Summary
The mechanisms that safeguard cells against chromosomal instability (CIN) are of great interest, as CIN contributes to tumorigenesis. To gain insight into these mechanisms, we studied the behavior of cells entering mitosis with damaged chromosomes. We used the endonuclease I-CreI to generate acentric chromosomes in Drosophila larvae. While I-CreI expression produces acentric chromosomes in the majority of neuronal stem cells, remarkably, it has no effect on adult survival. Our live studies reveal that acentric chromatids segregate efficiently to opposite poles. The acentric chromatid poleward movement is mediated through DNA tethers decorated with BubR1, Polo, INCENP, and Aurora-B. Reduced BubR1 or Polo function results in abnormal segregation of acentric chromatids, a decrease in acentric chromosome tethering, and a great reduction in adult survival. We propose that BubR1 and Polo facilitate the accurate segregation of acentric chromatids by maintaining the integrity of the tethers that connect acentric chromosomes to their centric partners.
Introduction
Although cells possess a number of mechanisms for repairing DNA damage, double-strand breaks (DSBs) are particularly troublesome (Su, 2006). Failure to repair DSBs results in chromosome fragments lacking either telomeres or centromeres. It is well established that without the protection of intact telomeres, end-to-end chromosome fusions produce dicentrics, extensive chromosome rearrangements, and aneuploidy (Tusell et al., 2008). The acentric fragments produced from unrepaired DSBs are equally problematic. Since acentric fragments lack kinetochores, one might expect acentrics to be incapable of poleward segregation. Surprisingly, however, poleward movement of acentrics has been observed in a number of systems. Efficient acentric segregation is demonstrated in budding yeast, where broken chromosomes transit through many generations before being repaired (Galgoczy and Toczyski, 2001; Malkova et al., 1996; Sandell and Zakian, 1993). In Drosophila, acentrics generated through the bridge-breakage-fusion cycle accumulate to high copy number, indicating efficient acentric segregation through several cell cycles (Titen and Golic, 2008). Further analysis in budding yeast shows that two chromosome fragments created by one irreversible DSB remain apposed throughout mitosis (Melo et al., 2001). Mechanisms of acentric segregation include neocentromere formation and association of acentrics with undamaged chromosomes (Ishii et al., 2008; Kanda et al., 2001; Platero et al., 1999).
Here, we describe a distinct tether-based mechanism facilitating acentric segregation. By studying the fate of acentrics in Drosophila larval neuroblasts, we discovered that acentric chromosome fragments lag on the metaphase plate but ultimately undergo poleward segregation during anaphase. Our data demonstrate that segregation is achieved through DNA tethers connecting the acentric and centric fragments. These tethers are decorated with Polo kinase, the spindle checkpoint component BubR1, and two chromosomal passenger complex proteins, INCENP and Aurora-B. In bubR1- and polo-compromised backgrounds, the efficiency of acentric poleward segregation is significantly diminished and the frequency of acentrics untethered to their centric partners is increased. These phenotypes are correlated with a dramatic decrease in survival upon DSB induction. These studies reveal a distinct mechanism by which acentric chromatids segregate accurately and suggest an additional role for BubR1 and Polo in the prevention of chromosomal instability (CIN).
Results
Efficient Generation of Acentric Chromosomes during Larval Development Does Not Affect Viability
To study the behavior of acentrics, we generated DSBs by expressing the endonuclease I-CreI. I-CreI recognizes a 20 nt sequence in the rDNA repeats located in the centromeric heterochromatin of the Drosophila sex chromosomes (Rong et al., 2002). The Drosophila X chromosome is acrocentric, and DAPI-stained mitotic X chromosomes form a “V” with the centromeric heterochromatin “head” more brightly stained that the two sister chromatid arms (Figure 1A). The I-CreI-induced breaks create two distinct chromosome fragments: a centric short “head” and an acentric long arm (Figure 1A) (Royou et al., 2005). A line bearing the I-CreI transgene with heat shock 70 promoter was used in our experiments. Late-third-instar larvae were heat shocked for 1 hr to induce I-CreI expression, followed by at least 1 hr of recovery. Heat shock itself provokes a dramatic reduction of the mitotic index of larval neuroblasts for up to 50 min after heat shock (Royou et al., 2005). I-CreI expression induced a DNA damage checkpoint-dependent delay in interphase, indicating that I-CreI had damaged the DNA (Royou et al., 2005).
Figure 1. I-CreI-Generated Double-Strand Breaks Create Chromosome X Fragments that Lag at Anaphase.
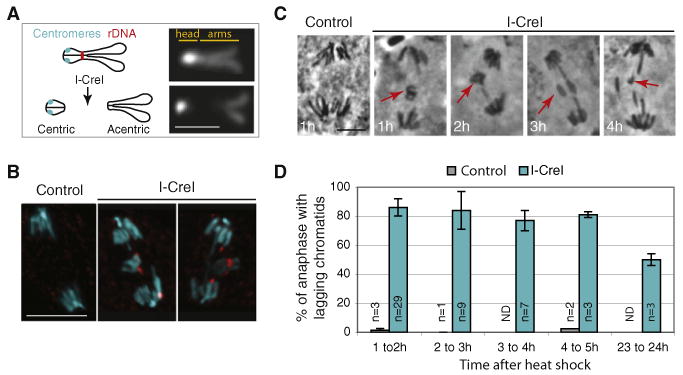
(A) Schematic illustration (left) and DAPI staining (right) of the Drosophila mitotic X chromosome. The centromeres (cyan circles) are very close to the telomeres in the heterochromatin “head” brightly stained with DAPI. The endonuclease I-CreI cuts at the rDNA locus located close to the centromeric heterochromatin. I-CreI generates two distinct chromosome fragments, a small heterochromatic fragment containing the centromeres (centric) and a long fragment containing the sister chromatid arms (acentric). The scale bar represents 2 μm.
(B) DAPI (cyan) and anti-γH2Av (red) fluorescent images of anaphase neuroblasts from heat-shocked third-instar larvae, with (I-CreI) or without (control) the I-CreI transgene. γH2Av that marks DSBs is detected on the lagging chromatids upon I-CreI induction. The images are maximum projections of deconvolved Z sections. The scale bar represents 10 μm.
(C) Orcein staining of anaphase neuroblasts from heat-shocked third-instar larvae with (I-CreI) or without (control) the I-CreI transgene. The time the larvae were dissected after heat shock is indicated on the bottom left. Lagging chromatids (red arrows) are observed after I-CreI induction. The scale bar represents 5 μm.
(D) Frequency of anaphase neuroblasts described in panel C with lagging chromatids (mean ± STD, n = number of brains).
To determine whether I-CreI efficiently creates DSBs, we stained neuroblasts with antibodies directed against γH2Av (Figure 1B). γH2Av accumulates specifically at the site of double-strand breaks (Vidanes et al., 2005). In heat-shocked neuroblasts from control larvae, no lagging chromatids were observed at anaphase and no γH2Av signal was detected. In contrast, heat-shocked neuroblasts from I-CreI larvae exhibited lagging chromatids at anaphase. A strong punctate γH2Av signal was detected on these lagging chromatids, indicating that I-CreI had created DSBs (Figure 1B).
