Summary
The polysaccharide β-1,6-glucan is a major component of the cell wall of Cryptococcus neoformans, but its function has not been investigated in this fungal pathogen. We have identified and characterized seven genes, belonging to the KRE family, which are putatively involved in β-1,6-glucan synthesis. The H99 deletion mutants kre5Δ and kre6Δskn1Δ contained less cell wall β-1,6-glucan, grew slowly with an aberrant morphology, were highly sensitive to environmental and chemical stress and were avirulent in a mouse inhalation model of infection. These two mutants displayed alterations in cell wall chitosan and the exopolysaccharide capsule, a primary cryptococcal virulence determinant. The cell wall content of the GPI-anchored phospholipase B1 (Plb1) enzyme, which is required for cryptococcal cell wall integrity and virulence, was reduced in kre5Δ and kre6Δskn1Δ. Our results indicate that KRE5, KRE6 and SKN1 are involved in β-1,6-glucan synthesis, maintenance of cell wall integrity and retention of mannoproteins and known cryptococcal virulence factors in the cell wall of C. neoformans. This study sets the stage for future investigations into the function of this abundant cell wall polymer.
Keywords: phospholipase, chitosan, chitin deacetylase
Introduction
Cryptococcus neoformans is an opportunistic fungal pathogen that causes potentially fatal meningoencephalitis in immunocompromised individuals, especially patients with AIDS. The cryptococcal cell wall is an essential organelle, vital for maintaining cell morphology and integrity. Since it is essential in fungi and its components are absent from mammalian host cells, the cell wall is an attractive target for antimicrobial therapies. The cryptococcal cell wall is composed of α- and β-linked glucans, chitin and its deacetylated derivative chitosan, and mannoproteins. In contrast to ascomycetes such as Saccharomyces cerevisiae and Candida albicans, in which β-1,3-glucan predominates (Klis et al., 2001; Klis et al., 2006), β-1,6-glucan is the most prevalent cell wall β-linked glucan molecule in C. neoformans (James et al., 1990). Despite its abundance, the function of β-1,6-glucan in the cryptococcal cell wall has not previously been investigated.
To date, no β-1,6-glucan synthase protein has been identified in any fungal species, which is in stark contrast to the well-characterized synthase enzymes of other major cell wall components (Araki and Ito, 1975; Shematek et al., 1980; Shaw et al., 1991; Ziman et al., 1996; Hochstenback et al., 1998). However, large-scale genetic screens have identified several S. cerevisiae proteins required for normal levels of cell wall β-1,6-glucan. These include Kre1p, Kre9p, Kre11p, Knh1p, Kre5p, and the functional homologs Kre6p and Skn1p. Several are localized along the secretory pathway, leading to the idea that β-1,6-glucan synthesis may involve intracellular events (reviewed in Shahinian and Bussey, 2000). A recent report using in silico analysis determined KRE1, KRE9, KRE11, and KNH1 are Ascomycete-specific, suggesting that these genes are not essential for the synthesis of β-1,6-glucan and are more likely involved in modification of the polymer (Ruiz-Herrera et al., 2008). KRE5 contains a COOH-terminal ER-retention sequence and shares some sequence homology to mammalian UDP-glucose glycoprotein glycosyltransferases (UGGTs) (Meaden et al., 1990) while the KRE6/SKN1 genes encode cis-Golgi type II transmembrane proteins sharing 66% amino acid identity (Roemer et al., 1994). In S. cerevisiae, deletion of KRE5 results in complete loss of alkali-insoluble cell wall β-1,6-glucan while disruption of KRE6 reduced β-1,6-glucan levels to half of that measured from wild type cells (Roemer and Bussey, 1991). Loss of SKN1 alone had no effect but its deletion further decreased the β-1,6-glucan levels in a kre6 null background (Roemer et al., 1993). The decrease in β-1,6-glucan in these mutants adversely affects growth, morphology and cell integrity (Meaden et al., 1990; Roemer and Bussey, 1991). Deletion of KRE5 in the fungal pathogen C. albicans had similar effects and resulted in decreased virulence, highlighting a potential role for this gene and β-1,6-glucan in fungal pathogenesis (Herrero et al., 2004).
In several fungal species, the majority of cell wall proteins are attached to β-1,6-glucan via a remnant of their glycosylphosphatidylinositol (GPI) anchor (Kapteyn et al., 1996; de Groot et al., 2007; Yin et al., 2005). Homology searches of the C. neoformans genome predicted that there are 55 GPI-anchored proteins (Levitz and Specht, 2006), 16 of which have been identified by mass spectrometry in the extracellular proteome (Eigenheer et al., 2007). Among these proteins were three chitin deacetylases (Cdas), responsible for the enzymatic conversion of chitin to chitosan (Baker et al. 2007). The cryptococcal virulence factor phospholipase B1 (Plb1) is also GPI-anchored and concentrated in the cell wall (Djordjevic et al. 2005; Siafakas et al. 2006; Siafakas et al. 2007). Digestion of wild type cryptococcal cells with β-1,3-glucanase released Plb1 linked to β-1,6-glucan polymer, suggesting covalent attachment between Plb1 and β-1,6-glucan. Plb1 is also secreted from the C. neoformans surface during infection and facilitates tissue invasion. The cell wall appears to be a source of Plb1 as β-1,6-glucan was detected on the secreted enzyme (Siafakas et al. 2007).
In this study we have used gene deletion to investigate the role of seven genes of the KRE family, for their potential role in β-1,6-glucan synthesis in C. neoformans. We demonstrate that deletion of KRE5 alone or KRE6 and SKN1 in combination results in the loss of cell wall β-1,6-glucan, accompanied by disruption of growth, morphology, cell wall integrity and virulence. Additionally, two important virulence factors, the exopolysaccharide capsule and the GPI anchored mannoprotein Plb1, as well as cell wall chitosan, are altered in these mutants.
RESULTS
C. neoformans encodes seven potential β-1,6-glucan synthesis-related genes
We used a homology-based approach to identify genes in C.neoformans potentially required for β-1,6-glucan synthesis, utilizing S. cerevisiae proteins cited in the literature as being involved in this process (Shahinian and Bussey, 2000). The proteins Kre1p, Kre11p and Kre9p/Knh1p paralogs, all associated with β-1,6-glucan synthesis in S. cerevisiae, had no apparent homologs in the H99 genome. This is in agreement with a similar analysis in the fungal plant pathogen Ustilago maydis that determined these genes are specific to ascomycetes and absent from basidiomycetes (Ruiz-Herrera et al. 2008). Our results provide further support for the proposal that the mechanism of β-1,6-glucan synthesis in C. neoformans and other basidiomycetes may be distinct from that in S. cerevisiae and ascomycetes, or that the proteins have significantly diverged throughout the evolutionary process.
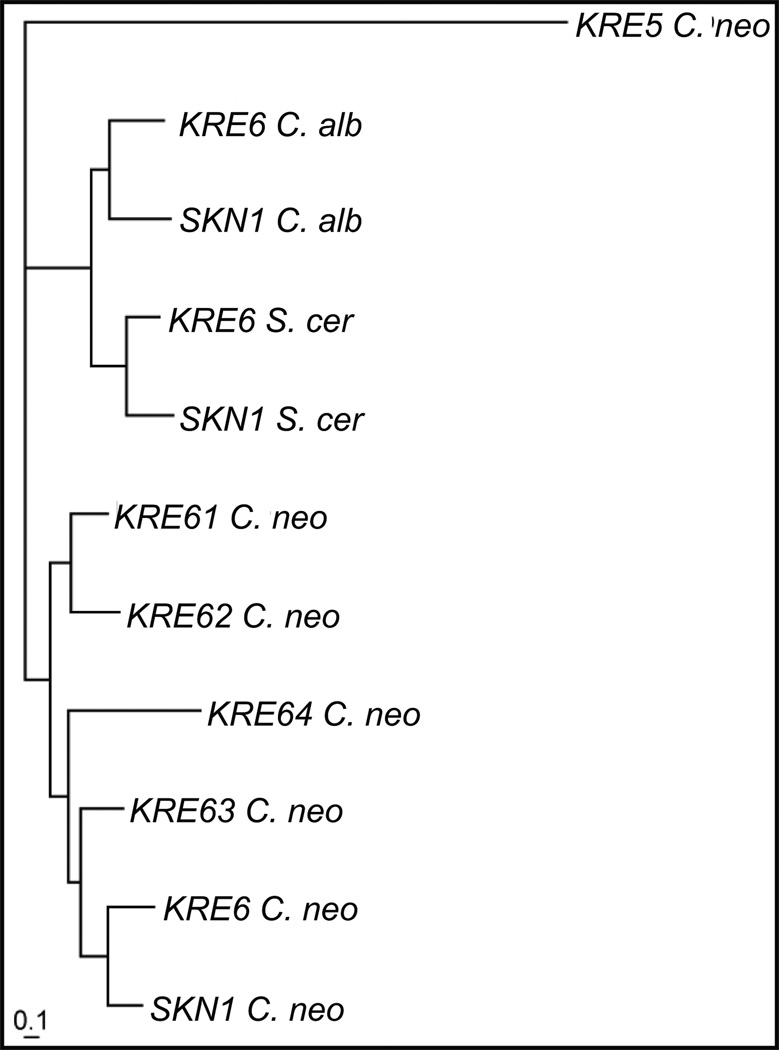
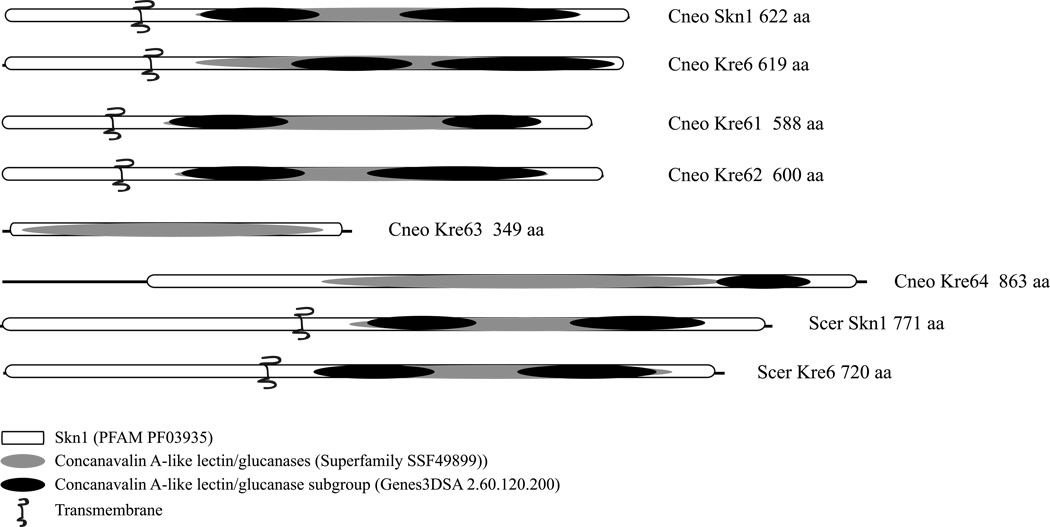
This search did yield one potential cryptotoccal homolog of S. cerevisiae Kre5p and six of the Kre6/Skn1 proteins (Table 1). The C. neoformans Kre6/Skn1 proteins were all quite similar to their S. cerevisiae homologs, varying from 38 – 48% identity and 53 – 61 % similarity at the amino acid level (Table 2). The C. neoformans homologs were more similar to each other, with 45 – 69% identity and 59 – 81 % similarity (Table 3). This clustering of the C. neoformans Kre6/Skn1 proteins is evident in the phylogenetic tree generated from an alignment of all the Kre6/Skn1 homologs from S. cerevisiae, C. neoformans and C. albicans, shown in Figure 1. A domain analysis using InterProScan revealed that the Kre6/Skn1 proteins from S. cerevisiae, C. albicans and the Skn1p, Kre6p, Kre61p and Kre62p from C. neoformans shared the same domains, domain number and domain organization (Fig. 2). The Kre63p and Kre64p have the major Skn1 domain, but neither has a transmembrane spanning domain and Kre64p has only 1 concanavalin A while Kre63p has none. This would suggest that the Kre6p, Skn1p, Kre61p and Kre62p may be functionally redundant in C. neoformans.
