Abstract
Considering the great physiological and behavioral similarities with humans, monkeys represent the ideal models not only for the study of complex cognitive behavior but also for the preclinical research and development of novel therapeutics for treating human diseases. Various powerful genetic technologies initially developed for making mouse models are being explored for generating transgenic primate models. We review the latest genetic engineering technologies and discuss the potentials and limitations for systematic production of transgenic primates.
Keywords: Non-human primate, Mouse, Transgene, Genetics, Learning, Memory, disease model
1 Introduction
The principal tool of biomedical research is to model human diseases in various animal systems. Considering the great genetic and physiological similarities with humans, non-human primates represent the most ideal experimental models for detailed analysis of biological processes under physiological or pathological conditions[1–4]. Consequently, new drug candidates are typically required to go through systematic assessments of drug efficacy, pharmacokinetics, and toxicity in monkeys prior to their evaluation in human clinical trials. Furthermore, because only monkeys have human phylogenetic analogs to most association neocortical areas, they are the most favored organisms for in-depth analysis of neural mechanisms underlying high cognition, and complex behavior, and diseases of age including age-related cognitive decline[5–16]. Increasing appreciation of this great value and the need to reduce clinical trial failure rates begins to drive the effort in exploring and optimizing genetic technologies for making transgenic monkey models.
Genetic technologies can be broadly divided into two major categories: gene knockout and transgenic overexpression. Gene knockout methods allow the researchers to assess gene function by the inactivation of a gene through homologous recombination in embryonic stem cells (gene targeting)[17,18]. The similar gene targeting strategies can be used to perform gene knock-in experiments in which the different gene or modified gene replaces the endogenous gene. On the other hand, transgenic overexpression introduces a foreign DNA into the fertilized eggs so that the organisms derived from these injected eggs are born with the transgene. Researchers can then study the consequence of transgene overexpression to assess and infer the function of the gene and its signaling pathways. Both gene knockout and transgenic overexpression provide valuable information about the function of the genes in the biological system when the system operates in the absence or overproduction of the gene of interest[19–28]. These various genetic technologies, mostly in mice, have revolutionized the way that biologists to study development, immunology, physiology, neurobiology, and human diseases.
2 Conditional gene knockout technologies
Conventional gene knockout technique is a powerful tool for studying the function of a particular gene. Since genetic deletions occur at the embryonic stage, the gene is inactivated throughout ontogenesis to the whole life in every cell type and every organ. However, such broad action often lead to severe developmental defects and/or premature death[29]. Even when the organism survived the development and born without apparent deformation, it is difficult to confidently exclude the possibility that any phenotype observed in adult animals is not due to abnormal development at prenatal and postnatal stages. Therefore, while conventional or global gene knockout can be used effectively to study development and immunology, it is not well suited to study gene function in many biological disciplines.
For example, it has been longed postulated that the NMDA receptor may be a crucial cellular device for controlling memory function. The conventional knockout of the NR1 gene which encoding the core subunit of the NMDA receptor resulted in neonatal lethality. The knockout pups usually died within 12–14 hours after birth, apparently with abnormal brain stem function such as lack of the suckling reflex[30,31]. Such a neonatal lethality prevented the analysis of the role of the NMDA receptor in adult brain functions such as learning and memory.
Recognizing the limitations of the conventional gene knockout, researchers in mid 1990s successfully developed region-specific gene knockout technique by employing the phage P1-derived Cre/loxP recombination system[22]. This Cre-LoxP-recombination-mediated gene knockout method is known as second-generation gene knockout technique or conditional gene knockout method. Using a brain-specific CaMKII promoter to drive the Cre transgene, the researchers were able to delete the NR1 gene specifically in the CA1 region of the hippocampus, the region known to be crucial for long-term memory formation[32]. These CA1-specific NR1 knockout mice were viable and developed normally (because the Cre-mediated gene knockout occurred at the fourth postnatal week and restricted to CA1 within the first two months). Interestingly, physiological and behavioral analyses reveal that the CA1-specific NMDA receptor knockout mice lack major forms of synaptic plasticity in the CA1 region and are profoundly impaired in many memory tests[32–34]. The successful development and demonstration of conditional gene knockout from these early studies have led to explosive growth in its application to many disciplines, and this technique is widely used by the biomedical research community.