Third-instar larvae brains were fixed and stained with Orcein at multiple time points after heat shock to score the frequency of anaphases with lagging chromatids (Figures 1C and 1D). Control larvae rarely exhibited anaphase with lagging chromatids. In contrast, lagging chromatids were observed in an average of 80% of neuroblasts for up to 5 hr after I-CreI induction (Figure 1D). Moreover, 1 day after I-CreI induction, half of the neuroblasts scored exhibited lagging chromatids (Figure 1D). Because the frequency of anaphases with acentrics remains high long after I-CreI induction, this suggests that the cells with acentrics are capable of multiple rounds of division. Alternatively, I-CreI may persist through multiple cycles, creating breaks that are constantly repaired.
Since 80% of the dividing cells exhibit lagging chromatids upon I-CreI induction, we assayed the effect of I-CreI expression on adult survival. Remarkably, no differences in the frequency of third-instar larvae developing into adults were observed between control and I-CreI-bearing larvae (Table 1).
Table 1.
Survival Rate of Late-Third-Instar Larvae to Adulthood
| I-CreI Expression | Number of Experiments | Total Number of Larvaea | Percent Survival into Adulthoodb | |
|---|---|---|---|---|
| Wild-type | No | 3 | 114 | 77.2 ± 8.4 |
| Wild-type | Yes | 3 | 125 | 78.0 ± 15.7 |
| mad2p | No | 2 | 33 | 73.5 ± 4.9 |
| mad2p | Yes | 7 | 116 | 72.9 ± 10.4 |
| bubR1–KEN | No | 4 | 51 | 72.1 ± 10.7 |
| bubR1–KEN | Yes | 9 | 187 | 18.4 ± 7.6 |
| polo1/polo10 | No | 6 | 117 | 80.9 ± 9.3 |
| polo1/polo10 | Yes | 6 | 126 | 0 |
Number of larvae per experiment is greater than nine.
Mean ± STD.
Acentric Sister Chromatids Lag during Anaphase but Ultimately Segregate Poleward
To understand how cells compensate for DSB-induced acentric chromosomes, we studied acentric behavior in living cells during mitosis using the GFP-H2Av marker (Clarkson and Saint, 1999). We examined neuroblasts from late-third-instar larvae as they entered the first mitosis after I-CreI induction (1–2 hr after heat shock). Because I-CreI creates long X and small Y acentric fragments, female larvae neuroblasts were studied exclusively. Control images of heat-shocked neuroblasts highlight alignment of the chromosomes during metaphase and rapid segregation of sister chromatids during anaphase (Figure 2A, top row; Movie S1 available online). After induction of I-CreI, acentric chromosomes aligned at the periphery of the metaphase plate, slightly separated from the main body of chromosomes (Figure 2A, cyan arrow; Movie S1). During anaphase the acentric fragments oriented on a plane parallel to the spindle and segregating toward the poles (Figure 2A, second row, cyan arrows; Movie S1). In this cell, the last acentric initiated its poleward movement 2.3 min after anaphase onset (yellow arrow). Remarkably, while acentric segregation was delayed, the acentrics underwent poleward movement in 95% of neuroblasts examined (n = 71). In 73% of anaphases, two acentric fragments segregated toward each pole (equal segregation), presumably giving rise to diploid daughter cells. In 27% of anaphases, we observed one, three, or four acentric fragments segregating to one pole (unequal segregation), thus creating aneuploid daughter cells (Figure 2A, third row, cyan arrowheads; Movie S1). In this cell, the last acentric initiated its poleward movement 5 min after anaphase onset (yellow arrow). Proper segregation of sister acentrics provides an opportunity for repair in the daughter cells and may account for the high survival rates. Because one-quarter of the cells going through mitosis become aneuploid at each round of division, we predict that I-CreI expression at an earlier stage of development will induce lethality. In fact, we found that I-CreI expression at second-instar larval stage produced a high rate of lethality (data not shown).
Figure 2. Acentrics Segregate toward the Poles and Remain Associated with their Centric Partners by DNA Tethers.
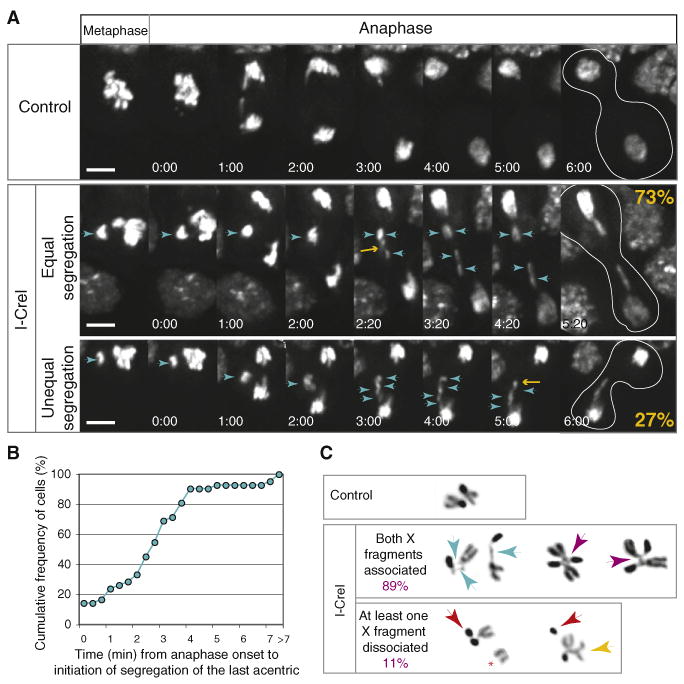
(A) Time-lapse images of mitotic GFP-H2Av-labeled neuroblasts from heat-shocked larvae with (I-CreI, second and third rows) or without (control, first row) the I-CreI transgene. Female larvae were selected in all these studies. First row images show normal chromosome segregation during anaphase (see Movie S1). Second-row images are representative of 73% mitotic neuroblasts in which acentrics segregate equally after I-CreI induction (n = 71). The acentrics (cyan arrowheads) are aligned at the metaphase plate slightly detached from the chromosome mass. At anaphase, they lag behind the main chromatids but eventually move toward the poles. Two acentric fragments are seen moving toward each pole (cyan arrowheads; the yellow arrow points to the last acentric initiating its poleward movement) (see Movie S1). Third-row images are representative of 27% mitotic neuroblasts scored in which acentric fragments segregate unequally toward the poles. In this example, four fragments segregate in one pole (cyan arrowheads, the yellow arrow points to the last acentric initiating its poleward movement) (see Movie S1). The white lines in the last column highlight the contour of the dividing cells. Time is presented as min:s. The scale bar represents 5 μm.