Table 1.
Identification of C. neoformans KRE5 and KRE6/SKN1 genes
|
S.cerevisiae Protein |
C. neoformans Broad Protein Match |
BLASTp e-value |
C. neoformans Proposed Gene Name |
|---|---|---|---|
| Kre5p | CNAG_03648.2 | 6.00E-24 | KRE5 |
| Skn1p | CNAG_00897.2 | 1.00E-114 | SKN1 |
| Kre6p | CNAG_00914.2 | 1.00E-113 | KRE6 |
| Kre6p | CNAG_06835.2 | 1.00E-116 | KRE61 |
| Kre6p | CNAG_06832.2 | 1.00E-109 | KRE62 |
| Kre6p | CNAG_06031.2 | 1.00E-73 | KRE63 |
| Kre6p | CNAG_05815.2 | 1.00E-90 | KRE64 |
Table 2.
Comparison of S. cerevisiae and C. neoformans Kre6/Skn1 proteins
| Protein 1 | Protein 2 | % identity | % similarity |
|---|---|---|---|
| S. cerevisiae Kre6p | C. neoformans Kre6 | 45 | 62 |
| S. cerevisiae Kre6p | C. neoformans Kre61 | 42 | 56 |
| S. cerevisiae Kre6p | C. neoformans Kre62 | 46 | 61 |
| S. cerevisiae Kre6p | C. neoformans Kre63 | 41 | 53 |
| S. cerevisiae Kre6p | C. neoformans Kre64 | 38 | 53 |
| S. cerevisiae Kre6p | C. neoformans Skn1 | 44 | 58 |
| S. cerevisiae Skn1p | C. neoformans Skn1 | 48 | 61 |
| S. cerevisiae Skn1p | C. neoformans Kre6 | 41 | 58 |
| S. cerevisiae Kre6p | C. albicans Kre6 | 65 | 78 |
| S. cerevisiae Skn1p | C. albicans Skn1 | 59 | 75 |
Table 3.
Comparison of C. neoformans Kre6/Skn1 proteins
| Protein 1 | Protein 2 | % identity | % similarity |
|---|---|---|---|
| C. neoformans Kre6 | C. neoformans Kre61 | 58 | 71 |
| C. neoformans Kre6 | C. neoformans Kre62 | 56 | 70 |
| C. neoformans Kre6 | C. neoformans Kre63 | 59 | 74 |
| C. neoformans Kre6 | C. neoformans Kre64 | 45 | 59 |
| C. neoformans Kre6 | C. neoformans Skn1 | 69 | 81 |
Fig. 1.
Phylogenetic tree of the Kre6/Skn1 proteins. The tree was created using the neighbor-joining algorithm in Phylip from a multiple alignment of the Kre6 and Skn1 protein sequences from S. cerevisiae, C.neoformans, and C. albicans, with the C. neoformans Kre5 protein used as an out group.
Fig. 2.
Domain organization of C. neoformans and S. cerevisiae Kre6/Skn1 homologs. Domains were identified using InterProScan.
Ruiz-Herrera et al. (2008) reported a similar expansion of the KRE6/SKN1 gene family in U. maydis. To determine whether this was true of other basiomycetes we conducted a BLASTp analysis of the NR database using C. neoformans Kre6 protein as the query and limiting the search to either basidiomycetes or ascomycetes species. We utilized the taxonomy report features to identify the number of possible Kre6 homologs in each species, using a cut-off of E ≤ 1e−100. Within the basidiomycetes, we found expanded families of Kre6/Skn1 like proteins in 3 species. Ustilago maydis had 7 homologs with E-values ranging from 6e−135 to 4e−116, all of which contained an Skn1 and concanavalin A-like lectin/glucanase domains. Laccaria bicolor had 6 homologs with E-values ranging from 8e−146 to 1e−119. Coprinopsis cinerea had 7 homologs with E-values ranging from 1e−139 to 3e−110. In all cases, the region of similarity covered the entire Skn1 domain of the C. neoformans Kre6 protein. We examined 25 ascomycetes species, almost all of which had only 1–2 potential Kre6/Skn1 homologs. This initial blast analysis suggests that an expanded family of Kre6-like proteins might be a distinguishing features of basidiomycetes compared to ascomycetes, although the number of basidomycete genomes is limited at this time.
KRE gene expression is temperature-regulated during vegetative growth
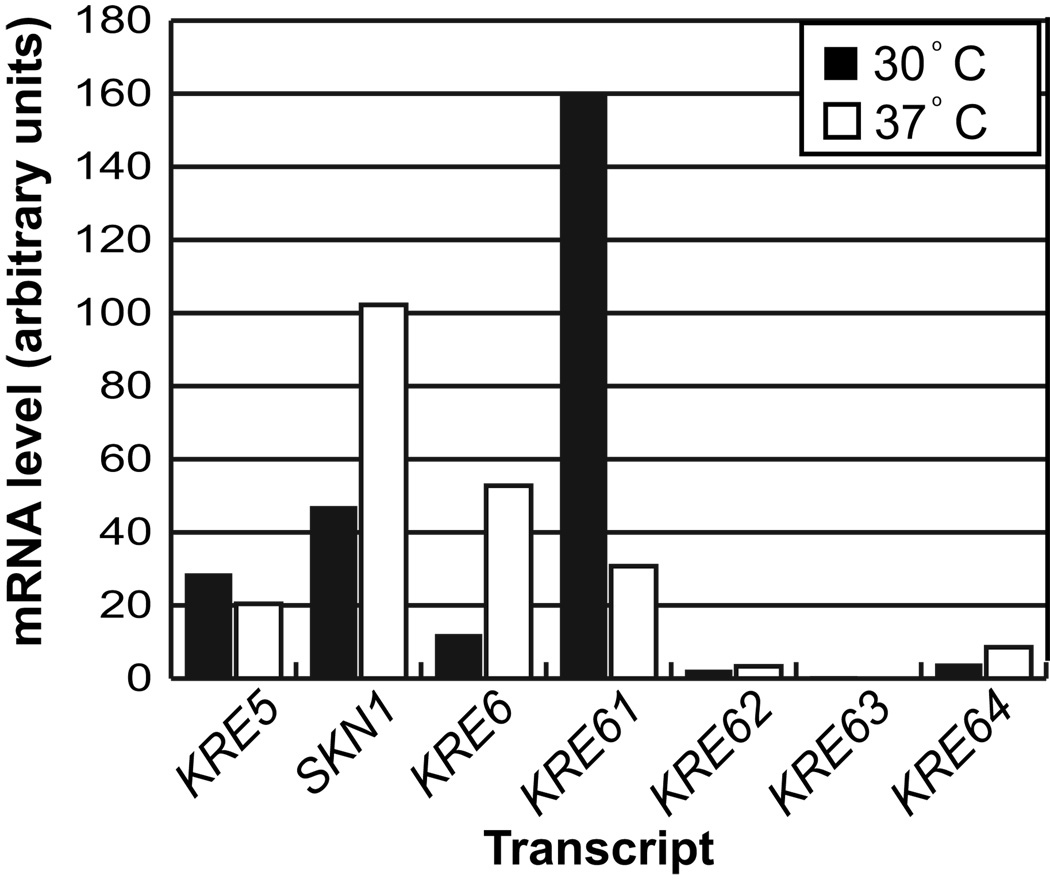
The existence of multiple KRE6 homologs suggests functional redundancy between these genes and that they may be differentially expressed under different biological conditions. We used northern blot analysis to determine the expression profiles of the putative KRE5 and the KRE6/SKN1 homologs during vegetative growth. KRE5, SKN1, KRE6 and KRE61 were expressed at 30°C (Fig. 3), whereas growth at 37°C showed comparatively higher expression of KRE6 and SKN1 but lower KRE61 expression. The remaining genes displayed no detectable expression at either temperature. These data lead to the hypothesis that KRE6 and SKN1 may be functionally redundant, especially during growth at 37°C.
Fig. 3.
Northern blot mRNA expression analysis of KRE5 and the KRE6/SKN1 homologs in C. neoformans. H99 was grown in liquid culture for 24 h at the indicated temperatures. Relative gene expression was determined utilizing ImageQuant (version 5.1) and was normalized to the level of actin. Values are the average of two independent biological replicates.
Generation of deletion strains
We generated targeted single deletions of KRE5, SKN1 and each of the KRE6/SKN1 genes using homologous recombination in the C. neoformans H99 strain. We also attempted to generate the various double deletion combinations of KRE6, KRE61 and SKN1, as they were the only KRE6/SKN1 genes with detectable mRNA expression during vegetative growth. Surprisingly, we were unable to obtain kre61Δskn1Δ isolates despite multiple attempts at deleting either gene in the opposite single deletion background. One transformation yielded viable colonies, but Southern blot analysis reveled the presence of both the NAT marker deletion cassette and the deleted region, indicating a gene duplication event may have occurred prior to transformation (data not shown). This evidence, although not conclusive, is consistent with synthetic lethality between KRE61 and SKN1, but such a possibility must be explored further in a mating competent background strain.