While conditional gene knockout technique can provide region- or tissue-specificity for the genetic inactivation, the temporal control of such deletion is limited by the promoter that drives the Cre transgene. This means that the conditional gene knockout still lacks inducible and reversible feature. As we all know, every biological process has its complexity in term of temporal stages. Those distinct temporal stages may involve different molecular mechanisms. For example, the memory process has at least four distinct temporal stages: acquisition, consolidation, storage, and retrieval. Thus, a problem in any of these four stages can be manifested as memory deficit. In addition, it is difficult to completely exclude the possible developmental or structural deficits that may be too subtle to detect since the conditional gene knockout occurred well before the behavioral experiments are performed (which may allow some genetic compensation within the system). Therefore, it is necessary to develop an new generation of conditional gene knockout that can be induced right before the experiment and can also be reversed so that the knockout event is restricted to a defined time period.
To achieve the flexible temporal control of genetic modification, Shimizu et al. developed an inducible, reversible, and region-specific knockout technique, also known as the third-generation gene knockout technique[24]. This inducible gene knockout system combines the Cre/loxP-mediated recombination system with the tetracycline-controlled transactivator (tTA) system[35]. The overall strategy is to use tTA/tetO system to achieve CA1-specific, tetracycline-regulated expression of the NR1-GPF transgene and consequently to rescue the CA1 NMDA receptor function in the CA1-specific NR1 knockout mice. Upon feeding of the inducible CA1-KO mice with the drinking water containing doxycycline, a tetracycline analog with higher permeability through the blood-brain-barrier, the researchers can switch off NR1-GFP transgene expression, thereby returning the mice to the NR1 knockout state in the CA1 region. Furthermore, removal of doxycycline from the water restores NR1-GFP expression in the CA1 region. Histological experiments suggest that the doxycycline treatment can inactivate the CA1 NMDA receptors within 3–5 days. This inactivation time course reflects the intrinsic turnover rate of the pre-existing NMDA receptor complex in vivo. This inducible gene knockout experiment has led to a new appreciation that long-term memory is not a static, single biochemical cascade, but rather a highly dynamic and reinforced process[11].
In the field of learning and memory, it has long been assumed that the NMDA receptor was required only for memory acquisition, and long-term memory consolidation and storage were the passive consequences of learning-initiated biochemical cascade which resulted in long-lasting structural changes. However, using this inducible, reversible, and region-specific gene knockout technology, the researchers found that switching off the NMDA receptor during the consolidation or storage stage would impair long-term memory[24,26,36], suggesting that long-term memory formation and storage are dynamic synaptic reentry-reinforcement processes controlled by repeated NMDA receptor reactivations. These findings provide an unexpected answer to a long-standing question as to how the brain maintains its synaptic efficacy and network stability in face of the routine metabolic turnovers of synaptic proteins[24,36,37]. The role of NMDA receptor reactivations in memory consolidation have been confirmed and extended by other laboratories in other memory tests using multiple animal species[38–41]. Peter Seeburg’s group has also used the similar tetracycline-based transgene system in knockout mice to rescue the AMAP receptor function, and they found that AMPA receptor play a specific role in synaptic plasticity and learning behavior[42]. Thus, inducible and region-specific gene knockout enables the researchers to elucidate novel insights into how the brain engages memory processes at the molecular and cellular levels[27].
3 Transgenic overexpression
While gene knockout approaches are powerful in revealing important aspects of genes’ biological functions, a full range assessment of their functions also requires the other manipulations such as by overexpressing the transgene or modified gene into the biological system of interest. For example, the conditional knockout of the NR1 gene which produced memory deficits disabled the entire NMDA receptor complex, therefore, it is not clear whether the observed memory deficits were due to lack of the NMDA receptor’s physical presence at the synapses, which interacts with many other synaptic signaling proteins and scaffolding proteins[43]. It may be the physical absence of the NMDA receptor at synapses, not necessarily the synaptic coincidence-detection function of the NMDA receptor, that has caused improper synaptic structural organization that in turn produced secondary effects on memory impairment. It is well known that activation of NMDA receptor requires two simultaneous events: pre-synaptic releases of glutamate which binds to the NMDA receptor and post-synaptic depolarization which relieves magnesium blockade of the NMDA receptor. Because of this unique cellular property, the NMDA receptor is also known as the coincidence detector. The voltage-dependent gating control of the NMDA receptor is regulated by NR2 subunits, mostly by interaction of NR2A and NR2B subunits with magnesium ions. The NR2A subunit-containing NMDA receptor has a shorter channel opening duration, whereas the NR2B NMDA receptor exhibit longer opening duration[44,45], thereby allowing greater influx of ion. Interestingly, NR2B expression is down-regulated during the transition period from juvenile to adulthood, correlating with the gradual shortening of the NMDA channel duration in the adult brain[46,47], and may contribute to reduced learning capacity. To fully demonstrate that the NMDA receptor acts as a gating switch for memory formation, researchers have used CaMKII promoter to overexpress the NR2B transgene in the mouse forebrain[23]. A series of analyses demonstrate that transgenic NR2B mice exhibit greater synaptic plasticity as well as remarkable improvement in variety of learning and memory tasks[23]. The follow-up studies further show that enhanced learning and memory are genetically stable even after fourteen generations of breeding[48] and are also well preserved into advanced ages[49]. The identification of the NR2B gene as the critical gating switch for NMDA receptor’s coincidence detection function has also led to other experiments in which enhanced learning and memory have been found when NR2B is upregulated by other molecules or pathways[50–53]. This NR2B transgenic overexpression experiment, although technically very simple, illustrates the power of transgenic approach can provide crucial information that otherwise was not readily revealed by gene knockout approach if used properly.