(B) Cumulative frequency of neuroblasts scored for the time elapsed between anaphase onset and initiation of poleward segregation of the last acentric fragment. In 38 out of 42 anaphases observed, the last acentric initiated its poleward movement within 4 min after anaphase onset.
(C) Inverted images of DAPI-stained mitotic neuroblast X chromosomes of colchicine-treated neuroblasts from control or I-CreI heat-shocked larvae. The control X chromosomes exhibit a typical V shape. After I-CreI induction, the X centric and acentric fragments remain associated by a thread (cyan arrows). In addition, X homologs are often interlinked (purple arrows). In 11 out of 103 neuroblasts observed, one X acentric chromosome is dissociated from its centric partner (red arrows). The dissociated acentric sisters always remain together (red star) or are associated with the X homolog (yellow arrows).
We analyzed the delay of acentric poleward segregation by calculating the time elapsed between the anaphase onset (segregation of centric chromosomes) and the initiation of poleward movement of the last acentric chromatid. Most of the acentrics segregated within 4 min following anaphase onset (90%, n = 42, Figure 2B).
Acentric and Centric X Chromosome Fragments Are Connected by DNA Tethers
In control heat-shocked neuroblasts, the DAPI-stained X chromosomes exhibited normal V shape (Figure 2C, control). After I-CreI induction, acentric and centric X chromosome fragments often remained associated with one another by a thin DNA thread (referred to as a “tether”) (Figure 2C, second row, cyan arrows). We also found 50% (n = 103) of acentric homologs interlinked (Figure 2C, second row, purple arrows). Only 11% (n = 103) of cells had at least one X chromosome where the acentric and centric fragments were dissociated (Figure 2C, third row, red arrows). In some instances, the two acentric fragments, one from each homolog, appeared to be associated with the same centric fragment (Figure 2C, third row, yellow arrow). When dissociated from the centric fragment, the acentric sister chromatids always remained paired (Figure 2C, third row, red star). Since we observe acentrics without tethers, this suggests that they are easily broken or do not always form. We cannot rule out that some tethers might be too thin to be detected.
The DNA Tethers Are Decorated with BubR1, Polo, Aurora-B, and INCENP
To determine whether neocentromeres play a role in acentric segregation, we examined the localization of Cid, the centromeric histone H3 variant, in live neuroblasts (Figure S1A). Functional kinetochores require Cid (Blower and Karpen, 2001). In control cells, Cid-GFP localized on centromeres throughout mitosis (data not shown) (Schuh et al., 2007). After I-CreI induction, no Cid-GFP signal was detected on lagging acentrics, suggesting that neocentromeres do not form on the acentrics (Figure S1A, white arrows).
To test whether the presence of an acentric activates the spindle checkpoint, we analyzed the localization of two components of the spindle checkpoint, Mad2 and BubR1, after I-CreI induction (Musacchio and Salmon, 2007). In vivo studies using GFP-Mad2 revealed faint Mad2 labeling at the kinetochore during anaphase (Figure S1B, yellow arrows) (Buffin et al., 2005). However, we did not observe signal of GFP-Mad2 in the vicinity of the lagging chromatids (Figure S1B, white arrowheads; Movie S1). In contrast, immunostaining of prometaphase neuroblasts with anti-BubR1 antibodies revealed BubR1 puncta along the DNA tethers that connect the centric and acentric fragments in addition to its normal kinetochore localization (Figure 3A, red arrows). A strong BubR1 signal was also observed at the broken end of each dissociated fragment (Figure 3A, yellow arrows). In anaphase, BubR1 labeled the DNA tether and lagging acentric fragments. Interestingly, immunostaining of anaphase neuroblasts with antibodies directed against INCENP and Aurora B, two components of the chromosomal passenger complex, found a robust staining of these proteins at the tips of the lagging acentrics and on the tether (Figure 3A, red arrows).
Figure 3. BubR1, Polo, INCENP, and Aurora-B Localize on Tethers that Connect the Two Chromosome Fragments.
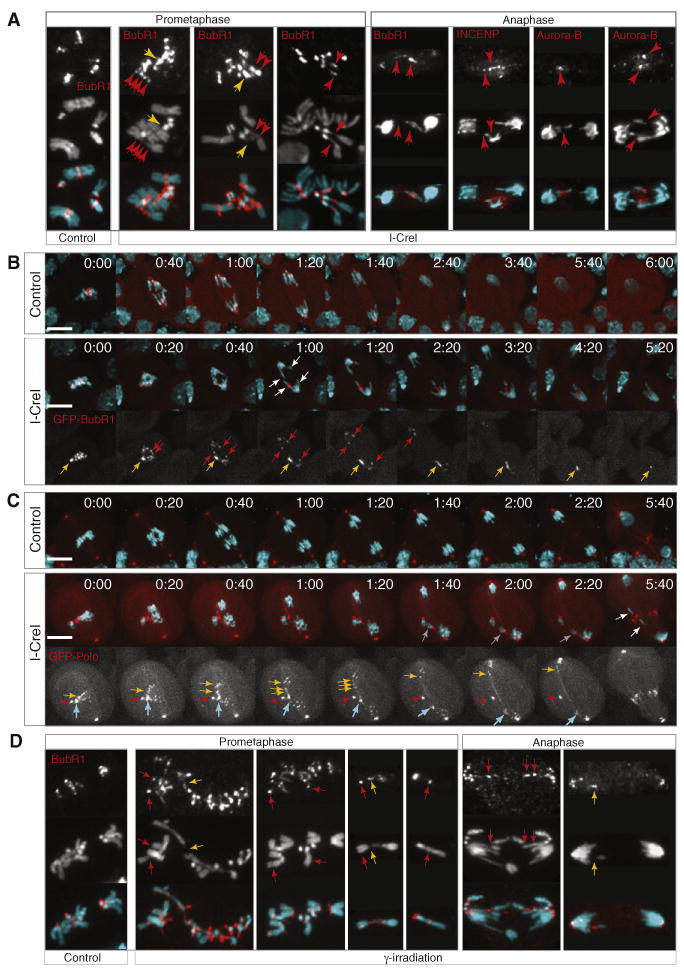
(A) Images of neuroblasts from control and I-CreI heat-shocked larvae stained with DAPI (cyan), anti-BubR1, anti-INCENP, or anti-Aurora B antibodies (red). BubR1 signal is detected at kinetochores in control and I-CreI prometaphase neuroblasts. Strong BubR1 staining is also detected on the broken end of the fragments (yellow arrows) and on the tether associating the two fragments (red arrows). BubR1 often forms puncta structures along the tether (red arrows, first panel). At anaphase, BubR1 signal is detected on the lagging chromatids and on the tethers that stretch from the lagging acentric to the segregated centric fragments (red arrows). INCENP and Aurora-B signals are also detected on the lagging acentrics in anaphase neuroblasts (red arrows).