The phenotypes of the kre61Δ, kre62Δ, kre63Δ, kre64Δ, and skn1Δ strains were indistinguishable from wild type in all experiments tested. This is somewhat unexpected for kre61Δ, as KRE61 mRNA was detectable at both 30 and 37 °C. However, the phenotypes obtained for the kre62Δ, kre63Δ and kre64Δ strains are consistent with the northern analysis, which detected no expression of KRE62, KRE63 or KRE64 mRNA.
The kre5Δ, kre6Δ, kre6Δkre61Δ and kre6Δskn1Δ mutants demonstrated abnormal phenotypes, as described below. The synthetic phenotypes displayed by the kre6Δskn1Δ mutant, although not conclusive evidence, suggests functional redundancy between these genes. In multiple attempts to complement the kre5Δ and kre6Δskn1Δ mutations, the mutant strains were completely recalcitrant to transformation either by biolistics or electroporation, yielding no viable colonies. Therefore, independent deletion isolates were used for analysis.
KRE5, KRE6 and SKN1 are involved in β-1,6-glucan synthesis in C. neoformans
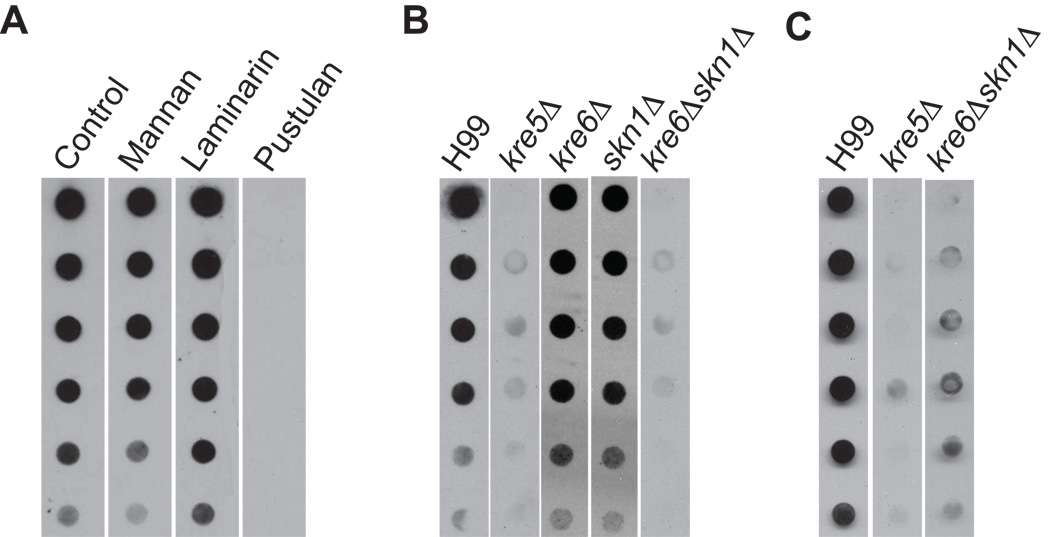
We further tested our hypothesis that proteins expressed from the KRE genes are involved in β-1,6-glucan synthesis in C. neoformans by analyzing β-1,6-glucan in cell walls prepared from each of the deletion mutants. Cell wall polysaccharides can be separated into an alkali-soluble fraction and a fraction that is alkali-insoluble due to the cross-linking of the polysaccharides to insoluble chitin polymers (Hartland et al. 1994). We initially analyzed the alkali-soluble cell wall material by dot blotting, using a polyclonal anti-β-1,6-glucan antiserum as described by Maneesri et al. (2005). Although this assay is not quantitative for the level of β-1,6-glucan in the cell wall, the assay does demonstrate qualitative differences between the overall levels of the polymer among the strains. We confirmed this antibody is specific for β-1,6-glucan in C. neoformans by demonstrating that pustulan, a purified β-1,6-glucan, was the only polysaccharide that significantly competed for antibody binding to wild type alkali-soluble cell wall material (Fig. 4A). Alkali-soluble β-1,6-glucan was absent from kre5Δ and kre6Δskn1Δ using this method while kre6Δ, skn1Δ and other deletion strains had levels similar to those of wild type (Fig. 4B, Suppl. Fig 1).
Fig. 4.
β-1,6-glucan dot blot assay. (A) Polysaccharide competition analysis. Alkali soluble cell wall material from H99 was spotted onto membrane. The primary anti-β-1,6-glucan antiserum was preincubated with the indicated purified polysaccharides for 30 min before probing the membrane. (B) Alkali-soluble cell wall β-1,6-glucan analysis of H99 and deletion strains. Cells were grown at 30°C to mid-log phase and alkali-soluble polysaccharides were isolated and spotted in 1:2 serial dilutions onto nitrocellulose membrane. Membranes were probed with polyclonal anti-β-1,6-glucan antiserum. (C) Alkali-insoluble, chitinase-released cell wall β-1,6-glucan analysis of H99 and deletion strains.
These results indicated that kre5Δ and kre6Δskn1Δ are deficient in cell wall β-1,6-glucan. An alternative explanation was that β-1,6-glucan levels are unchanged in the mutants, but the polymer had shifted to the insoluble fraction. Such phenomena have been observed for other cell wall mutants (Reese et al., 2007). Therefore, we further analyzed the alkali-insoluble fractions by digesting with purified chitinase to release glucans into the soluble fraction. As shown in Fig. 4C, this treatment released β-1,6-glucan from the alkali-insoluble fraction of wild type cells. However, no β-1,6-glucan was released from the corresponding fractions of kre5Δ or kre6Δskn1Δ cells. These results indicated that loss of KRE5 or KRE6 and SKN1 causes a decrease in the level of cell wall β-1,6-glucan.
Disruption of KRE5 or KRE6 and SKN1 alters cell morphology
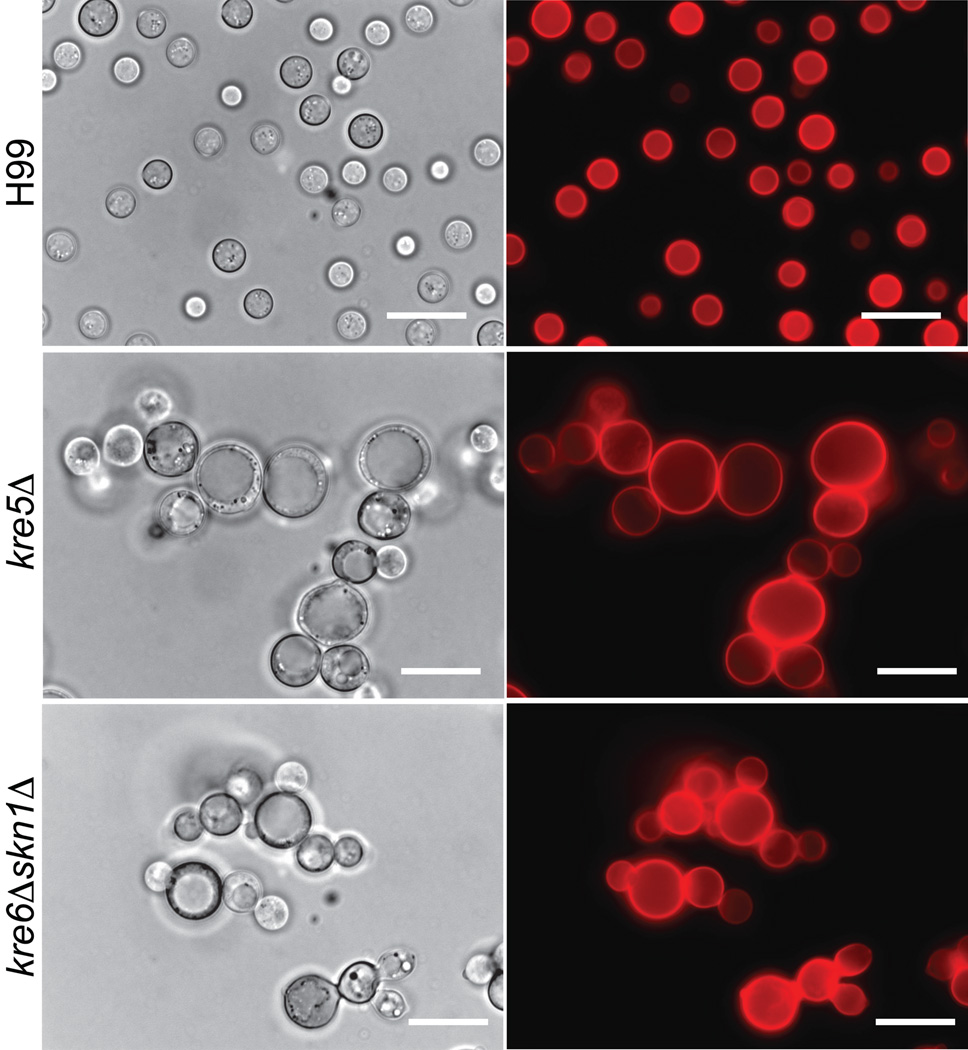
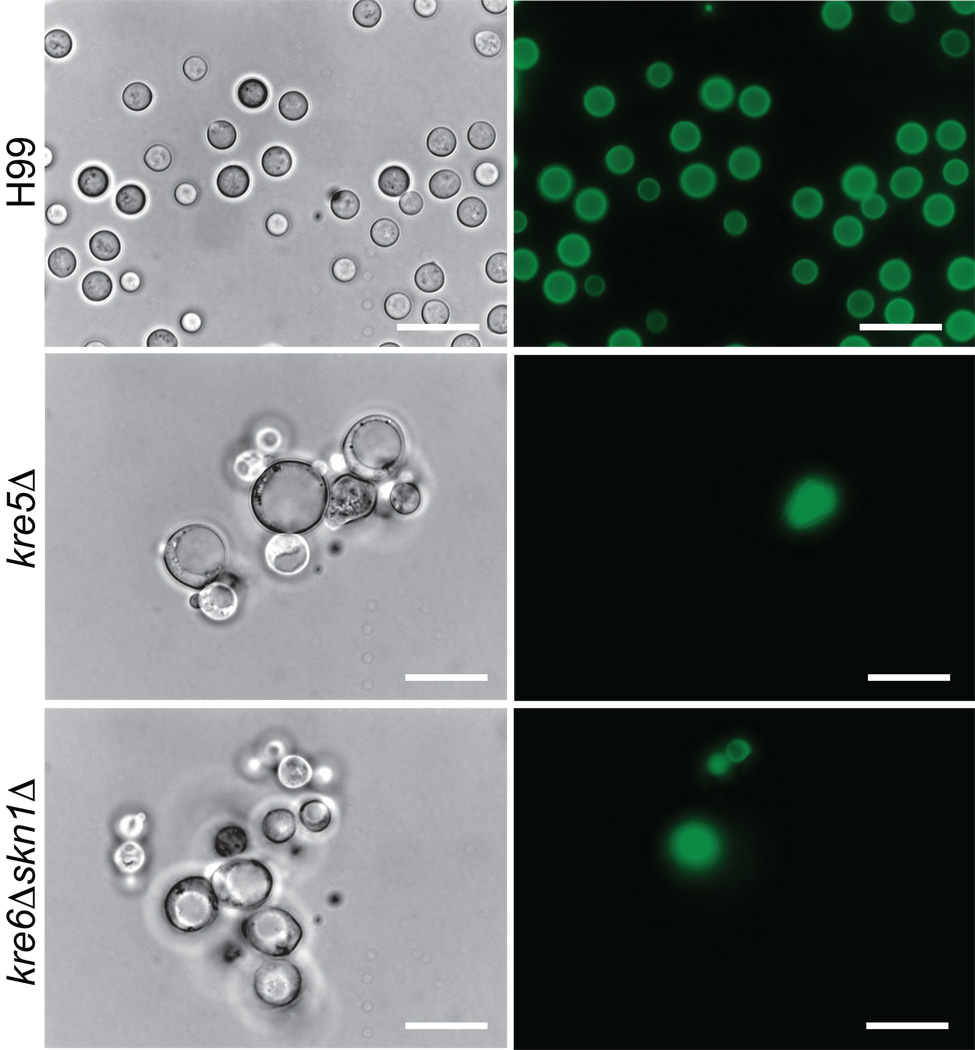
Since the cell wall is a primary determinant of cell shape, we established whether alterations in β-1,6-glucan levels in kre5Δ or kre6Δskn1Δ would alter cell morphology. Mutant strains were stained with the general cell wall stain Pontamine Fast Scarlet (Hoch et al., 2005), and examined by both light and fluorescent microscopy. The cell morphology and pattern of pontamine staining obtained for the skn1Δ, kre6Δ, kre61Δ, kre62Δ, kre63Δ, kre64Δ and kre6Δkre61Δ deletion strains were indistinguishable from that of wild type (Suppl. Fig. 2). However, Fig. 5 shows the kre5Δ and kre6Δskn1Δ cells were enlarged compared to wild type cells, formed clumps and contained large intracellular vacuole/vesicle structures. Multiple buds were present, resulting in chains of cells formed by incomplete separation of daughter cells from the mother cell.