Transgenic overexpression can also be manipulated in an inducible and reversible manner. For example, by using tetracycline transactivator-based system to overexpress CaMKII specifically in the forebrain regions or striatum, Mayford et al. have established evidence for the role of CaMKII in learning behaviors[54,55]. In addition, temporally regulated overexpression of transgene can be also achieved by using a tamoxifen (TAM)-dependent mutant of an estrogen receptor ligand-binding domain[56]. In comparison to gene knockout techniques, transgenic overexpression is relatively simple and less time consuming.
4 Inducible protein knockout technique
Inducible gene knockout or transgenic overexpression approaches have come a long way since the conventional gene knockout was first reported. However, because the inactivation event occurs at the DNA level, manifestation of any phenotype depends on the turnover rate of the existing protein, which may take days depending on the turnover rate of the protein. Therefore, there is still strong need to develop new types of genetic techniques that can direct the knockout event at the protein level, rather than at the DNA level, for achieving almost instantaneous effects.
By integrating convergent protein engineering and rational inhibitor design, Tsien and Shokat have developed an inducible protein knockout technology[25]. This method is based on the creation of a specific interaction interface (bump-and-hole) between a modified protein domain and sensitized inhibitors. By introducing this bump-and-hole system into genetically modified mice (Figure 1), the researchers were able to switch on or off the transgenic alpha-CaMKII kinase activity rapidly within minutes as the mice underwent various memory tests[25].
Figure 1.
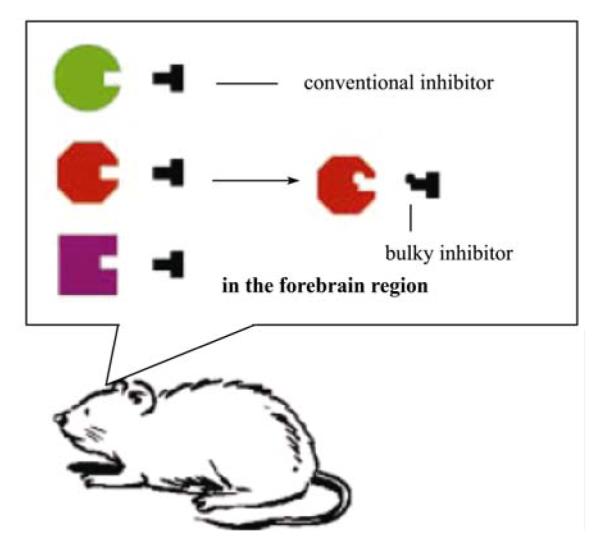
The bump-and-hole-based chemical genetic strategy for achieving inducible protein knockout in mice. The ATP-binding pocket of a protein kinase (e.g., alpha-CaMKII) is enlarged via silent mutation such that a bulky inhibitor can be rationally designed, synthesized, and identified to fit only the enlarged pocket but not the unmodified pocket. Once this system is introduced into transgenic mice, researchers can then use the bulky inhibitor to specifically and rapidly control the kinase activity at a time scale of minutes.
The systematic temporal manipulation of alpha-CaMKII activity during the memory consolidation period suggests that the first post-learning week is the critical time-window during which changes in CaMKII activity level disrupt the consolidation of one-month-old fear memories. This work for the first time suggests that in addition to the reactivations of the NMDA receptor, alpha-CaMKII reactivations are also important for memory consolidation[25]. More recently, alpha-CaMKII reactivation in the entorhinal cortex has also been shown to be necessary for memory consolidation[57].
Using the same inducible protein manipulation strategy, the researchers further examined the role of beta-CaMKII during various distinct memory stages. Beta-CaMKII is a co-constituent of the CaMKII holoenzyme, but has not been studied much. With this inducible chemical-genetic method, they demonstrated that beta-CaMKII reactivations in the dentate gyrus play an important role in the consolidation of long-term memory[58].