(B) Time-lapse images of live neuroblasts from control and I-CreI heat-shocked larvae double labeled with GFP-BubR1 (red) and RFP-Histone (cyan). Each image is a maximum-intensity projection of deconvolved Z series. The first row shows typical BubR1 and chromosome dynamics from metaphase through telophase in control neuroblasts. BubR1 kinetochore signal disappears during late anaphase (see Movie S2). The second row shows BubR1 and chromosome dynamics from metaphase through anaphase in neuroblasts after I-CreI induction. The third row shows only the BubR1 signal. BubR1 localizes at the kinetochore and at the tip of each acentric fragment (yellow and red arrows). While BubR1 signal at kinetochores becomes faint as the cell progress through anaphase, a strong BubR1 signal remains at the tip of one of the acentric fragments (yellow arrow). Notice the change in length of the BubR1 signal on the tether as the cell progress through anaphase. It stretches half way through anaphase and then regresses (yellow arrows) (see Movie S2). Time is presented as min:s. The scale bar represents 10 μm.
(C) Time-lapse series of neuroblasts from control and I-CreI heat-shocked larvae double labeled with GFP-Polo (red) and RFP-Histone (cyan). Each image is a maximum-intensity projection of deconvolved Z series. The first row shows typical GFP-Polo and chromosome dynamics from metaphase through telophase in a control neuroblast (see Movie S2). The second row shows GFP-Polo and chromosome dynamics from metaphase through anaphase in neuroblasts after I-CreI induction. GFP-Polo alone is shown in the third row. Notice the strong GFP-Polo signal at the tip of the acentrics from metaphase through anaphase (red, cyan, and yellow arrows). GFP-Polo signal on the tethers stretches from the lagging acentrics through the main chromosome mass (yellow arrows). The segregation of the acentrics are unequal in this example, with two sister acentrics segregating together in one daughter cell (gray arrow, second row) and the two other sisters porperly segregating each to one daughter cell (white arrows) (see Movie S2). Time is presented as min:s. The scale bar represents 10 μm.
(D) BubR1 localizes on chromosome breaks in euchromatin. Images of neuroblasts from grp(chk1) mutant larvae untreated (control) or treated with γ-irradiation, stained with DAPI (cyan) and anti-BubR1 antibodies (red). BubR1 localized at the kinetochore in control and γ-irradiated neuroblasts. In γ-irradiated neuroblasts, ectopic BubR1 signal was also detected on chromosome arms (red arrows) and tether-like structures (yellow arrows). See also Figure S1.
Our live analysis of neuroblasts expressing RFP-Histone and GFP-BubR1 confirmed the localization of BubR1 on the DNA tether. In control neuroblasts, GFP-BubR1 stained only the kinetochores (Figure 3B and Movie S2) (Buffin et al., 2005). Upon I-CreI induction, in addition to kinetochore staining, strong BubR1 puncta were observed in nonkinetochore regions (Figure 3B, yellow arrow; Movie S2). This strong BubR1 staining persisted through anaphase and colocalized with the tip of the acentric (white and yellow arrows). BubR1 signal stretched from the tip of the lagging acentric to the bulk of chromatids as the cell progressed though anaphase (Figure 3B, yellow arrow; Figure S1C). The signal decreased in length during late anaphase as the acentric reached the main mass of chromosomes (Figure 3B, yellow arrows). Three other unconventional BubR1 localizations were detected during anaphase. These were correlated with the tip of each lagging acentric fragment (white and red arrows). During metaphase and anaphase, the BubR1 signal on the tether was stronger than the BubR1 kinetochore signal (Figure 3B and Figure S1C).
Because Polo kinase interacts with BubR1 (Elowe et al., 2007; Wong and Fang, 2007), we used a GFP-Polo construct to determine whether Polo localized on the tethers as well. GFP-Polo localized on centrosomes, spindle microtubules, and kinetochores during early anaphase and accumulated on the central spindle at cytokinesis in control neuroblasts (Figure 3C and Movie S2) (Moutinho-Santos et al., 1999). Upon I-CreI induction, GFP-Polo localized at the tip of the acentric fragments and along the length of the DNA tether (Figure 3C and Movie S2). The GFP-Polo signal became punctate as the tethers stretched from the acentric to the bulk of chromatids during anaphase (Figure 3C, yellow arrows, 2:00 min).
To investigate whether similar BubR1-decorated tethers form following DBSs created in euchromatin, we exposed larval neuroblasts to 680 RADS γ-irradiation. To increase the frequency of cells entering mitosis with DBSs, we irradiated neuroblasts from third-instar larvae mutant for the DNA damage checkpoint gene grp(chk1) (Royou et al., 2005). In nontreated and treated grp(chk1) neuroblasts, BubR1 localized at the kinetochore, indicating that, in contrast with what has been observed in some mammalian cell types (Zachos et al., 2007), Grp(Chk1) is not required for proper BubR1 accumulation at the kinetochore in Drosophila (Figure 3D, control). After γ-irradiation, tether-like structures were detected on chromosome arms (Figure 3D, yellow arrows). Significantly, ectopic BubR1 signals were clearly visible on chromosome arms and on tethers (Figure 3D, red and yellow arrows). These results indicate that tethers form at both euchromatic and heretochromatic DNA breaks. In addition, they show that BubR1 accumulates on DNA breaks regardless of the chromatin state.
BubR1 Spindle Checkpoint Activity Is Not Required to Delay Anaphase Onset upon I-CreI Induction
We tested the possibility that BubR1 tether association activates the spindle checkpoint. In fixed cells, we previously demonstrated that anaphase onset was delayed upon I-CreI induction (Royou et al., 2005). Grapes (Grp), the Drosophila Chk1 homolog, but not BubR1, was required for this delay. To confirm that BubR1 spindle checkpoint activity is not required for the delay, we examined living cells in mitosis and timed the interval from nuclear envelope breakdown (NEB) to anaphase onset (Figures S2A and S2B). In control neuroblasts, the average transit time from NEB to anaphase onset was 7.8 min (standard deviation [STD] = 1.8, n = 18). This timing increased significantly upon I-CreI induction (mean = 13.4 min, STD = 4.4, n = 44, Wilcoxon rank-sum test p < 0.0001). To test whether this delay required BubR1 spindle checkpoint function, we performed this analysis on homozygous strong hypomorph bubR11 mutant neuroblasts expressing one copy of BubR1 mutated in one of its KEN boxes and fused to RFP (thereafter referred to as bubR1-KEN mutant; see the Extended Experimental Procedures) (Rahmani et al., 2009). Mutations in the KEN box impair BubR1 spindle checkpoint function (Burton and Solomon, 2007; Davenport et al., 2006; King et al., 2007; Malureanu et al., 2009; Rahmani et al., 2009; Sczaniecka et al., 2008). Examination of live neuroblasts revealed that, after I-CreI induction, anaphase onset was delayed in bubR1-KEN mutant relative to non-I-CreI bubR1-KEN mutant (11.9 ± 4.0 versus 6.4 ± 1.4, mean ± STD, n = 38 and 23, respectively, Wilcoxon rank-sum test, p < 0.0001) (Figure S2B). These results suggest that BubR1 spindle checkpoint activity is not required to delay anaphase entry upon I-CreI induction. We further verified this result by comparing the anaphase index in heat-shocked bubR1-KEN mutants with or without the I-CreI transgene (control and I-CreI respectively, Figure S2C). The anaphase index was significantly lower in bubR1-KEN after I-CreI induction (Student's t test, p < 0.001).