Fig. 5.
Morphology of C. neoformans H99, kre5Δ and kre6Δskn1Δ. Cells were grown on YPD medium at 30°C for 72h and stained with Pontamine FastScarlet. All images are at 1000X magnification. Scale bar = 25µm.
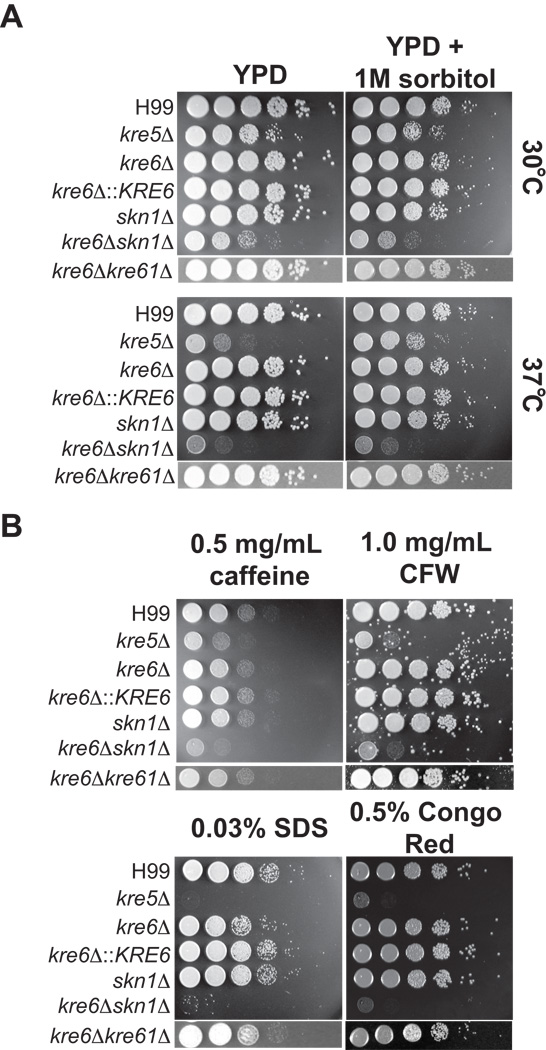
Loss of KRE5 or KRE6 and SKN1 affects sensitivity to high temperature and cell wall inhibitors but not antifungal agents
The deletion mutants were tested for sensitivity to conditions that challenge cell wall integrity, including increased temperature and various cell wall inhibiting agents. Calcofluor white binds chitin fibrils and disrupts their assembly (Roncero and Durán 1985); caffeine interferes with cell wall integrity-related signal transduction pathways (López-Sáez et al., 1982); sodium dodecyl sulfate (SDS) disrupts the plasma membrane and lyses cells with membrane or cell wall defects; Congo red binds β-glucans and inhibits the microfibril assembly of the cell wall (Roncero and Durán, 1985). The kre5Δ and kre6Δskn1Δ strains grew slowly at 30°C and growth was further inhibited at 37°C (Fig. 6A). This temperature sensitivity was partially rescued by the addition of 1M sorbitol, commonly used as an osmotic stabilizer, in the medium, with the growth of kre5Δ being more fully restored than that of kre6Δskn1Δ (Fig. 6A). Loss of KRE5 or both KRE6 and SKN1 resulted in increased sensitivity to all cell wall inhibitors tested (Fig. 6B). The kre6Δ mutant was mildly sensitive to 0.03% SDS, and this sensitivity was rescued by complementation with the wild type gene (Fig. 6B). The kre6Δkre61Δ double deletion strain displayed only the SDS sensitivity observed for kre6Δ (Fig. 6B). Deletion of the five remaining KRE6/SKN1 homologs individually did not cause an observable increase in sensitivity under any of the conditions tested (Suppl. Fig. 3).
Fig. 6.
Sensitivity of mutants to temperature (A) and cell wall inhibitors (B). Strains were grown overnight in YPD medium, then diluted to an OD650 of 1.0. Tenfold serial dilutions were made in PBS and 5 µl of each was spotted on medium containing the indicated inhibitors. Plates were incubated at the indicated temperatures for 2 days.
The kre5Δ and kre6Δskn1Δ mutants were also tested for sensitivity to the antifungal compounds fluconazole, voriconazole and caspofungin. Fluconazole and voriconazole disrupt production of ergosterol in the fungal plasma membrane and caspofungin inhibits β-1,3-glucan synthesis. Neither mutant had altered sensitivities to any of these drugs compared to wild type H99 (data not shown).
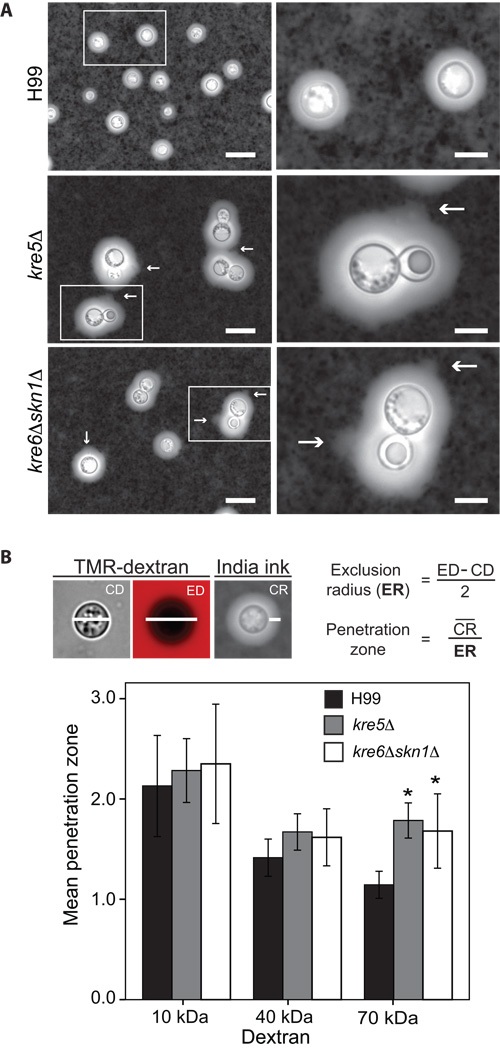
kre5Δ and kre6Δskn1Δ cells have enlarged capsules with altered architectures
The C. neoformans cell wall is associated with two prominent virulence factors; the exopolysaccharide capsule and melanin (Chang and Kwon-Chung, 1994; Casadevall et al., 2000). Alterations in cell wall composition have been shown to adversely affect their production or attachment (Baker et al., 2007; Reese et al., 2007; Reese and Doering, 2003). Therefore, we examined both capsule and melanin in the deletion strains. Interestingly, when grown under capsule-inducing conditions and stained with India ink, the kre5Δ and kre6Δskn1Δ cells had enlarged capsules compared with those of wild type cells. A minimum of 200 cells from each strain were examined and the average capsule radius of wild type cells was 2.28 µm while the average radii of kre5Δ and kre6Δskn1Δ capsules were 4.49 µm and 4.24 µm, respectively. Furthermore, the mutant capsules had a fainter, more diffuse appearance with rough edges, in contrast to the smooth ring of wild type capsule (Fig. 7A). There were several instances where the capsular polysaccharides appeared to slough from the capsule perimeter (Fig. 7A, arrows). The capsules of the remaining deletion strains had an appearance indistinguishable from wild type capsules (data not shown).
Fig. 7.
Capsule analysis (A) India ink staining of capsule of H99 and deletion strains. Strains grown for 3 days on DME medium at 30°C with 5% CO2 to induce capsule production were stained with India ink and visualized by light microscope. Arrows point to the rough appearance of the polysaccharide sloughing from capsular edge. Scale bars in the left and right panels are 25 and 10 µm, respectively. (B) Penetration of the cryptococcal capsule by TMR-dextrans. Top panel shows representative images of wild type cells stained with India ink or incubated with 70 kDa TMR-dextran and illustrates the measurements taken and the equations used to calculate the penetration zone. CD = capsule diameter; ED = exclusion diameter; CR = capsule radius. Strains were grown as in (A) and incubated 60 min with TMR-dextrans with average molecular masses of 10, 40 and 70 kDa, and the radius of penetration was determined by examination of digital images. Results shown are mean ± SD for at least 24 cells in two independent biological replicates. * denotes p≤ 0.02 compared to wild type H99 with 70 kDa dextran.
