki for PAGs was estimated by fitting the data to the following equation by a procedure detailed in the reference (Knight, 1995):
where Eo and Io are the initial concentration of enzyme and inhibitor respectively, νo, is the initial rate of the reaction without inhibitor and ‘ν’ is the observed velocity.
Accurate measurement of kcat/Km was obtained from single progress curves collected under pseudo first-order conditions of [S] ≪ Km and [S] ≫ [E], as illustrated by the slower curves in Fig. 5, fitted to the following equation:
where y is the relative fluorescence intensity, Fmax is the maximum change in fluorescence intensity during the reaction, Et is the enzyme active site concentration in the assay, x is time in seconds, and B is a Y axis offset correction (Orsi and Tipton, 1979; Niedzwiecki et al., 1992; Neumann et al., 2004). Steady-state kinetics parameters, kcat and Km, were determined from two progress curves, one of which was under pseudo first-order conditions of [S] ≪ Km and [S] ≫ [E]. Origin Pro 7.5 (Microcal) was used to obtain kcat and Km from global, nonlinear fitting of the pairs of progress curves as described (Palmier and Van Doren, 2007). All kinetic experiments were performed in 0.1 M sodium citrate-citric acid, 0.1 M NaCl (pH 4.0) buffer at 37 °C with the fluorescent (FRET) Cathepsin D/E substrate in 3×3 mm cuvettes to reduce the path length and inner filter effect (Matayoshi et al., 1990; Lakowicz, 1999; Y. Liu et al., 1999). The data were obtained by monitoring product development in a SLM Aminco fluorometer (model 8100) with PC1™ Photon counting upgrade (ISS Inc., IL, USA).
Comparison of Aspartic Protease Specificity by using a FRET Peptide Substrate Library
To gain insight into differences in substrate preferences, if any, between full-length boPAG-2 and -12, a synthetic 25-x FRET substrate library (Peptides International, KY, USA) was employed. The library consisted of 5 different amino acids at position Y (Pro, Tyr, Lys, Ile and Asp) and Z (Phe, Ala, Val, Glu and Arg) for each residue at position X [D-A2pr(Nma)-Gly-(Zaa)5-(Yaa)5-Xaa-Ala-Phe-Pro-Lys(Dnp)-D-Arg-D-Arg. There are 19 different primary substrate libraries with each natural amino acid at position X with the exception of cysteine. Therefore, the libraries represented 475 distinct peptides. The library is useful for delineating optimal substrate sequence for a given peptidase, based on initial rates for the 19 different primary libraries by the peptidase, followed by more refined analysis (Tanskul et al., 2003; Hiraga et al., 2005; Oda et al., 2005; Namwong et al., 2006). Pre-extracted native preparations of cathepsin D (bovine spleen) and pepsin (porcine stomach) were obtained (Sigma) and used in the assay. All the preparations were active site titrated, as for the kcat and KM determinations. Equal amounts (5 nM) of active enzyme and 20 μM of substrate were used in the assay. The assay was performed in duplicate under conditions described for the kinetic analysis [0.1 M sodium citrate -citric acid, 0.1 M NaCl, pH 4.0 at 37°C]. The initial velocities were obtained against the primary substrate libraries for first 10 min of the reaction using the fluorescent plate reader.
Computational methods for generating homology models of boPAG-2 and -12
Predicted 3-D models for boPAGs-2 and -12 were constructed by using SWISS-MODEL (http://swissmodel.expasy.org//SWISS-MODEL.html) in automated mode (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006). The crystal structure of human pepsin conjugated with pepstatin A (1psn.pdb) was used as a template to generate the model (Fujinaga et al., 1995; Westbrook et al., 2003). Multiple sequence alignments of boPAGs -2, -12, boPAG-1 and human pepsin were generated by using CLUSTALW (http://align.genome.jp). The template structure of human pepsin shares sequence identity of 50.6% with boPAG-2 and 53.4% with boPAG-12. Specific residues predicted to constitute the substrate binding pockets for boPAG-1 were reported elsewhere (Guruprasad et al., 1996). The residues contributing to specific substrate binding pockets in boPAG-2 and -12 were identified from the alignments with boPAG-1. The structural views of the homology models in Figure 6 were generated by using Pymol (DeLano).