Reduction of BubR1 and Polo Function Results in Acentric Segregation Defects
To decipher the function of BubR1 and Polo on tethers, we examined the segregation of I-CreI-induced acentrics in live bubR1-KEN and bubR1-KD (kinase dead) mutant and transheterozygote polo1/polo10 mutants (see the Extended Experimental Procedures). It has recently been shown that bubR1-KD retains spindle checkpoint activity but exhibits defects in spindle morphology. (Rahmani et al., 2009). Live analysis of bubR1-KD neuroblasts revealed a slight increase in the frequency of unequal segregation of acentrics compared to the wild-type (37.5% and 26.7%, respectively; Figure 4C). However, the frequency of unequal segregation of acentrics was dramatically higher in the bubR1-KEN mutant (62.5% versus 26.7% for the wild-type, Pearson χ2 test, p < 0.001; Figures 4A and 4C and Movie S3). Similarly, the polo1/polo10 mutant exhibited a high frequency of improper acentric segregation (56.3% versus 26.7% for the wild-type, Pearson χ2 test, p < 0.004; Figure 4C).
Figure 4. Defective Acentric Segregation in bubR1-KD, bubR1-KEN, and polo1/polo10 Mutants.
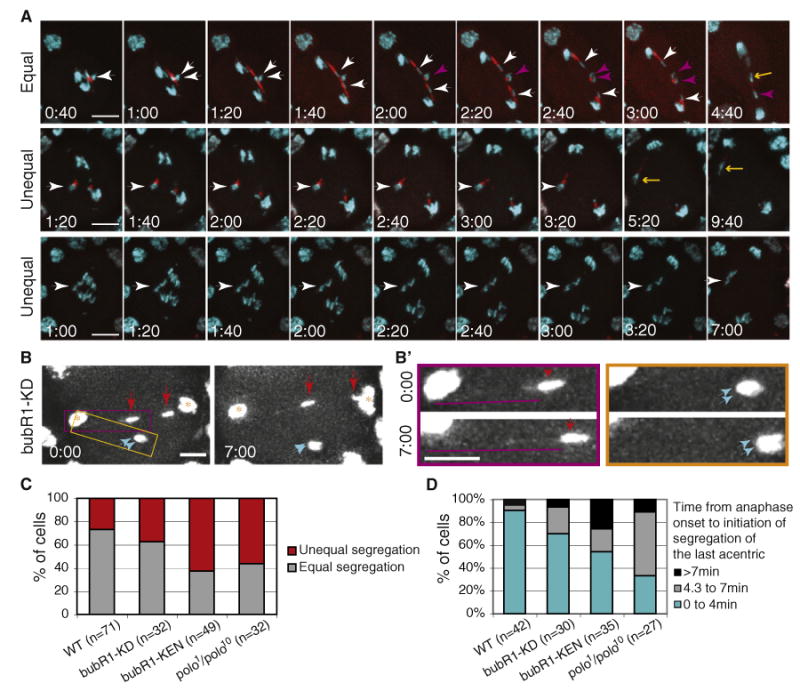
(A) Time-lapse images of anaphase neuroblasts from heat-shocked bubR1-KEN mutant larvae bearing the I-CreI transgene and double labeled with GFP-H2Av (cyan) and RFP-BubR1-KEN (red). The time (min:sec)in the first column represents the time elapsed since anaphase onset. The first row shows an example of equal segregation of acentrics to opposite poles (see Movie S3). Strong RFP-BubR1-KEN signal is present along the length of the tether. The white arrowheads highlight two sister acentrics segregating to opposite poles. The purple arrowheads highlight the other two sister acentrics that will move to opposite poles. The yellow arrow points to the last acentric initiating its poleward movement more than 4 min after anaphase onset. The second and third rows provide examples of defects in acentric segregation. In the second row, two acentrics associated with each other stay at the metaphase plate (white arrowhead) before initiating as low poleward movement toward the same pole (yellow arrow) (see Movie S3). In the third row, the acentrics have not yet initiated poleward movement 7 min after anaphase onset (white arrowhead) (see Movie S3). Each image is a maximum-intensity projection of deconvolved Z series. Time is presented as min:s. The scale bar represents 10 μm.
(B) Early (0:00) and late (7:00) anaphase time points of a bubR1-KD mutant neuroblast after I-CreI induction. GFP-H2Av labels the main mass of segregated chromatids (yellow star) as well as the two pairs of acentrics (red and cyan arrows). One sister acentric rejoined the main chromosomes after 7 min (right red arrow), while the other three sisters remained at the metaphase plate (left red and cyan arrows). The scale bar represents 5 μm.
(B′) The images are enlargements of the yellow and purple highlighted regions in (B). The images surrounded by the purple rectangle show an acentric (red arrows) that remains at the metaphase plate during anaphase. A DNA tether (purple lines) can be detected connecting the acentric to the main mass of chromatids. The tether increases in length over time. The images from the yellow rectangle depict sister acentrics that also linger on the metaphase plate and remain closely associated with one another (cyan arrows). No DNA tether can be detected connecting them to the main mass of segregating chromatids. Images are maximum-intensity projections of deconvolved Z series. Time is presented as min:s. The scale bar represents 5 μm.
(C) Frequency of unequal versus equal segregation of acentrics in wild-type, bubR1-KD, bubR1-KEN, and polo1/polo10 mutant neuroblasts.
(D) Frequency of neuroblasts in which the last acentric initiates its poleward movement within 4 min, from 4.3 to 7 min or after 7 min following the onset of anaphase in the wild-type and bubR1-KD, bubR1-KEN, and polo1/polo10 mutants. The last category (>7 min) includes neuroblasts where acentrics never initiate poleward movement within the time frame of the movie. n = number of neuroblasts.
See also Figure S2.
We also examined the influence of BubR1 and Polo on the timing of acentric segregation. In bubR1-KD, bubR1-KEN, and polo1/polo10 mutants, the frequency of anaphases in which the last acentric initiated its poleward movement beyond 4 min after anaphase onset increased more than 3-, 5-, and 7- fold, respectively, relative to the wild-type (Pearson χ2 test, p < 0.04, p < 0.002, and p < 0.001, respectively; Figure 4D). Interestingly, the frequency of acentrics that initiated poleward movement more than 7 min after anaphase onset (or did not initiate poleward movement within the time frame of the movie) was higher in the bubR1-KEN mutant compared to the wild-type and BubR1-KD and polo1/polo10 mutants (26% versus 5% for the wild-type, Pearson χ2 test, p < 0.009; Figure 4D).