The altered appearance of the kre5Δ and kre6Δskn1Δ capsules may have been the result of increased penetration of the India ink particles into the capsule interior, possibly due to alterations in capsular architecture. To test this idea, we measured the permeability of wild type and mutant capsules to TMR-dextrans of various sizes. The 70-kDa dextrans diffused further into the capsules of kre5Δ and kre6Δskn1Δ cells than into wild type capsules, as indicated by a significant increase in the penetration zone (Fig. 7B). These data indicate that the capsules of the kre5Δ and kre6Δskn1Δ mutants are more diffuse than the capsules of wild type cells.
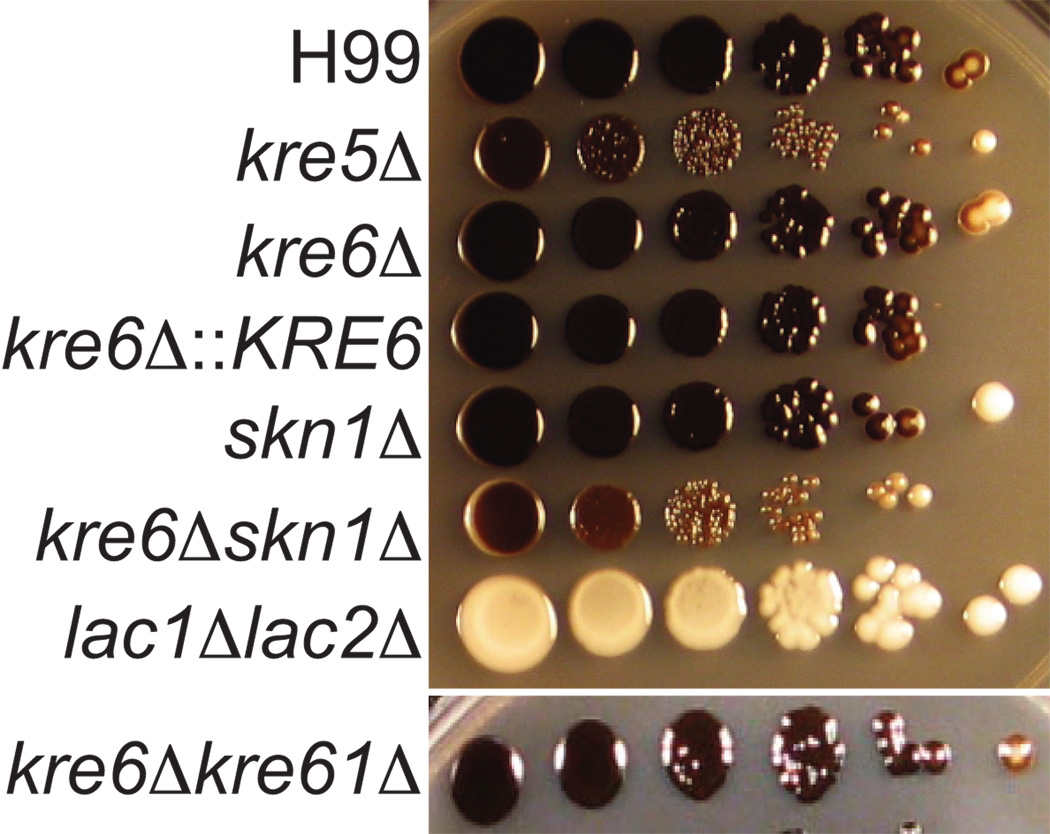
Melanin production was assessed by growing strains on media containing the substrate L-DOPA (Polacheck et al., 1982; Williamson, 1994). All of the deletion strains produced a dark melanin pigment that remained associated with the cell, in contrast to the “leaky melanin” phenotype observed for other cell wall deficient mutants (Baker et al., 2007; Banks et al., 2005). However, both the kre5Δ and kre6Δskn1Δ cells appeared slightly lighter in color compared to wild type cells (Fig. 8), indicating a possible decrease in total melanin production. As expected, a lac1Δlac2Δ mutant (Missall et al., 2005) missing the enzymes responsible for melanin production produced no pigment.
Fig. 8.
Melanin production. Strains were grown overnight in YPD medium, diluted to an OD650 of 1.0. and 5 µl of each tenfold serial dilution (in PBS) and was plated on medium containing 1 mM L-DOPA. Plates were incubated at 30°C for 7 days.
Virulence analysis
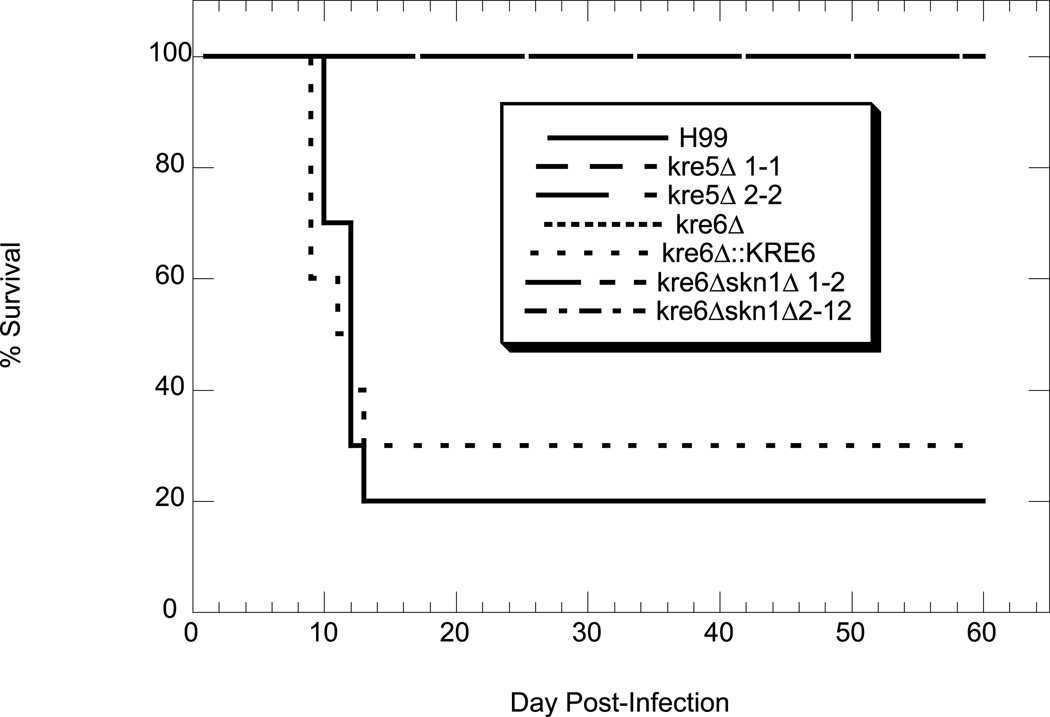
The single deletion mutants were initially screened for alterations in virulence using STM analysis in a mouse tail vein model of infection (Nelson et al., 2001). Results from these preliminary experiments indicated that only kre5Δ and kre6Δ had consistent differences in virulence compared to wild type H99 (data not shown). Since these were also the only two single mutants to demonstrate altered phenotypes in vitro we analyzed them independently using a mouse inhalation model of infection. The kre6Δskn1Δ double mutant and kre6Δ::KRE6 complemented strain were also tested. We hypothesized that kre5Δ and kre6Δskn1Δ would have decreased ability to cause mammalian infection, especially due to their inability to grow at 37°C. As anticipated, kre5Δ and kre6Δskn1Δ were completely avirulent (Fig. 9). The kre6Δ mutant was also avirulent and this phenotype was rescued by reconstitution of the wild type gene. This result is somewhat surprising, as this mutant has wild type levels of β-1,6-glucan and displayed only a moderate sensitivity to SDS (Fig. 6B).
Fig. 9.
Virulence analysis. Mice (10 per strain) were inoculated with 1×106 cryptococcal cells, dripped into the nares. Mice were weighed before infection and sacrificed once they reached 80% of their original body weight.
Cellular distribution of Phospholipase B1 is altered in kre5Δ and kre6Δskn1Δ
Plb1 is present in the cryptococcal cell wall and this localization requires the presence of its GPI-anchor (Djordjevic et al.,2005; Siafakas et al., 2006, Siafakas et al., 2007). Secreted Plb1 contains β-1,6-glucan (Siafakas et al., 2007). These data indicate that the cell wall is a source of Plb1 and that β-1,6-glucan is important for its attachment via a remnant of its GPI-anchor. To further explore this possibility, we next investigated whether reduced β-1,6-glucan in the kre5Δ and kre6Δskn1Δ mutants coincided with altered Plb1 localization. Plb1 is a multifunctional enzyme displaying three activities (phospholipase B: PLB; lysophospholipase: LPL and lysophospholipase acyltransferase :LPTA). Almost all of the cell-associated and secreted PLB activity in Cryptococcus is attributable to the Plb1 protein (Cox et al. 2001; Wright et al. 2007), whereas the LPL and LPTA activities could also be contributed by other enzymes (Coe et al. 2003; Wright et al. 2004). Therefore, we focused on the PLB activity.