The major advantage of the inducible, reversible, and region-specific protein knockout technique is to allow researchers to be able to control the transgenic kinase activity specifically and rapidly at a time scale of minutes. It has been shown that a single i.p. injection of NM-PP1 can suppress the transgenic alpha-CaMKII level to bring the total level of aCaMKII back to normal from 10 to 40 min after the injection[59]. With this rapid temporal control, Wang et al. examined the molecular basis of short-term memory, a topic that has been proven to be difficult to study using other genetic methods. They found that the rapid upregulation of alpha-CaMKII activity in the mouse forebrain within the immediate 10 min after learning dramatically disrupted the short-term memory representation. The same manipulation beyond 15 min post-learning has no effect[59]. Similar, long-term potentiation also exhibits the same 10-minute time window during which potentiated synapses are subjected to alpha-CaMKII manipulation[59].
Interestingly, this inducible protein manipulation method has been applied to erase memories in the mouse brain[28]. Cao et al. showed that temporarily boosting the level of CaMKII at the time of memory retrieval can rapidly and specifically erase memories from the mouse brain[28]. This inducible and selective memory erasure was effective in newly-formed memories, or old memories. Further experiment suggested that the erasure was selective to the memories undergoing retrieval at the time of excessive CaMKII activity. Therefore, those rapid and selective memory erasure experiments have nicely demonstrated the power of this protein-based genetic technology.
The ‘bump-and-hole’-based inducible protein knockout has been also applied to study other protein functions in mice, including the function of neurotrophin receptor (TrkB)[60,61] and the role of PKA in sperm capacitation[62]. It is foreseeable that this strategy will be increasingly used to address the functional significance of many proteins with high temporal resolution. However, the current strategy is limited to one class of proteins that contain ATP-binding pockets (myosin, tyrosine kinase receptor, kinases, etc.). It will be extremely important to develop various other kinds of small molecule-protein motif interface techniques to manipulate multiple of protein classes.
5 Towards transgenic primates
Because of the close evolutionary relationship, non-human primates parallel humans with regard to genetic, anatomic, cognitive, and behavioral characteristics. The similarity in blood pressure, heart rate, menstrual cycle, and total body oxygen consumption between monkeys and human makes non-human primates more reliable human disease models for pre-clinical study of new drugs and therapeutics. While mice have been proven to be a valuable model system for molecular and neural understanding of many basic brain functions, many sophisticated behavior and cognition still need to be investigated in non-human primates. For example, like humans, monkeys possess imitation learning skills, that is, they can learn tasks or skills by watching how others perform. In fact, in the macaque monkey researchers have discovered the mirror neuron which would respond to not only its own motor action but also to the same action performed by other monkeys or humans[63–65]. Therefore, these mirror neurons may play a very important role in perception action coupling and imitation learning[66]. It has been postulated that dysfunction of mirror system in human may be related to some cognitive deficit, especially in autism[67]. Thus, one may create autism monkey models by genetic manipulation of the mirror neuron circuits. This is just one example where genetics can bring clear benefits to the scientific quest of high brain functions. On the other hand, it is always important to realize that such hypotheses need to be tested and carefully examined by actual experiments.
However, the manipulations of genes in the monkeys or other non-human primates are still at its infancy. In 2001, Chan and his colleagues succeeded producing a genetically modified monkey, ANDi, carrying the jellyfish gene for green fluorescent protein (GFP)[68]. The report of birth of ANDi took an important step toward the development of genetic non-human primate model. More recently, Chan and his colleagues have also described transgenic monkeys that expresse polyglutamine-expanded human huntingtin (HTT). Careful analyses revealed many hallmark features of Hungtinton Disease (HD), including nuclear inclusions and neuropil aggregates, in the brains of the HD transgenic monkeys which died within initial months. Moreover, the transgenic monkeys also exhibit clinical features of Huntington syndromes, including dystonia and chorea. This landmark work opens the way not only to better understanding of the pathogenesis of Huntington disease, but also to the development of potential therapies[69].
In another example, researchers have directly applied the lentiviral vector onto the monkey brain[70]. Although this approach did not produce transgenic monkeys, it can be highly useful in terms of perturbing precisely the activity of specific cell types and pathways in the nonhuman primate nervous system. Desimone and Boyden used lentivirus to target the light-activated cation channel channelrhodopsin-2 (ChR2) specifically to excitatory neurons of the macaque frontal cortex. Using a laser-coupled optical fiber in conjunction with a recording microelectrode, they reported that activation of excitatory neurons caused well-timed excitatory and suppressive effects on neocortical neural networks. ChR2 was safely expressed, and could mediate optical neuromodulation in primate neocortex over many months. These new findings highlight additional methodology studying nonhuman primate cognition and behavior. It may also open up the possibility of a new generation of precise neurological and psychiatric therapeutics via cell-type-specific optical neural control prosthetics.