Some acentrics that failed to segregate within 7 min after anaphase onset were still connected to the bulk of chromatids by a GFP-histone positive tether (Figures 4B and 4B′, purple square, red and cyan arrows). In other cases, no GFP-histone-labeled tethers were detected between the inert acentric fragments and the segregated chromatids (Figures 4B and 4B′, yellow square, cyan arrows). In these cases, tethers may be present but not detectable by the GFP-histone signal.
Reduction of BubR1 or Polo Function Results in a Decrease of Acentric Chromosome Tethering
The I-CreI-induced acentrics congressed and aligned properly on the metaphase plate in all wild-type neuroblasts examined (Figure 2). In bubR1-KEN and bubR1-KD mutants, we occasionally observed acentrics detaching from the metaphase plate. In the example shown in Figure 5A (purple circle and arrows), one acentric fragment was attached to the bulk of chromosomes by what appears to be the telomeric end of one sister chromatid. It detached during the prolonged metaphase but eventually reattached to the main chromosome mass (Figure 5A, purple arrow). Anaphase was subsequently abnormal, giving rise to aneuploid daughter cells. Thus, tethers are unstable when BubR1 function is reduced.
Figure 5. Reduced Tether Stability in bubR1 and polo Mutants.
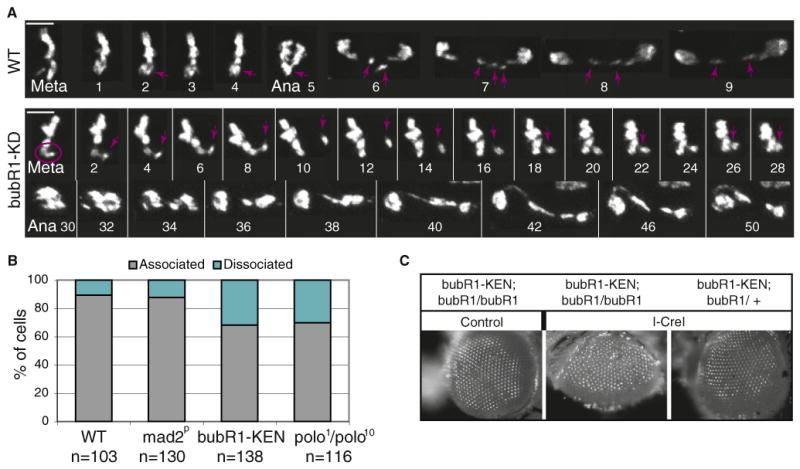
(A) Time-lapse imaging of mitotic GFP-H2Av-labeled neuroblasts from wild-type and bubR1-KD mutant larvae expressing I-CreI. In the wild-type, the acentrics align at the metaphase plate and segregate toward each pole during anaphase (purple arrow). In the bubR1-KD mutant, we observe instances in which the acentric (purple circle) is associated with the main chromosome mass, and then detaches and migrates away (purple arrow). The acentric eventually reassociates with the chromosome mass. Anaphase is triggered soon after but is abnormal. Time is presented as min:s. The scale bar represents 5 μm.
(B) Frequency of acentrics that are associated or dissociated from their centric partners. The quantification was done as described in Figure 2C. n = number of neuroblasts. Neuroblasts were scored in two independent experiments for each genotype (two to four brains per experiment).
(C) Images of adult eyes from bubR1-KEN mutant, homozygote (bubR1/bubR1), or heterozygote (bubR1/CyO) for bubR11 with (I-CreI) or without (control) the I-CreI transgene. The third-instar larvae were heat shocked and allowed to develop into adults. Note the roughness of the eye of bubR1-KEN mutant after I-CreI induction suggesting a loss of omatidia during development.
Examination of chromosomes from prometaphase arrested cells in fixed bubR1-KEN mutant neuroblasts confirmed this result. The frequency of acentrics not tethered to their centric partner was significantly higher in the bubR1-KEN mutant compared to the wild-type (32% versus 11%, n = 138 and 103, respectively, Pearson χ2 test, p < 0.001; Figure 5B). In addition, the frequency of dissociated acentrics more than doubled in the polo1/polo10 mutant relative to the wild-type (31% versus 11%, n = 116 and 103, respectively, Pearson χ2 test, p < 0.001; Figure 5B). We also examined the frequency of dissociated acentrics in mad2p null mutants. Mad2 is a core component of the spindle checkpoint but does not localize on tethers. In mad2p mutants, we observed similar frequencies of dissociated fragments as in the wild-type (12% versus 11%,n = 130 and 103, respectively; Figure 5B). These results, combined with the live analysis, suggest a model in which BubR1 and Polo are required for the integrity of the tethers in a Mad2-independent manner.
Reduction of BubR1 or Polo Function Results in a Great Reduction of the Survival Rate
Disruption of BubR1 function dramatically decreased the frequency of I-CreI larvae developing into adults (18.4% ± 7.6% versus 72.1% ± 10.7% in control larvae) (Table 1). In addition, bubR1-KEN larvae produced adults with rough eyes, defective wings, and missing bristles (Figure 5C and data not shown). These defects are likely the result of significant cell loss during development. We also observed a great sensitivity of polo1/polo10 mutants to I-CreI induction (Table 1). No viable adults developed from larvae in which I-CreI was induced. In some instances, adults died while enclosing. Unlike polo- and bubR1-compromised mutants, mad2p mutant survival rate was similar to the control (Table 1).
Discussion
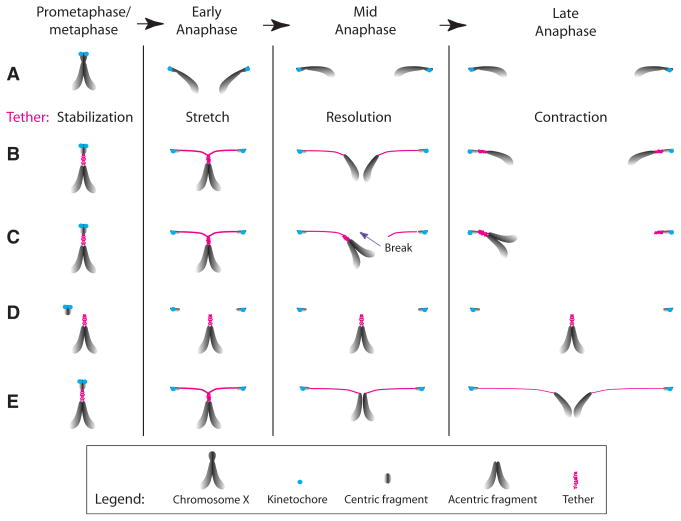
While the canonical mechanism driving chromosome segregation is via kinetochore-microtubule interactions, studies have demonstrated efficient segregation of chromosomes lacking centromeric DNA (Ishii et al., 2008; Kanda et al., 2001; Kaye et al., 2004; Platero et al., 1999). This occurs either through the formation of neocentromere or direct association of the acentric to intact chromosomes. Our analysis of I-CreI-induced acentrics reveals a distinct tether-based mechanism by which acentrics are efficiently segregated to daughter cells. These acentrics rely on DNA threads decorated with BubR1, Polo, INCENP, and Aurora-B to segregate equally toward the poles (Figure 6).