Acknowledgments
This work was supported by funding from Monsanto Co. (St Louis, Missouri) and the Animal Reproductive Biology Group of the University of Missouri, Food for the Twenty-first Century Program. Additional support came from National Institutes of Health grants R01 GM57289 (SRV) and training grant GM008396 for MOP.
Footnotes
References
- Abad-Zapatero C, Rydel TJ, Erickson JW. Revised 2.3 A0 structure of porcine pepsin: evidence for a flexible subdomain. Proteins. 1990;8:62–71. doi: 10.1002/prot.340080109. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Barrett A, Rawlings N, Woessner J. Hanbook of proteolytic enzymes. Elsevier Academic Press; San Diego: 2004. [Google Scholar]
- Beyer BB, Johnson JV, Chung AY, Li T, Madabushi A, Agbandje-McKenna M, McKenna R, Dame JB, et al. Active-Site Specificity of Digestive Aspartic Peptidases from the Four Species of Plasmodium that Infect Humans Using Chromogenic Combinatorial Peptide Libraries. Biochemistry. 2005;44:1768–1779. doi: 10.1021/bi047886u. [DOI] [PubMed] [Google Scholar]
- Brandt GA, Parks TE, Killian G, Ealy AD, Green JA. A cloning and expression analysis of pregnancy-associated glycoproteins expressed in trophoblasts of the white-tail deer placenta. Mol Reprod Dev. 2007;74:1355–1362. doi: 10.1002/mrd.20669. [DOI] [PubMed] [Google Scholar]
- Cody RJ. The clinical potential of renin inhibitors and angiotensin antagonists. Drugs. 1994;47:586–598. doi: 10.2165/00003495-199447040-00003. [DOI] [PubMed] [Google Scholar]
- Cooper JB, Khan G, Taylor IJ, Tickle IJ, Blundell TL. X-ray analyses of aspartic peptidases II. Three dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3A° resolution. J Mol Biol. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- Copeland RA. Enzymes: a Practical Introduction to Structure, Mechanism, and Data Analysis. 2. Wiley-VCH, Inc; New York: 2000. [Google Scholar]
- Davies DR. The structure and function of the aspartic proteinases. Annu Rev Biophys Biophys Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: [Google Scholar]
- Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- Fujinaga M, Chernaia MM, Tarasova NI, Mosimann SC, James MNG. Crystal structure of human pepsin and its complex with pepstatin. Protein Sci. 1995;4:960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbayo JM, Remy B, Alabart JL, Folch J, Wattiez R, Falmagne P, Beckers JF. Isolation and partial characterization of a pregnancy-associated glycoprotein family from the goat placenta. Biol Reprod. 1998;58:109–15. doi: 10.1095/biolreprod58.1.109. [DOI] [PubMed] [Google Scholar]
- Garbayo JM, Green JA, Mannekin M, Beckers J-F, Kiesling DO, Ealy AD, Roberts RM. Caprine pregnancy-associated glycoproteins (PAG): their cloning, expression and evolutionary relationship to other PAG. Mol Reprod Devel. 2000;57:311–322. doi: 10.1002/1098-2795(200012)57:4<311::AID-MRD2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Godbold GD, Ahn K, Yeyeodu S, Lee LF, Ting JP, Erickson AH. Biosynthesis and intracellular targeting of the lysosomal aspartic proteinase cathepsin D. Adv Exp Med Biol. 1998;436:153–162. doi: 10.1007/978-1-4615-5373-1_21. [DOI] [PubMed] [Google Scholar]
- Green JA, Xie S, Roberts RM. Pepsin-related molecules secreted by trophoblast. Rev Reprod. 1998;3:62–69. doi: 10.1530/ror.0.0030062. [DOI] [PubMed] [Google Scholar]
- Green JA, Xie S, Quan X, Bao B, Gan X, Mathialagan N, Beckers J-F, Roberts RM. Pregnancy-Associated Bovine and Ovine Glycoproteins Exhibit Spatially and Temporally Distinct Expression Patterns During Pregnancy. Biol Reprod. 2000;62:1624–1631. doi: 10.1095/biolreprod62.6.1624. [DOI] [PubMed] [Google Scholar]