6 Future perspective
Scientific progress is the perpetual cycles of new technologies to new knowledge and questions and back-and-forth again and again. Biomedical/behavioral research is no exemption. Maturation of genetic technologies in mice now allows us to start approaching more complicated cognitive functions such as attention, decision making, and vocabulary learning in primates. Those higher functions are associated more closely with the frontal cortex and enlarged high-association cortical structure that become much more evolved in human or nonhuman primates. These intellectual interests beg development of new genetic methodologies in non-human primate, a new frontier for genetic manipulations.
The emerging transgenic monkey models certainly mark the first step of this exciting journey. Yet clearly there are a lot of hurdles. The first hurdle in systematic production of transgenic nonhuman primates has been the low efficiency of live birth baby monkeys, which limits the scale of transgenic efforts. The second hurdle is related with the transgene delivery method. Lentivirus has so far been the main vehicle due to its low toxicity, the moderate insert size, and its readily availabilities. However there are a few caveats. One caveat roots from high copy numbers of lentivirus integrations. This very same feature, which is useful to increase the transgenic rate, may potentially cause future complication if the transgenic monkeys are to be used for breeding. At the moment, it is not clear whether ANDi has achieved any germ line transmission. Most recently, the first germline transmission of GFP trangene in transgenic monkeys has been recently reported[71]. In this case, the authors used marmosets which are a small New World primate rather than Old World primates, such as the rhesus monkey (Macaca mulatta) and cynomolgus monkey (Macaca fascicularis). Marmosets are much smaller in their body size and have relatively short reproductive cycles and can be highly valuable for better modeling many human diseases. However, marmosets are less closely related to humans than Old World primates are. Some diseases such as HIV/AIDS, macular degeneration, and tuberculosis can be explored only in these Old World monkey[72].
Typically, unless all copies of virus integrated onto one same chromosome, Mundelein Segregation would predict different genotypes among progenies of the same founder. Moreover, in most cases because the F1 often harbors multiple integrations of the transgene, random segregation will likely lead to segregated transgene distributions among F2s and weaker phenotypes in offsprings. It is conceivable that due to position effects often seen with transgene mice, same transgene placed into different genomic locations in monkeys can result in different expression levels and population patterns which are different from the original founders.
In addition, the insert size of a lentiviral vector limits the transgene construct size no larger than 8 kb. This can be an issue for delivering transgene to specific tissue or cell types, as often such promoters are fairly large in size. In fact, on many occasions mouse geneticists had to employ large vector such as BAC and PAC to cover enough transcription regulating sequence for achieving tissue-specificity. Thus, other alternative methods in delivering transgene need to be explored. One alternative choice is to take advantage of the cloning technology. Since Dolly sheep, somatic cell nucleus transfer (SCNT) technique has been applied to different animal species including mice, cat, horse and Rhesus monkeys. Cloning with transfected monkey cells following thorough characterizations of the transgene integration could allow control of integration copy and also eliminate the size restriction on transgenes. Conceivably, transfection and subsequent selection of cultured cells could allow introduction of many subtle mutations via homologous recombination. Therefore, cloning technology can allow generation of not only transgenic but also knockout monkeys. Obviously, SCNT is a very demanding technique, and prolonged in vitro cell culture may also induce gene deletion or amplification thus limiting the utility of SCNT in generating transgenic monkeys. Nevertheless, when a complicated genetic manipulation needs to be introduced into monkeys, cloning is definitely a valid option.
The third hurdle in producing transgenic monkeys is the financial resources since the sustained and large-scale production of various transgenic monkeys can be cost-prohibitive. This would require strong investment of precious research support from governments, pharmaceutical industry, as well as private foundations. Primate researchers will also need to bring down the management cost and increase procedural efficiency. Recently, the researchers of Yunnan Banna Primate Disease Model Research Center and Shanghai Institute of Brain Functional Genomics at East China Normal University have reported a high efficient method for achieving higher live birth rate[69]. Although this technical improvement may be a very small step, it is welcoming news for those who are interested in seeing systematic and large-scale generation of various primate models.
Finally, various ethical and cultural issues will also be carefully considered before, during, and after the production of transgenic primate disease models. The broader support of primate research by public and society will also require scientists to engage general public and governments for informative communication and education. Despite the above discussed issues and hurdles, the clear need to better understand complex behaviors and to develop more effective means for new drug testing will ensure that the primate transgenics have a bright future ahead.
Acknowledgments
Supported by Funding from GRA, NIMH and NIA.