Figure 6. Model for the Role of Tether in Acentric Segregation.
(A) In metaphase, X sister chromatids are held together by cohesins at the pericentric region. During anaphase, sister chromatids separate and segregate via kinetochore (blue circle)/spindle attachment.
(B) Upon I-CreI induction, the centric and acentric sister chromatids fragments remain associated via BubR1- and Polo-decorated DNA tethers (pink). At anaphase onset, the centric sisters separate and initiate poleward movement via kinetochore/spindle attachment. The acentric sisters stay together at the metaphase plate, while the tether stretches to compensate for the increased distance between the moving centrics and the inert acentrics. Tether formation may be the consequence of unreplicated DNA or unresolved repair intermediates in the region of the DSBs. Entangled tethers may serve to hold sister acentrics together until early anaphase. We speculate that the tether is elastic and that once the entanglement is resolved, the tethers contract, pulling the sister acentrics apart from each other and toward their segregating centric partner. This scenario explains the predominantly (73%) equal segregation of acentrics observed in wild-type mitotic neuroblasts. In the remaining neuroblasts, unequal segregation of sister acentricsis observed.
(C–E) Prolonged entanglement of sister tethers through anaphase could result in the breakage of one of the tether (C). Reduced BubR1 and Polo function increases the frequency of acentrics lacking tethers (D), results in slower poleward movement of the acentric (E), and increases the frequency of unequal acentric segregation (C). These phenotypes suggest that these proteins play a role in tether stability and contraction and may be involved in the resolution of entangled tethers. Whether these phenotypes are a consequence of a global role that BubR1 and Polo may play on chromatin architecture remains to be elucidated.
The observation that segregating acentrics possess a DNA tether connecting them to their centric partner suggests that tethers facilitate acentric segregation. During the period in which segregation of the acentric is delayed while the main mass of chromosomes has fully segregated, the length of the tether increases to accommodate the increased distance between the segregating centric fragment and the inert acentric. This increase could occur either through a spooling-out to create a longer tether, or stretching of the tether. We favor the latter alternative, as this readily explains the delay in acentric segregation followed by prompt segregation to the poles until they reach the main mass of chromatids. That is, the tether may be elastic, and as tension builds, the tether stretches ultimately resulting in rapid tether contraction (Figure 6). Elastic forces have been proposed in other instances in which chromatin tethers have been observed. Severing of crane-fly meiotic chromosomes during anaphase results in the acentric chromosome fragments moving backward across the equator (LaFountain et al., 2002). This finding led to the conclusion that sister telomeres are connected by an elastic tether that exerts a force opposing poleward forces. In Drosophila, heterochromatic threads connect achiasmatic chromosome homologs during meiosis (Hughes et al., 2009). It is proposed that these threads mediate congression of nonexchange chromosomes via their elastic properties.
Reduction of BubR1 or Polo activity results in an increase in the frequency of abnormal acentric segregation and a decrease in acentric chromosome tethering. These observations indicate that these tether-associated kinases are involved in tether function. They may play a role in generating tether-elastic forces driving acentric segregation. This idea is supported by the observation that, during anaphase, in bubR1- and polo-compromised mutants, acentrics linger at the metaphase plate much longer than acentrics in wild-type cells. We also find instances in these mutants where tethers stretch from the inert acentrics to the segregating chromosome mass without initiating acentric poleward movement. This suggests a failure in tether contraction.
Although a large number of cells in larval brains exhibit lagging acentric chromosomes after I-CreI induction, there is no effect on adult survival. Insight into the high survival rates comes from the finding that in a wild-type background, sister acentrics segregate accurately to opposing poles with a high frequency. Thus, if a cell enters anaphase with a DSB, a final option may be to properly segregate acentrics enabling reassociation of the centric and acentric chromosome fragments and repair of the DSB in the daughter cells. This is supported by the observation that in bubR1 and polo mutants, the frequency in which acentrics segregate equally to opposing poles is significantly decreased and there is a corresponding reduction in adult survival. We cannot rule out that spindle defects inherent to the polo mutants underly the synthetic lethality of polo mutants with I-CreI expression. However, we found that mutations disrupting BubR1 kinase activity, which alters spindle structure, produce a less dramatic defect in acentric segregation than mutations in the BubR1 KEN box that impair BubR1 checkpoint function. The fact that spindle integrity was not detectably altered in a previous analysis of bubR1-KEN mutant neuroblasts (Rahmani et al., 2009) indicates that abnormal spindle structure is not the primary cause of acentric segregation defects in BubR1-compromised mutants and its corresponding synthetic lethality with I-CreI expression. Moreover, this suggests that BubR1 spindle checkpoint function plays a role in efficient acentric segregation. In undamaged cells, BubR1 localizes at kinetochores and inhibits the anaphase-promoting complex/cyclosome (APC/C) until all chromosomes are properly attached to the spindle (Musacchio and Salmon, 2007). Recently, BubR1 has been found to accumulate on unprotected telomeres and is thought to activate the spindle checkpoint (Musarò et al., 2008). We found that I-CreI-generated acentrics delay anaphase onset via activation of the DNA damage checkpoint Grp(Chk1) but independently of the BubR1 spindle checkpoint activity (Royou et al., 2005). We speculate that some as-yet unidentified APC/C substrates are associated with the tether and are important for tether function. Since BubR1 remains strongly associated with the tether well into anaphase, it may efficiently inhibit the APC/C activity locally on the tether, thus preserving tether integrity throughout mitosis. On the other hand, BubR1 KEN box may have a role in addition to APC/C inhibition that is important for BubR1 function on the tether.
We currently do not know the complete nature of the DNA tethers reported here and the mechanisms by which they form. The fact that tether can form in euchromatin as well as heterochromatin indicates they are a general feature of Drosophila chromosomes. We speculate that the presence of DBSs may result in the cell entering mitosis with unresolved replication intermediates that promote tether formation. Support for this idea comes from recent studies reporting that replication stress results in the formation of BLM (Bloom syndrome, RecQ helicase-like)-associated ultrafine DNA bridges linking homologs at fragile loci during mitosis (Chan et al., 2009). An alternative possibility is that the presence of DSBs even after replication has terminated necessitates the long-term recruitment of the repair machinery. The cell may enter mitosis with repair intermediates hampering chromatin condensation at the site of DSBs, thus creating the tethers. Interestingly, DNA tethers form between chromosome homologs or heterologs during meiosis when condensin complex function is impaired (Hartl et al., 2008). The observation that BubR1 accumulates on DNA breaks regardless of the chromatin state suggests a more direct role of BubR1 on DNA repair in mitotic cells. It might, for instance, stabilize the repair machinery that keeps the DNA fragments apposed.