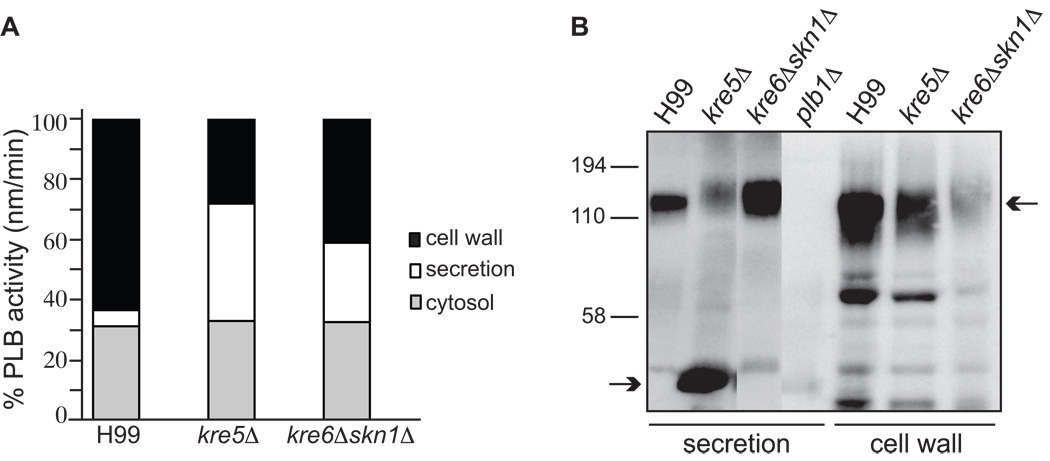
Subcellular fractions were prepared from concentrated cryptococcal cell suspensions that had been allowed to secrete Plb1 overnight. The cell wall, ccrude membranes, cytosol and secretions were assayed for PLB activity using a radiometric enzyme assay. Overall, the mutants produced 2–3 times more total PLB activity (secreted and cell-associated) than the wild type (1705, 6348 and 4209 units for H99, kre5Δ and kre6Δskn1Δ, respectively). Therefore, we compared PLB activity in corresponding fractions of wild type and each mutant strain by expressing the activity in each fraction as a percentage of the total activity in that strain (Fig. 10A). The percentage of cell wall-associated PLB activity was reduced in both the kre5Δ and kre6Δskn1Δ mutants, with the reduction being greatest in kre5Δ (H99 183 units; kre5Δ, 1792 units; kre6Δskn1Δ, 1734 units). This reduction in activity correlated with reduced retention of Plb1 protein in the cell wall fraction, as detected by Plb1 western blotting (Fig. 10B). The ratio of cell wall-associated/secreted PLB activity was 12, 0.72 and 1.6 for wild type, kre5Δ and kre6Δskn1Δ, respectively, in this experiment, and 7.88, 0.88 and 2.35 in a second independent experiment. For each strain, PLB activity associated with the crude membrane fraction was negligible (H993; kre5Δ, 2; kre6Δskn1Δ, 2 units). In strains with reduced cell wall-associated PLB activity, secretion of PLB activity (H99, 91 units; kre5Δ, 2474 units; kre6Δskn1Δ, 1109 units) and Plb1 protein was concomitantly increased (Fig. 10A and 10B, respectively). Interestingly, Plb1 protein secreted by kre5Δ exhibited extensive breakdown (Fig. 10B, bottom arrow) despite the presence of protease inhibitors during sample preparation for SDS PAGE.
Fig. 10.
Cellular distribution of PLB activity (A) and the Plb1 protein (B). (A) Subcellular fractions were prepared from concentrated cell suspensions after an overnight incubation period and harvesting of the secreted proteins. PLB activity in the crude membrane fraction was negligible. * denotes p<0.005 (unpaired 2-tail t-test) compared to the corresponding fraction from H99. Similar results for all three strains were obtained in a second independent experiment (not shown). (B) Plb1 protein in the secreted and cell wall fractions were detected by western blotting with an anti-Plb1 peptide antibody. Top and bottom arrows indicate the position of full-length, fully glycosylated Plb1 and a breakdown product (most prominent in kre5Δ secretions), respectively.
kre5Δ and kre6Δskn1Δ have altered chitosan staining but normal chitin and chitosan levels
Chitosan is an abundant polysaccharide in the vegetative cell wall of C. neoformans, and is important for maintaining cell wall integrity (Baker et al., 2007; Banks et al., 2005). Chitosan is produced by the removal of acetyl groups from nascent chitin polymers by chitin deacetylase (Cda) enzymes. The Cdas contain a putative GPI-anchor attachment motif (Baker et al., 2007) and have been identified in the extracellular proteome of C. neoformans, indicative of cell wall localization (Eigenheer et al., 2007). The loss of β-1,6-glucan in kre5Δ and kre6Δskn1Δ may result in mis-localization or secretion of the cell wall associated Cdas, as was seen for Plb1. Such altered Cda localization could potentially result in reduced or altered cell wall chitosan. Therefore, we assessed cell wall chitosan by staining strains with the polyanionic dye eosin Y, which binds specifically to the positively charged chitosan in the C. neoformans cell wall under mildly acidic conditions (Baker et al., 2007). The kre5Δ and kre6Δskn1Δ strains did not stain with eosin Y after 72 hours of growth (Fig. 11). This alteration in staining was specific for chitosan and not the result of a general cell wall defect, as pontamine staining produced the characteristic bright ring appearance (Fig. 5). The eosin Y staining patterns of the remaining single deletion strains were indistinguishable from that of wild type cells (data not shown). It has previously been shown that chs3Δ and cda1Δcda2Δcda3Δ mutants, which lack cell wall chitosan, do not stain with eosin Y (Baker et al., 2007). The loss of eosin Y staining observed for kre5Δ and kre6Δskn1Δ suggested that levels of chitosan may be reduced. However, there was surprisingly no significant difference in the levels of either chitin or chitosan between wild type and kre5Δ or kre6Δskn1Δ when measured biochemically (data not shown). Chitin/chitosan content of the remaining single deletion strains also did not deviate significantly from wild type levels (data not shown). We tested whether the apparent disparity between the eosin Y staining and biochemical measurement data could be due to differing phases of growth between wild type and the slow-growing mutant strains, as chitosan content increases during vegetative growth. In addition, the interaction between eosin Y and chitosan (pKa 6.3) is highly dependent upon charge and a decrease in staining could be caused by changes in the cell wall microenvironment that decrease the binding efficiency between eosin Y and chitosan. Both of these possibilities are unlikely, as both kre5Δ and kre6Δskn1Δ cultures reached stationary phase by 24 h and staining at both pH 5.0 and pH 6.0 did not result in eosin Y binding to the mutants (data not shown). Together these data indicate that the cell wall organization of kre5Δ and kre6Δskn1Δ may be altered in such a way that chitosan is no longer accessible for binding by eosin Y.
Fig. 11.
Eosin Y staining of H99 and kre5Δ and kre6Δskn1Δ. Strains were incubated at 30 °C for 72 hours and stained with eosin Y. Images were captured at 1000X magnification. Bright cells in the eosin Y panels of the mutants are presumably dead cells that have taken up dye, as seen previously (Gerik et al. 2008). Scale bar = 25µm.
DISCUSSION
The fungal cell wall is an ideal drug target, as it is essential for viability and absent from mammalian cells. The polysaccharide β-1,6-glucan is the most abundant β-glucan in the cell wall of the opportunistic pathogen C. neoformans. This work is the first report of the function of this polymer in Cryptococcus. We demonstrated that β-1,6-glucan, and the genes involved in its synthesis, namely KRE5, KRE6 and SKN1, are critical for maintaining growth, morphology and cell wall integrity in C. neoformans. Their disruption also results in striking alterations to cell wall chitosan and important virulence factors including the exopolysaccharide capsule, the GPI-anchor mannoprotein Plb1 as well as loss of virulence in a mammalian host. These data indicate that β-1,6-glucan, along with being abundant, has in important function in the maintenance of cell wall organization and integrity in C. neoformans.
We sought first to determine whether the KRE genes identified were involved in β-1,6-glucan synthesis in C. neoformans. The data from dot blot analyses using polyclonal anti-β-1,6-glucan antiserum clearly demonstrated that disruption of KRE5 or KRE6 and SKN1 in combination resulted in obvious, qualitative decreases in the level of this cell wall polymer. Although these results are dramatic, the actual levels of β-1,6-glucan cannot be quantified by these means. An alternative method for analyzing β-1,6-glucan levels would be to digest the cell wall with recombinant β-1,6-glucanase and measure the amount of reducing sugars released. However, the results garnered through this approach may still not necessarily reflect the true composition of the cryptococcal wall. There are only a few β-1,6-glucanases from different organisms which have been cloned and/or purified (de la Cruz et al., 1995, Boisramé and Gaillardin, 2009), but their specificities or activities have not been completely characterized, especially in the context of a complex mixture of polysaccharides present in a cell wall extract. Knowledge of specificity is crucial because, following glucanase treatment, free sugar ends are measured, therefore monomers, dimers, trimers, etc. are all detected. The β-1,6-glucanase from Trichoderma harzianum described by de la Cruz et al. (1995), although effective on purified β-1,6-glucan, was ineffective on cell wall preparations from two different fungal species. Only one β-1,6-glucanase has been used to characterize S. cerevisiae and C. albicans walls, and it is becoming clearer from our work and others (Banks et al., 2005, Reese et al., 2003) that the cryptococcal wall is more complex (containing chitosan and α-glucan) and likely organized very differently than the ascomycete wall. Although it is clear that the cryptococcal wall contains higher levels of β-1,6-glucan (James et al., 1990) the molecular organization of this polymer and the linkages with other cell wall components have not been defined biochemically. A further complication is the presence of the adjacent capsular polysaccharide of C. neoformans, which interferes with cell wall analysis, requiring the use of an acapsular background strain (James et al., 1990, Reese et al. 2007) that may not truly reflect the composition of a wild type cell wall. Future investigations of the biochemical composition and organization of the cryptococcal cell wall will certainly prove to be informative, providing that interpretations carefully consider these caveats.
Expansion of the KRE6/SKN1 gene family in C. neoformans
An interesting feature of β-1,6-glucan synthesis in Cryptococcus and other basidiomycetes is the expansion of the KRE6/SKN1 gene family to multiple members. There are several possible explanations for the retention of such apparent redundancy within the cryptococcal genome. One reason may be that these genes function under various growth conditions or during different stages of the life cycle, such as during mating. Consistent with this idea, microarray analysis of RNA isolated from a C. neoformans serotype D strain revealed differential expression of the KRE6/SKN1 genes during different stages of mating (Christina Hull, personal communication). It remains to be determined whether this phenomenon holds true for serotype A. Another interesting aspect is the correlation between the expansion of the KRE6/SKN1 genes with the predominance of β-1,6-glucan polymer over that of β-1,3-glucan in C. neoformans. To date, the identity of the β-1,6-glucan synthase enzyme has remained elusive. There is no direct biochemical evidence that any of the KRE genes encode the protein required for enzymatic catalysis of β-1,6-glucan. The localization of Kre5p to the ER and the fact that a kre5/kre5 mutant of C. albicans contains residual levels of β-1,6-glucan make it an unlikely candidate. The absence of apparent KRE1, KRE11 or KRE9/KNH1 homologs in basidiomycetes known to have β-1,6-glucan, including C. neoformans, argues against these genes filling the role of the synthase. It is tempting to speculate that the increased number of KRE6/SKN1 genes coupled with the high levels of β-1,6-glucan suggest that this family could encode the synthases. Consistent with this idea, Kitamura et al. (2009) recently reported the identification of Kre6p as the target of a β-1,6-glucan small molecule inhibitor. Although β-1,6-glucan is still found in a S. cerevisiae kre6Δskn1Δ strain, this polymer is smaller and has an altered structure (Roemer et al., 1993). Conversely, the presence of a Kre6p-related gene in Aspergillus fumigatus, which lacks β-1,6-glucan in its cell wall, appears to be evidence contradictory to this idea (Aimanianda et al., 2009). However, a similar condition exists in Schizosaccharomyces pombe where a chitin synthase is present in the genome but little to no chitin was found in the cell wall (Sietsma and Wessels, 1990, Bush et al., 1974). Studies using the recently published radiometric assay for β-1,6-glucan synthesis (Aimanianda et al., 2009) make the prospect of identification of the synthase enzyme(s) optimistic.