References
- 1.Bavister BD, Boatman DE, Collins K, et al. Birth of rhesus monkey infant after in vitro fertilization and nonsurgical embryo transfer. Proc Natl Acad Sci USA. 1984;81(7):2218–2222. doi: 10.1073/pnas.81.7.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner DK, Lane M. Culture and selection of viable blastocysts: a feasible proposition for human IVF? Hum Reprod Update. 1997;3(4):367–382. doi: 10.1093/humupd/3.4.367. [DOI] [PubMed] [Google Scholar]
- 3.Schramm RD, Paprocki AM, Watkins DI. Birth of MHC-defined rhesus monkeys produced by assisted reproductive technology. Vaccine. 2001;20(3-4):603–607. doi: 10.1016/s0264-410x(01)00336-x. [DOI] [PubMed] [Google Scholar]
- 4.Hara K, Yasuhara T, Matsukawa N, et al. Hippocampal CA1 cell loss in a non-human primate model of transient global ischemia: a pilot study. Brain Res Bull. 2007;74(1-3):164–171. doi: 10.1016/j.brainresbull.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Jolly A. Lemur social behavior and primate intelligence. Science. 1966;153(735):501–506. doi: 10.1126/science.153.3735.501. [DOI] [PubMed] [Google Scholar]
- 6.Gross CG, Rocha-Miranda CE, Bender DB. Visual properties of neurons in inferotemporal cortex of the Macaque. J Neurophysiol. 1972;35(1):96–111. doi: 10.1152/jn.1972.35.1.96. [DOI] [PubMed] [Google Scholar]
- 7.Desimone R, Albright TD, Gross CG, et al. Stimulus-selective properties of inferior temporal neurons in the macaque. J Neurosci. 1984;4(8):2051–2062. doi: 10.1523/JNEUROSCI.04-08-02051.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanemaru K, Iwatsubo T, Ihara Y. Comparable amyloid beta-protein (A beta) 42(43) and A beta 40 deposition in the aged monkey brain. Neurosci Lett. 1996;214(2-3):196–198. doi: 10.1016/0304-3940(96)12893-7. [DOI] [PubMed] [Google Scholar]
- 9.Jentsch JD, Redmond DE, Jr., Elsworth JD, et al. Enduring cognitive deficits and cortical dopamine dysfunction in monkeys after long-term administration of phencyclidine. Science. 1997;277(5328):953–955. doi: 10.1126/science.277.5328.953. [DOI] [PubMed] [Google Scholar]
- 10.Zola SM, Squire LR. Relationship between magnitude of damage to the hippocampus and impaired recognition memory in monkeys. Hippocampus. 2001;11(2):92–98. doi: 10.1002/hipo.1027. [DOI] [PubMed] [Google Scholar]
- 11.Miller EK, Nieder A, Freedman DJ, et al. Neural correlates of categories and concepts. Curr Opin Neurobiol. 2003;13(2):198–203. doi: 10.1016/s0959-4388(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 12.Kimura N, Nakamura S, Ono F, et al. Presenilin-2 in the cynomolgus monkey brain: investigation of age-related changes. Primates. 2004;45(3):167–175. doi: 10.1007/s10329-004-0076-x. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki WA, Amaral DG. Functional neuroanatomy of the medial temporal lobe memory system. Cortex. 2004;40(1):220–222. doi: 10.1016/s0010-9452(08)70958-4. [DOI] [PubMed] [Google Scholar]
- 14.Gross CG. Processing the facial image: a brief history. Am Psychol. 2005;60(8):755–763. doi: 10.1037/0003-066X.60.8.755. [DOI] [PubMed] [Google Scholar]
- 15.Penn DC, Povinelli DJ. Causal cognition in human and nonhuman animals: a comparative, critical review. Annu Rev Psychol. 2007;58:97–118. doi: 10.1146/annurev.psych.58.110405.085555. [DOI] [PubMed] [Google Scholar]
- 16.Buccafusco JJ. Estimation of working memory in macaques for studying drugs for the treatment of cognitive disorders. J Alzheimers Dis. 2008;15(4):709–720. doi: 10.3233/jad-2008-15414. [DOI] [PubMed] [Google Scholar]
- 17.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244(4910):1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 18.Capecchi MR. The new mouse genetics: altering the genome by gene targeting. Trends Genet. 1989;5(3):70–76. doi: 10.1016/0168-9525(89)90029-2. [DOI] [PubMed] [Google Scholar]
- 19.Grant SG, O’Dell TJ, Karl KA, et al. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258(5090):1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 20.Silva AJ, Stevens CF, Tonegawa S, et al. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257(5067):201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 21.Silva AJ, Paylor R, Wehner JM, et al. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257(5067):206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 22.Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87(7):1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 23.Tang YP, Shimizu E, Dube GR, et al. Genetic enhancement of learning and memory in mice. Nature. 1999;401(6748):63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu E, Tang YP, Rampon C, et al. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290(5494):1170–1174. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Shimizu E, Tang YP, et al. Inducible protein knockout reveals temporal requirement of CaMKII reactivation for memory consolidation in the brain. Proc Natl Acad Sci USA. 2003;100(7):4287–4292. doi: 10.1073/pnas.0636870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Z, Wang H, Tan Y, et al. Inducible and reversible NR1 knockout reveals crucial role of the NMDA receptor in preserving remote memories in the brain. Neuron. 2004;41(5):781–793. doi: 10.1016/s0896-6273(04)00072-8. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Hu Y, Tsien JZ. Molecular and systems mechanisms of memory consolidation and storage. Prog Neurobiol. 2006;79(3):123–135. doi: 10.1016/j.pneurobio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Cao X, Wang H, Mei B, et al. Inducible and selective erasure of memories in the mouse brain via chemical-genetic manipulation. Neuron. 2008;60(2):353–366. doi: 10.1016/j.neuron.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyner AL, Guillemot F. Gene targeting and development of the nervous system. Curr Opin Neurobiol. 1994;4(1):37–42. doi: 10.1016/0959-4388(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 30.Forrest D, Yuzaki M, Soares HD, et al. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13(2):325–338. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Erzurumlu RS, Chen C, et al. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell. 1994;76(3):427–437. doi: 10.1016/0092-8674(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 32.Tsien JZ, Chen DF, Gerber D, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87(7):1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 33.McHugh TJ, Blum KI, Tsien JZ, et al. Impaired hippocampal representation of space in CA1-specific NMDAR1 knockout mice. Cell. 1996;87(7):1339–1349. doi: 10.1016/s0092-8674(00)81828-0. [DOI] [PubMed] [Google Scholar]
- 34.Rampon C, Tang YP, Goodhouse J, et al. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci. 2000;3(3):238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- 35.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittenberg GM, Tsien JZ. An emerging molecular and cellular framework for memory processing by the hippocampus. Trends Neurosci. 2002;25(10):501–5. doi: 10.1016/s0166-2236(02)02231-2. [DOI] [PubMed] [Google Scholar]
- 37.Cui Z, Lindl KA, Mei B, et al. Requirement of NMDA receptor reactivation for consolidation and storage of nondeclarative taste memory revealed by inducible NR1 knockout. Eur J Neurosci. 2005;22(3):755–763. doi: 10.1111/j.1460-9568.2005.04257.x. [DOI] [PubMed] [Google Scholar]
- 38.McDonald RJ, Hong NS, Craig LA, et al. NMDA-receptor blockade by CPP impairs post-training consolidation of a rapidly acquired spatial representation in rat hippocampus. Eur J Neurosci. 2005;22(5):1201–1213. doi: 10.1111/j.1460-9568.2005.04272.x. [DOI] [PubMed] [Google Scholar]
- 39.Winters BD, Bussey TJ. Glutamate receptors in perirhinal cortex mediate encoding, retrieval, and consolidation of object recognition memory. J Neurosci. 2005;25(17):4243–4251. doi: 10.1523/JNEUROSCI.0480-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takehara-Nishiuchi K, Nakao K, Kawahara S, et al. Systems consolidation requires postlearning activation of NMDA receptors in the medial prefrontal cortex in trace eyeblink conditioning. J Neurosci. 2006;26(19):5049–5058. doi: 10.1523/JNEUROSCI.4381-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu CL, Xia S, Fu TF, et al. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10(12):1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mack V, Burnashev N, Kaiser KM, et al. Conditional restoration of hippocampal synaptic potentiation in Glur-A-deficient mice. Science. 2001;292(5526):2501–2504. doi: 10.1126/science.1059365. [DOI] [PubMed] [Google Scholar]
- 43.Husi H, Ward MA, Choudhary JS, et al. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 44.Monyer H, Sprengel R, Schoepfer R, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256(5060):1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 45.Monyer H, Burnashev N, Laurie DJ, et al. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12(3):529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 46.Carmignoto G, Vicini S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science. 1992;258(5084):1007–1011. doi: 10.1126/science.1279803. [DOI] [PubMed] [Google Scholar]
- 47.Hestrin S. Developmental regulation of NMDA receptor-mediated synaptic currents at a central synapse. Nature. 1992;357(6380):686–689. doi: 10.1038/357686a0. [DOI] [PubMed] [Google Scholar]
- 48.Tang YP, Wang H, Feng R, et al. Differential effects of enrichment on learning and memory function in NR2B transgenic mice. Neuropharmacology. 2001;41(6):779–790. doi: 10.1016/s0028-3908(01)00122-8. [DOI] [PubMed] [Google Scholar]