We find that tethered and untethered acentric sister chromatids remain tightly apposed well into anaphase. The mechanism by which these sisters are held together is unclear as cohesins are removed from chromosome arms as early as prophase in Drosophila (Warren et al., 2000). Similar observations have been shown in yeast in which centric and acentric fragments were created by the HO endonuclease (Kaye et al., 2004; Melo et al., 2001). The authors found instances where acentric sister chromatids remain linked. This association depends partially on repair machinery components and impairs their proper segregation. Based on these findings, we speculate that I-CreI-generated acentric sister chromatids are held together by the entanglement of their respective tethers generated by repair mechanisms. In most instances, this entanglement is resolved during progression through anaphase. Failure to resolve entanglement would result in the unequal segregation of acentrics (Figure 6).
Recent studies have reported DNA tether-like structures that connect achiasmate chromosomes in Drosophila meiosis (Hughes et al., 2009). These threads contain the passenger proteins INCENP and Aurora B. INCENP-coated DNA tethers are also present during anaphase in mammalian cells (Baumann et al., 2007; Wang et al., 2008). Significantly Aurora B and INCENP decorate I-CreI-induced tethers. This implies that all tethers share similar properties and their origin and structure are conserved features of the eukaryotic cell cycle.
Experimental Procedures
Fly Stocks
All stocks were raised on standard medium at 25°C. The bubR11 (Basu et al., 1999; Logarinho et al., 2004), bubR1-KEN, bubR1-KD (Rahmani et al., 2009), mad2P (Buffin et al., 2007), polo1 (Llamazares et al., 1991), and polo10 (Donaldson et al., 2001) mutations were previously described. Additional stocks and crosses are described in the Extended Experimental Procedures.
Cytology and Microscopy
Crawling third-instar larvae were placed in a vial containing small amount of standard media and heat shocked in a 37°C water bath for 1 hr or irradiated for 680 rads with a Torrex 120D machine (Astrophysics Research Corporation). The larvae were dissected 1–2 hr after treatment, unless otherwise indicated. Methods for measuring survival rate and lagging chromosome frequency are described in the Extended Experimental Procedures. For immunostaining, the brains were dissected in PBS and fixed as described in Williams and Goldberg (1994). The anti-γH2Av (Kim McKim) and anti-BubR1 (Claudio Sunkel) antibodies were used at 1/1000. Anti-INCENP and anti-Aurora B (William Earnshaw) antibodies were used at 1/500. Secondary alexa 488 anti-rabbit antibodies (Molecular Probes) were used at 1/500. The preparations were mounted with Vectashield containing DAPI (Vector Laboratories, Inc.) and observed with a wide-field fluorescence inverted Leica DMI6000B microscope equipped with a Hamamatsu ORCA C9100 EM-CCD camera and 100× (NA 1.4) lens and 1 × binning. Z series images of 0.2 μm intervals were acquired, processed, and deconvolved with LAS AF6000 software. Additional image processing was done with Adobe Photoshop. All images are maximum-intensity projections.
Live Analysis of Larval Neuroblasts
Preparation of third-instar larval brains for live analysis were dissected in PBS and transferred in a drop of PBS on a coverslip. The brain was slightly squashed as described in Buffin et al. (2005). The brains were observed with the wide-field fluorescent microscope described above. Z series of 0.5 μm steps were acquired every 20 s for a maximum time of 25 min. All movie frames are maximum-intensity projections.
Supplementary Material
Acknowledgments
We thank Kent Golic (University of Utah, UT), Kim McKim (Rutgers University, NJ), Claudio Sunkel (University of Porto, Portugal) and William Earnshaw (University of Edinburgh, UK) for sharing reagents, the Sullivan lab members and Derek McCusker (Institut Européen de Chimie et Biochimie, Bordeaux, France) for critical reading of the manuscript. We also thank Alfredo Villasante (Universidad Autonoma de Madrid, Spain) for sharing unpublished data. A.R. and W.S. were supported by grants from the California Institute for Regenerative Medicine and the National Institutes of Health (GM046409), respectively. M.E.G. was supported by a training and mobility grant from the Research Training Network program of the European Union. R.K. was supported by the Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche (ANR-08-BLAN-0006-01).
Footnotes
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, two figures, and three movies and can be found with this article online at doi:10.1016/j.cell.2009.12.043.
References
- Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, Sunkel CE, Goldberg ML. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- Buffin E, Emre D, Karess RE. Flies without a spindle checkpoint. Nat Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM. Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol. 2001;153:663–676. doi: 10.1083/jcb.153.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy DJ, Toczyski DP. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol Cell Biol. 2001;21:1710–1718. doi: 10.1128/MCB.21.5.1710-1718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Sweeney SJ, Knepler PJ, Bosco G. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 2008;4:e1000228. doi: 10.1371/journal.pgen.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Gilliland WD, Cotitta JL, Takeo S, Collins KA, Hawley RS. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain JR, Jr, Cole RW, Rieder CL. Partner telomeres during anaphase in crane-fly spermatocytes are connected by an elastic tether that exerts a backward force and resists poleward motion. J Cell Sci. 2002;115:1541–1549. doi: 10.1242/jcs.115.7.1541. [DOI] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5(12A):2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, Antunes-Martins A, Sunkel CE. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T, Sampaio P, Amorim I, Costa M, Sunkel CE. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol Cell. 1999;91:585–596. [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Musarò M, Ciapponi L, Fasulo B, Gatti M, Cenci G. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat Genet. 2008;40:362–366. doi: 10.1038/ng.2007.64. [DOI] [PubMed] [Google Scholar]
- Platero JS, Ahmad K, Henikoff S. A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol Cell. 1999;4:995–1004. doi: 10.1016/s1097-2765(00)80228-2. [DOI] [PubMed] [Google Scholar]
- Rahmani Z, Gagou ME, Lefebvre C, Emre D, Karess RE. Separating the spindle, checkpoint, and timer functions of BubR1. J Cell Biol. 2009;187:597–605. doi: 10.1083/jcb.200905026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A, Macias H, Sullivan W. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr Biol. 2005;15:334–339. doi: 10.1016/j.cub.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sczaniecka M, Feoktistova A, May KM, Chen JS, Blyth J, Gould KL, Hardwick KG. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J Biol Chem. 2008;283:23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Titen SW, Golic KG. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics. 2008;180:1821–1832. doi: 10.1534/genetics.108.093625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell L, Soler D, Agostini M, Pampalona J, Genescà A. The number of dysfunctional telomeres in a cell: one amplifies; more than one translocate. Cytogenet Genome Res. 2008;122:315–325. doi: 10.1159/000167818. [DOI] [PubMed] [Google Scholar]
- Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Williams BC, Goldberg ML. Determinants of Drosophila zw10 protein localization and function. J Cell Sci. 1994;107:785–798. doi: 10.1242/jcs.107.4.785. [DOI] [PubMed] [Google Scholar]
- Wong OK, Fang G. Cdk1 phosphorylation of BubR1 controls spindle checkpoint arrest and Plk1-mediated formation of the 3F3/2 epitope. J Cell Biol. 2007;179:611–617. doi: 10.1083/jcb.200708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, Gillespie DA. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.