Loss of β-1,6-glucan disrupts chitosan localization
Alterations in the level of one cell wall polysaccharide are often accompanied by compensatory changes in the levels of other cell wall components. In S. cerevisiae, decreases in β-glucan levels can lead to increased chitin deposition in the cell wall to maintain cell integrity (Popolo et al. 1997; Bulik et al. 2003; Lesage et al. 2005; Reese et al. 2007). The C. neoformans cell wall is distinct from that of S. cerevisiae in that it has significant levels of chitosan, the deacetylated derivative of chitin. Both kre5Δ and kre6Δskn1Δ had markedly decreased staining intensity with the chitosan-specific dye eosin Y. However, the levels of both chitin and chitosan in kre5Δ and kre6Δskn1Δ were not significantly different from that in wild type cells grown under the same conditions. One possible explanation for this disparity is that chitosan distribution in the cell wall is altered in such a way that it is “masked” from eosin Y binding. Three chitin deacetylases (Cdas) are responsible for the production of cell wall chitosan by the enzymatic deacetylation of nascent chitin polymers (Baker et al. 2007). The Cdas contain a predicted GPI-anchor attachment motif and were identified in the C. neoformans extracellular proteome (Eigenheer et al. 2007; Baker et al. 2007). We hypothesize that altered chitosan distribution in kre5Δ and kre6Δskn1Δ could result from altered localization of the Cdas, as was observed for Plb1. If the Cdas are absent from the cell wall of kre5Δ and kre6Δskn1Δ but remain in the plasma membrane, chitosan could still be produced, but would now be generated in the interior of the cell wall, hidden from eosin Y binding.
Cryptococcus has seven chitin synthase (Chs) genes, but Chs3 is primarily responsible for the production of the chitin from which chitosan is derived. An alternative explanation for the loss of eosin Y staining is that Chs3 localization may be altered in kre5Δ and kre6Δskn1Δ, possibly trapped in the membrane-bound "chitosomes" which contain chitin synthase activity (reviewed in Bartnicki-Garcia, 2006). Chitosomes are proposed sites of chitin microfibril synthesis, but trafficking of these vesicles is not understood. It is also not known whether the Cdas co-localize to these structures. The potential existence of secretory pathway defects in kre5Δ and kre6Δskn1Δ could affect trafficking of Chs3 via chitosomes and hence prevent the chitin that is deacetylated to chitosan from reaching the cell wall. Investigation of the localization and interaction of the Cdas and Chs3 in wild type cells compared to kre5Δ and kre6Δskn1Δ will provide further insight into the mechanism of chitosan production in C. neoformans.
KRE genes are important for maintaining capsule architecture
We have shown that disruption of KRE5 or KRE6 and SKN1 in combination affects the size and architecture of the exopolysaccharide capsule, a primary C. neoformans virulence factor. The capsules of the kre5Δ and kre6Δskn1Δ cells are enlarged, appear diffuse and are more permeable to TMR-labeled dextrans than the capsules of wild type cells. There are several possible explanations for these data, which are not mutually exclusive. First, Yoneda and Doering (2006) demonstrated that the capsular polysaccharide building block, GXM, is synthesized in vesicles within the cell and secreted at the cell surface, where it is attached to the cell wall via an unknown mechanism. The appearance of enlarged intracellular vesicular structures within the kre5Δ and kre6Δskn1Δ cells, and the proposed localization of KRE5 to the endoplasmic reticulum (Meaden et al., 1990) and KRE6 and SKN1 to the Golgi (Roemer et al., 1994) indicates potential disruption of the secretory pathway. Such disruption could account for alterations in capsule formation. It has also been shown that cell wall α-1,3-glucan is required for capsule attachment to the C. neoformans cell surface, although it is unknown whether this interaction is direct (Reese and Doering, 2003). It is possible that the diffuse appearance of the kre5Δ and kre6Δskn1Δ capsule could result from decreased levels or altered distribution of α-1,3-glucan throughout the cell wall. Another study has implicated β-1,4-N-acetylglucosamine (GlcNAc) oligomers, which include both chitin and chitosan in C. neoformans, in the association of capsule with the cell wall (Rodrigues et al. 2008). Furthermore, mutants with decreased cell wall chitosan, particularly chs3Δ and cda1Δcda2Δcda3Δ, have enlarged capsules. The apparent alteration in chitosan localization in both kre5Δ and kre6Δskn1Δ, indicated by the loss of eosin Y staining, is consistent with the idea that chitosan may play an important role in attachment or assembly of the capsule.
Loss of β-1,6-glucan results in secretion of GPI-anchored cell wall proteins, including Plb1
Another important cryptococcal virulence factor is Plb1. Previous studies have demonstrated that Plb1 is GPI-anchored and localized both in the plasma membrane and covalently linked to β-1,6-glucan in the cell wall (Siafakas et al., 2006; Siafakas et al., 2007). In S. cerevisiae, the majority of cell wall proteins are attached to β-1,6-glucan via a remnant of their GPI-anchor (Kapteyn et al., 1996; Yin et al., 2005). We hypothesized that the loss of β-1,6-glucan in kre5Δ and kre6Δskn1Δ would result in decreased Plb1 retention in the cell wall and increased Plb1 secretion. Our results demonstrate that kre5Δ and kre6Δskn1Δ have decreased Plb1 protein and activity associated with the cell wall, with a concomitant increase in the secretion of PLB activity and Plb1 protein (Fig. 9). Some PLB activity was still released from mutant cell walls by treatment with β-1,3-glucanse. This enzyme is unlikely to have been non-covalently associated with the cell wall, as extensive washing with saline was performed prior to treatment with β-1,3-glucanse. More likely, it represents enzyme that is covalently-associated to a different cell wall polysaccharide as a compensatory mechanism for the absence of β-1,6-glucan. Such altered cross-linking of cell wall mannoproteins to cell wall polysaccharides has been observed in mutant strains with aberrant cell walls in S. cerevisiae (Kapteyn et al., 1997). The low PLB activity in the crude membrane preparations is most likely due to the sequestration of the Plb1 protein within the raft portion of these membranes, which were found previously to create a suppressive environment for PLB activity (Siafakas et al., 2006).
In summary, we have identified three genes, KRE5, KRE6 and SKN1 involved in β-1,6-glucan synthesis in C. neoformans. Our data demonstrate an important role for these genes and β-1,6-glucan in cell wall integrity, capsular architecture, virulence and the retention of cell wall mannoproteins. They are also important for cell wall organization, particularly chitosan distribution. This is the first investigation of β-1,6-glucan in C. neoformans. These findings provide the framework for future investigations into cell wall protein attachment as well as the mechanism of chitosan production in this important fungal pathogen.
EXPERIMENTAL PROCEDURES
Strains and media
The C. neoformans serotype A clinical isolate H99 was used as the wild type strain. C. neoformans was grown on YPD (1% yeast, 2% bacto-peptone, 2% dextrose), with solid media containing 2% bacto-agar. Selective YPD contained 200 units ml−1 hygromycin (Calbiochem, San Diego), 100 mg ml−1 nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) or 200 µg ml−1 gentamicin (G418) (Invitrogen, Carlsbad, CA).
Identification of β-1,6-glucan synthesis-related protein sequences
The sequences of the S. cerevisiae Kre5p, Kre6p, Skn1p, Kre1p, Kre9p, Kre11p and Knh1p were used as a query sequences in BLASTp searches of the predicted proteins from the C. neoformans H99 genome (http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans/MultiHome.html). Seven proteins with an E-value < 1E−6 were identified. These were used in a reciprocal BLASTp analysis of the S. cerevisiae predicted proteins. The proposed name of the predicted C. neoformans protein was based on the reciprocal BLASTp analysis as well as the clustering observed in the phylogenetic analysis. The suggested names for the C. neoformans KRE6 genes are based on the level of similarity to the known S. cerevisiae genes. For example, CNAG_00897 is most similar to the the S. cerevisiae SKN1 gene and thus was designated SKN1 whereas the remaining C. neoformans genes had a higher level of similarity to the S. cerevisiae KRE6 gene and thus are designated KRE6, KRE61, KRE62, KRE63 and KRE64. It is accepted practice to append a number onto the gene name when multiple C. neoformans genes share the highest level of similarity to a single S. cerevisiae gene.
RNA extraction and poly (A) RNA purification
H99 was grown for 24 h at 30°C or 37°C in liquid YPD medium and total RNA isolated as described (Baker et al., 2007). All samples had an OD 260/280 ratio of 1.8–2.0. Poly(A) RNA was purified from the total RNA sample by using an oligo-Tex RNA purification kit (QIAGEN, Valencia, CA) following the manufacturer's specifications.
Northern hybridizations
Analysis of mRNA levels by Northern blot was performed as described (Baker et al., 2007). The procedure used was adapted from that of Sambrook and Russell (2001). All gene-specific DNA probes were generated using H99 cDNA as template. Probes were designed to span an intron, to ensure specificity for RNA, not genomic DNA. Primers 17 and 18 were used to amplify the KRE5 probe while the remaining probes were amplified with primers 13 and 14 (Table S1).
Generation of deletion strains
All constructs used to generate single deletion mutants were made using overlap PCR gene deletion technology (Davidson et al., 2002) and were biolistically transformed (Toffaletti et al., 1993; Hua et al., 2000) into C. neoformans H99 to generate single deletion strains as described (Baker et al., 2007). Primer sequences are listed in Table S2. Each deletion construct included a hygromycin resistance cassette (Hua et al., 2000) for selection. Transformants that grew on selective media were passaged 5x on YPD then screened for homologous recombination at both the 5’ and 3’ end using PCR (Nelson et al., 2003) and a single site of integration was confirmed by Southern blot analysis as described (Baker et al., 2007). Isolates recovered from separate transformation plates were considered independent and used when complementation could not be acheived. The kre6Δskn1Δ and kre6Δkre61Δ double deletion strains were generated by biolistic transformation of a KRE6 deletion construct containing a nourseothricin cassette (McDade and Cox, 2001) into the skn1Δ or kre61Δ backgrounds, respectively.