- 49.Cao X, Cui Z, Feng R, et al. Maintenance of superior learning and memory function in NR2B transgenic mice during ageing. Eur J Neurosci. 2007;25(6):1815–1822. doi: 10.1111/j.1460-9568.2007.05431.x. [DOI] [PubMed] [Google Scholar]
- 50.Wong RW, Setou M, Teng J, et al. Overexpression of motor protein KIF17 enhances spatial and working memory in transgenic mice. Proc Natl Acad Sci USA. 2002;99(22):14500–14505. doi: 10.1073/pnas.222371099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White TL, Youngentob SL. The effect of NMDA-NR2B receptor subunit over-expression on olfactory memory task performance in the mouse. Brain Res. 2004;1021(1):1–7. doi: 10.1016/j.brainres.2004.05.114. [DOI] [PubMed] [Google Scholar]
- 52.Coultrap SJ, Bickford PC, Browning MD. Blueberry-enriched diet ameliorates age-related declines in NMDA receptor-dependent LTP. Age (Dordr) 2008;30(4):263–272. doi: 10.1007/s11357-008-9067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng D, Pitcher GM, Szilard RK, et al. Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol. 2009;7(2):e41. doi: 10.1371/journal.pbio.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayford M, Bach ME, Huang YY, et al. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 55.Mayford M, Mansuy IM, Muller RU, et al. Memory and behavior: a second generation of genetically modified mice. Curr Biol. 1997;7(9):R580–589. doi: 10.1016/s0960-9822(06)00287-9. [DOI] [PubMed] [Google Scholar]
- 56.Kida S, Josselyn SA, de Ortiz SP, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5(4):348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda M, Mayford MR. CaMKII activation in the entorhinal cortex disrupts previously encoded spatial memory. Neuron. 2006;50(2):309–318. doi: 10.1016/j.neuron.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 58.Cho MH, Cao X, Wang D, et al. Dentate gyrus-specific manipulation of beta-Ca2+/calmodulin-dependent kinase II disrupts memory consolidation. Proc Natl Acad Sci USA. 2007;104(41):16317–16322. doi: 10.1073/pnas.0703344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Feng R, Wang L Phillip, et al. CaMKII activation state underlies synaptic labile phase of LTP and short-term memory formation. Curr Biol. 2008;18(20):1546–1554. doi: 10.1016/j.cub.2008.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, Ye H, Kuruvilla R, et al. A chemical-genetic approach to studying neurotrophin signaling. Neuron. 2005;46(1):13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 61.Johnson AW, Chen X, Crombag HS, et al. The brain-derived neurotrophic factor receptor TrkB is critical for the acquisition but not expression of conditioned incentive value. Eur J Neurosci. 2008;28(5):997–1002. doi: 10.1111/j.1460-9568.2008.06383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan DJ, Weisenhaus M, Shum S, et al. Tissue-specific PKA inhibition using a chemical genetic approach and its application to studies on sperm capacitation. Proc Natl Acad Sci USA. 2008;105(52):20740–20745. doi: 10.1073/pnas.0810971105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.di Pellegrino G, Fadiga L, Fogassi L, et al. Understanding motor events: a neurophysiological study. Exp Brain Res. 1992;91(1):176–180. doi: 10.1007/BF00230027. [DOI] [PubMed] [Google Scholar]
- 64.Rizzolatti G, Fadiga L, Gallese V, et al. Premotor cortex and the recognition of motor actions. Brain Res Cogn Brain Res. 1996;3(2):131–141. doi: 10.1016/0926-6410(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 65.Rizzolatti G, Craighero L. The mirror-neuron system. Annu Rev Neurosci. 2004;27:169–192. doi: 10.1146/annurev.neuro.27.070203.144230. [DOI] [PubMed] [Google Scholar]
- 66.Dinstein I, Thomas C, Behrmann M, et al. A mirror up to nature. Curr Biol. 2008;18(1):R13–18. doi: 10.1016/j.cub.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oberman LM, Hubbard EM, McCleery JP, et al. EEG evidence for mirror neuron dysfunction in autism spectrum disorders. Brain Res Cogn Brain Res. 2005;24(2):190–198. doi: 10.1016/j.cogbrainres.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 68.Chan AW, Chong KY, Martinovich C, et al. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291(5502):309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- 69.Yang SH, Cheng PH, Banta H, et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han X, Qian X, Bernstein JG, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62(2):191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sasaki E, Suemizu H, Shimada A, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459(7246):523–528. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 72.Schatten G, Mitalipov S. Developmental biology: Transgenic primate offspring. Nature. 2009;459(7246):515–516. doi: 10.1038/459515a. [DOI] [PMC free article] [PubMed] [Google Scholar]