- Grundker C, Kirchner C. Influence of uterine growth factors on blastocyst expansion and trophoblast knob formation in the rabbit. Early Pregnancy. 1996;2:264–270. [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guruprasad K, Blundell TL, Xie S, Green J, Szafranska B, Nagel RJ, McDowell K, Baker CB, et al. Comparative modelling and analysis of amino acid substitutions suggests that the family of pregnancy-associated glycoproteins includes both active and inactive aspartic proteinases. Protein Eng. 1996;9:849–56. doi: 10.1093/protein/9.10.849. [DOI] [PubMed] [Google Scholar]
- Hartsuck JA, Koelsch G, Remington SJ. The high-resolution crystal structure of porcine pepsinogen. Proteins. 1992;13:1–25. doi: 10.1002/prot.340130102. [DOI] [PubMed] [Google Scholar]
- Hazel HB, Kielland-Brandt MC, Winther JR. Autoactivation of proteinase A initiates activation of yeast vacuolar zymogens. European Journal of Biochemistry. 1992;207:277–283. doi: 10.1111/j.1432-1033.1992.tb17048.x. [DOI] [PubMed] [Google Scholar]
- Hiraga K, Nishikata Y, Namwong S, Tanasupawat S, Takada K, Oda K. Purification and Characterization of Serine Proteinase from a Halophilic Bacterium, Filobacillus sp. RF2-5. Biosci Biotechnol Biochem. 2005;69:38–44. doi: 10.1271/bbb.69.38. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Garbayo JM, Roberts RM. Adaptive Diversification within a Large Family of Recently Duplicated, Placentally-Expressed Genes. Proc Natl Acad Sci. 2000;97:3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Piontkivska H, Roberts RM. Aspartic Proteinase Phylogeny and the Origin of Pregnancy-Associated Glycoproteins. Mol Biol Evol. 2003;20:1940–1945. doi: 10.1093/molbev/msg217. [DOI] [PubMed] [Google Scholar]
- Ishidoh K, Kominami E. Processing and activation of lysosomal proteinases. Biol Chem. 2002;383:1827–1831. doi: 10.1515/BC.2002.206. [DOI] [PubMed] [Google Scholar]
- Kay J, Dunn BM. Substrate specificity and inhibitors of aspartic proteinases. Scand J Clin Lab Invest Suppl. 1992;210:23–30. [PubMed] [Google Scholar]
- Knight CG. Active-site titration of peptidases. Methods Enzymol. 1995;248:85–101. doi: 10.1016/0076-6879(95)48008-0. [DOI] [PubMed] [Google Scholar]
- Ko Y, Lee CY, Ott TL, Davis MA, Simmen RC, Bazer FW, Simmen FA. Insulin-like growth factors in sheep uterine fluids: concentrations and relationship to ovine trophoblast protein-1 production during early pregnancy. Biol Reprod. 1991;45:135–42. doi: 10.1095/biolreprod45.1.135. [DOI] [PubMed] [Google Scholar]
- Koelsch G, Mares M, Metcalf P, Fusek M. Multiple functions of pro-parts of aspartic proteinase zymogens. FEBS Lett. 1994;343:6–10. doi: 10.1016/0014-5793(94)80596-2. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Specroscopy. Kluwer Academic/Plenum; New York: 1999. [Google Scholar]
- Matayoshi ED, Wang GT, Krafft GA, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- Mathialagan N, Roberts RM. A role for cytokines in early pregnancy. Indian J Physiol Pharmacol. 1994;38:153–62. [PubMed] [Google Scholar]
- Medina M, Lopez-Rivas A, Zuidema D, Belsham GJ, Domingo E, Vlak JM. Strong buffering capacity of insect cells. Implications for the baculovirus expression system. Cytotechnology. 1995;17:21–26. doi: 10.1007/BF00749217. [DOI] [PubMed] [Google Scholar]
- Munger JS, Harpel JG, Giancotti FG, Rifkin DB. Interactions between growth factors and integrins: latent forms of transforming growth factor-beta are ligands for the integrin alphavbeta1. Molecular Biology of the Cell. 1998;9:2627–38. doi: 10.1091/mbc.9.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namwong S, Hiraga K, Takada K, Tsunemi M, Tanasupawat S, Oda K. A halophilic Serine proteinase from Halobacillus sp. SR%-3 isolated form fish sauce: purification and characterization. Biosci Biotechnol Biochem. 2006;70:1395–1401. doi: 10.1271/bbb.50658. [DOI] [PubMed] [Google Scholar]
- Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem. 2004;328:166–173. doi: 10.1016/j.ab.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki L, Teahan J, Harrison RK, Stein RL. Substrate specificity of the human matrix metalloproteinase stromelysin and the development of continuous fluorometric assays. Biochemistry. 1992;31:12618–12623. doi: 10.1021/bi00165a011. [DOI] [PubMed] [Google Scholar]
- O’Reilly DR, Miller AK, Luckow VA. A Laboratory Manual. W. H. Freeman and Company; New York: 1992. Baculovirus Expression Vectors. [Google Scholar]
- Oda K, Takahashi T, Takada K, Tsunemi M, Ng K, Hiraga K, Harada S. Exploring the subsite structure of vimelysin and thermolysin using FRETS-libraries. FEBS Lett. 2005;579:5013–5018. doi: 10.1016/j.febslet.2005.07.089. [DOI] [PubMed] [Google Scholar]
- Orsi BA, Tipton KF. Kinetic analysis of progress curves. Methods Enzymol. 1979;63:159–183. doi: 10.1016/0076-6879(79)63010-0. [DOI] [PubMed] [Google Scholar]
- Palmier MO, Van Doren SR. Rapid determination of enzyme kinetics from fluorescence: Overcoming the inner filter effect. Analytical Biochemistry. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroutis P, Touret N, Grinstein S. The pH of the Secretory Pathway: Measurement, Determinants, and Regulation. Physiology. 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- Patel OV, Yamada O, Kizaki K, Takahashi T, Imai K, Hashizume K. Quantitative analysis throughout pregnancy of placentomal and interplacentomal expression of pregnancy-associated glycoproteins-1 and -9 in the cow. Mol Reprod Dev. 2004;67:257–263. doi: 10.1002/mrd.20017. [DOI] [PubMed] [Google Scholar]
- Punturieri A, Filippov S, Allen E, Caras I, Murray R, Reddy V, Weiss SJ. Regulation of Elastinolytic Cysteine Proteinase Activity in Normal and Cathepsin K-deficient Human Macrophages. J Exp Med. 2000;192:789–800. doi: 10.1084/jem.192.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Tanaka T, Yada RY. Mechanism of activation of the gastric aspartic proteinases: pepsinogen, progastricsin and prochymosin. Biochem J. 1998;335:481–490. doi: 10.1042/bj3350481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin DB, Mazzieri R, Munger JS, Noguera I, Sung J. Proteolytic control of growth factor availability. APMIS. 1999;107:80–5. doi: 10.1111/j.1699-0463.1999.tb01529.x. [DOI] [PubMed] [Google Scholar]
- Scarborough PE, Dunn BM. Redesign of the substrate specificity of human cathepsin D: the dominant role of position 287 in S2 subsite. Protein Eng. 1994;7:495–502. doi: 10.1093/protein/7.4.495. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Bazer FW. Uterine and placental factors regulating conceptus growth in domestic animals. J Anim Sci. 2004;82:E4–13. doi: 10.2527/2004.8213_supplE4x. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Xie S, Green J, Roberts RM. Porcine pregnancy-associated glycoproteins: new members of the aspartic proteinase gene family expressed in trophectoderm. Biol Reprod. 1995;53:21–8. doi: 10.1095/biolreprod53.1.21. [DOI] [PubMed] [Google Scholar]
- Szecsi PB. The aspartic proteases. Scand J Clin Lab Invest Suppl. 1992;210:5–22. [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tang J, Sepulvelda P, Marciniszyn J, Chen KCS, Huang WY, Tao N, Liu D, Lainier JP. Proc Natl Acad Sci U S A. 1973;70 doi: 10.1073/pnas.70.12.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanskul S, Oda K, Oyama H, Noparatnaraporn N, Tsunemi M, Takada K. Substrate specificity of alkaline serine proteinase isolated from photosynthetic bacterium, Rubrivivax gelatinosus KDDS1. Biochem Biophys Res Commun. 2003;309:547–551. doi: 10.1016/j.bbrc.2003.08.035. [DOI] [PubMed] [Google Scholar]
- Telugu BP, Green JA. Characterization of the Peptidase Activity of Recombinant Porcine Pregnancy-Associated Glycoprotein-2. J Biochemistry. 2008;144:725–732. doi: 10.1093/jb/mvn127. [DOI] [PubMed] [Google Scholar]
- Telugu BP, Walker A, Green J. Characterization of the bovine pregnancy-associated glycoprotein gene family - analysis of gene sequences, regulatory regions within the promoter and expression of selected genes. BMC Genomics. 2009;10:185. doi: 10.1186/1471-2164-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J, Feng Z, Chen L, Yang H, Berman HM. The Protein Data Bank and structural genomics. Nucl Acids Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittlin S, Rosel J, Hofmann F, Stover DR. Mechanisms and kinetics of procathepsin D activation. European Journal of Biochemistry. 1999;265:384–393. doi: 10.1046/j.1432-1327.1999.00747.x. [DOI] [PubMed] [Google Scholar]
- Wooding FBP, Roberts RM, Green JA. Light and electron microscope immunocytochemical studies of the distribution of Pregnancy-Associated Glycoproteins (PAGs) throughout pregnancy in the cow: possible functional implications. Placenta. 2005;26:807–827. doi: 10.1016/j.placenta.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Beckers J-F, Roberts R. The gene encoding bovine pregnancy-associated glycoprotein-1, an inactive member of the aspartic proteinase family. Gene. 1995;159:193–197. doi: 10.1016/0378-1119(94)00928-l. [DOI] [PubMed] [Google Scholar]
- Xie S, Low BG, Nagel RJ, Beckers JF, Roberts RM. A novel glycoprotein of the aspartic proteinase gene family expressed in bovine placental trophectoderm. Biol Reprod. 1994;51:1145–1153. doi: 10.1095/biolreprod51.6.1145. [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Bao B, Beckers J-F, Valdez K, Hakami L, Roberts R. Multiple pregnancy-associated glycoproteins are secreted by day 100 ovine placental tissue. Biology of Reproduction. 1997;57:1384–1393. doi: 10.1095/biolreprod57.6.1384. [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Bixby JB, Szafranska B, DeMartini JC, Hecht S, Roberts RM. The diversity and evolutionary relationships of the pregnancy-associated glycoproteins, an aspartic proteinase subfamily consisting of many trophoblast-expressed genes. Proc Natl Acad Sci USA. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SC, Low BG, Nagel RJ, Kramer KK, Anthony RV, Zoli AP, Beckers JF, Roberts RM. Identification of the major pregnancy-specific antigens of cattle and sheep as inactive members of the aspartic proteinase family. Proc Natl Acad Sci USA. 1991;88:10247–10251. doi: 10.1073/pnas.88.22.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kati W, Chen CM, Tripathi R, Molla A, Kohlbrenner W. Use of a fluorescence plaer reader for measuring kinetic parameters with inner filter effect correction. Anal Biochem. 1999;267:331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. Characterization of New Fluorogenic Substrates for the Rapid and Sensitive Assay of Cathepsin E and Cathepsin D. J Biochem (Tokyo) 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- Yelich JV, Pomp D, Geisert RD. Detection of transcripts for retinoic acid receptors, retinol-binding protein and transforming growth factors during rapid trophoblastic elongation in the porcine conceptus. Biol Reprod. 1997;57:286–294. doi: 10.1095/biolreprod57.2.286. [DOI] [PubMed] [Google Scholar]