Generation of complemented deletion strain
The kre6Δ deletion strain was reconstituted using a method described previously (Goins et al., 2006). A PCR product containing the wild-type KRE6 coding region plus 1000 bp of the upstream and 500 bp of the downstream untranslated regions was amplified from H99 genomic DNA using primers KRE6-1 and KRE6-2 listed in Table S1. This linear fragment along with a second, unlinked PCR product containing the G418 resistance cassette, were was biolistically co-transformed into kre6Δ. The transformants were screened by PCR for the presence of KRE6 at the wildtype locus and for the absence of the deletion cassette. The same method was utilized without success to attempt to reconstitue kre5Δ with KRE5 and kre6Δskn1Δ with KRE6.
Isolation of cell wall material
Alkali-soluble cell wall material was prepared from H99 and the deletion strains using a modification of the procedures described by Maneesri et al. (2005) and Page et al. (2003). Strains were grown in 25 ml YPD at 30°C overnight with shaking. Cells were pelleted by centrifugation (650 g for 10 min), washed with 5 ml sterile water and resuspended in 0.5–1.0 ml sterile water. Cells were added to 1 volume glass beads (425–600 micron) and then lysed using a mini bead beater. Samples were beaten five times for 30 sec at 5K rpm, with intermittent incubation on ice. Lysate was removed from beads and protein content determined using the QuickStart Bradford assay kit (BioRad) with BSA as a standard. Volumes of lysate containing 8 µg protein were alkali-extracted with 50 µL 1.5N NaOH (total volume 100 µL) at 75°C for 1h. Samples were centrifuged 5 min at 11 000 g and the supernatants containing alkali-soluble glucans were removed. The alkali-insoluble pellets were resuspended in 50 µL of 100 mM K2PO4 buffer, pH 6.0 containing 0.2 U chitinase from Serratia marcescens (C1650; Sigma) and incubated at 37°C for 72h.
Dot blot assay
Serial two-fold dilutions of alkali-soluble material or the alkali-insoluble, chitinase-digested material were spotted onto nitrocellulose membranes. Spots were allowed to dry completely, and then the membrane was blocked with 10 ml TBST (25 mM Tris-HCl, pH 8.1, 0.15 M NaCl, 0.1% Tween-20) containing 5% non-fat milk powder for 1h at room temperature. Membranes were washed 3x with TBS (25 mM Tris-HCl, pH 8.1, 0.15 M NaCl) and then probed with rabbit polyclonal anti-β-1,6-glucan antiserum (1:1000) overnight at 4°C. Membranes were washed as before and incubated with HRP-conjugated goat anti-rabbit secondary antibody for 1h at room temperature. Secondary antibody was detected with ECL reagent and signal analyzed by autoradiography.
For competition analysis, H99 alkali-soluble cell wall material was spotted onto membrane, as described above. The primary rabbit polyclonal anti-β-1,6-glucan antiserum working solution was preincubated with 5 µg/ml purified pustulan from Umbilicaria pustulata (400507; CarboMer, Inc) laminarin from Laminaria digitata (61430; Fluka), or mannan from Saccaromyces cerevisiae (M7504; Sigma) for 30 min with shaking prior to probing the blot as described above.
Fluorescence microscopy
For assessing morphology, cells were streaked on YPD plates and grown on YPD agar plates for 2 days at 30 °C. A loop of cells was then were resuspended in 180 µl PBS, and 20 µl of a 1:1,000 dilution of pontamine in PBS solution (Fast Scarlet 4B; Bayer Corp.) was added. Incubation was carried out and incubated at room temperature in the dark for 10 min. For assessing cell wall chitosan, strains were grown in 25 ml of YPD broth at 30°C for 24, 48 and 72 hours with shaking. At each time pointthe given timepoints, 0.5 ml of each culture was pelleted by centrifugation and washed with 1 ml McIlvaine’s buffer [0.2 M Na2HPO4 and 0.1 M citric acid pH 6.0]. Cells were then resuspended in 0.5 ml McIlvaine’s buffer containing with 30 µl eosin Y (5 mg ml−1 stock; Sigma, St. Louis, MO), incubated at room temperature in the dark for 10 minutes and washed twice, as before. Cells were examined with an Olympus Vanox AHBT3 microscope at 1000× magnification. Excitation wavelengths were 488 nm and 548 nm for eosin Y and pontamine, respectively.
Cell wall inhibitor assays
Strains were grown in 2 ml of YPD broth overnight at 30°C with shaking and then diluted to 1 × 107 cells ml−1. Tenfold serial dilutions were made and 5 µl of each were plated onto solid YPD agarmedium without and with the designated amounts of calcofluor white (Fluorescent Brightner 28, Sigma F-3543), caffeine (Sigma, C-0750), SDS (AnalaR Biochemical, BDH Chemicals, Poole, UK) NaCl, sorbitol and Congo red (Sigma, C-6767). Plates were incubated at 30°C or 37°C and photographed after 2 and 7 days.
Antifungal testing
Etest strips (AB Biodisk) containing a gradient of fluconazole, voriconazole or caspofungin were used. Strains were grown in YPD overnight at 30°C, diluted to an OD650 of 0.1 and streaked in triplicate on RPMI-1640 (Sigma, R6504) agar to create even lawns. Antifungal strips were placed on the center of each plate and incubated at 30°C for 72h. MIC values were determined from two independent experiments according to the manufacturer’s instructions.
Analysis of capsule production and architecture
Strains were streaked on YPD plates and grown on YPD agar plates for 2 days at 30°C. Cells from these plates were subcultured onto DME plates [13.4 g l−1 Dulbecco’s modified Eagle’s medium, 25mM MOPS pH 7.0, 1.8% agar] and incubated for 3 days at 30°C with 5% CO2 to induce capsule production. Cells were resuspended in a 1:4 India ink:H2O solution to visualize the capsule and observed through an Olympus AHBT3 microscope at 1000× magnification. Capsule radii were measured from digital images using Olympus Microsuite 5 software.
Capsule permeability assays using tetramethyl-rhodamine (TMR) conjugated dextrans were performed as described by Gates et al. (2004). Cells were diluted to 8×106 cells mL−1 in PBS and 25 µL of each suspension was added to an equal volume of 10 kDa, 40 kDa or 70 kDa TMR-dextran solution (2 mg mL−1 in PBS). Samples were incubated in the dark at room temperature for 60 min and then observed through an OlympusVanox AHB2 microscope at 1000× magnification. Digital images were captured with an Olympus FVII digital camera and processed and analyzed with Olympus Microsuite 5 software. Since the kre5Δ and kre6Δskn1Δ cells and capsules were enlarged compared to wild type, we modified the analysis to determine the “penetration zone,” which is a measure of how far the TMR-dextrans diffused into the capsule (see Fig, 7B). Aliquots of the starting cultures were stained with India ink to measure capsule radius. The radius of exclusion of each cell was determined by subtracting the diameter of the cell (measured on the bright field image) from the diameter of the exclusion zone (measured on the fluorescent image) and dividing by 2. The radius of penetration is defined as: average capsule radius (measured by India ink staining)/radius of exclusion.
Analysis of melanin production
Strains were grown and diluted as per for the cell wall inhibitor assays, plated on L-DOPA-containing agar plates medium (13 mM glycine, 15 mM glucose, 29.4 mM KH2PO4, 10mM MgSO4*7H2O, 3.0 µM thiamine, 5.0 µM D-biotin, 1.0 mM L-DOPA, 2% bacto-agar) and assayed for melanin production by noting the color of the cells that grew after 2–7 days at 30°C.
Pooled mutant STM virulence analysis
Virulence of the single deletion strains was analyzed by signature tagged mutagenesis using a mouse tail vein model of infection, as described previously (Nelson et al., 2001).
Inhalation mouse model
Cryptococcus neoformans strains were grown at 30°C with shaking for 2 nights in YPD broth. The cells were centrifuged, washed, and resuspended in endotoxin-free PBS. The cells were counted on a hemocytometer and diluted to 2×107 cells/ml. CBA/J female mice (Jackson Laboratories) were anesthetized and allowed to inhale 1×106 (50 µl) cells that were dripped into the nares. Mice were weighed before and during the course of infection and sacrificed by CO2 asphyxiation once their body weight had decreased to 80% of the pre-infection body weight. At this point, the mice showed signs of being morbidly ill, including a ruffled coat, lethargy, a hunched posture, unstable gait, and loss of appetite.
Phospholipase B1 analysis
Cryptococcal secretions and subcellular fractions were prepared essentially as described (Siafakas et al., 2007) with minor modifications. Cryptococcal cells, grown as SAB agar lawns for 72 h, were harvested, washed with saline and resuspended in secretion buffer (10 mM Immidazole pH 5.0, 2% glucose) as a concentrated suspension (approximately 1 ml buffer : 5 ml packed cell volume). Incubation was carried out overnight at 30°C. Cells were pelleted by centrifugation and the secretions collected. Cell pellets were washed once with saline and once with MES-buffered saline (MBS; 25 mM MES, 150 mM NaCl, 2 mM EDTA [pH 6.5]) and frozen at −80°C. Following thawing on ice, pellets were resuspended in 5 ml of pre-chilled MBS containing 0.1% [vol/vol] Triton X-100 [TX100] and a fungal-specific protease inhibitor cocktail [Sigma P8215], to achieve a 1:1 (buffer: packed cell volume) suspension, and 1 ml aliquots of the suspension were transferred to prechilled, 2 ml screw-capped eppendorf tubes containing 1 ml of 0.5 mm zirconia/silica beads (Biospec Products, Inc 11079105z). Suspensions were homogenized using a MiniBeadbeater-8 cell disrupter (Daintree Scientific) for three cycles of 1 min, with alternating 1-min cooling periods on ice and the cell lysates were transferred to a fresh prechilled eppendorf tube. The beads were washed with 0.5 ml MBS containing 0.1% Triton X-100 [TX100] and protease inhibitors, and the wash was combined with the lysate to maximise protein recovery. Cell lysates were centrifuged at 3,500 × g for 10 min at 4°C and the supernatants collected and set aside on ice. The 3,500 × g cell wall-containing pellets were washed three times with 30 ml saline and resuspended in 1.5–2.0 ml of β-1,3-glucanase lysing enzyme (Sigma L1412; 20 mg/ml made up in water and protease inhibitors). Incubation was carried out for 1 h with agitation at 37°C and the supernatant, containing released Plb1 and defined as the cell wall-fraction, was collected by centrifugation (14,000 × g for 15 min at 4°C). The 3,500 × g supernatants were centrifuged at 135,000 × g (45,000 rpm) for 1 h at 4°C in a Beckman bench-top ultracentrifuge fitted with a TLA45 rotor, to separate the pellet (crude membranes) and the supernatant (cytosol). Crude membranes were resuspended in 200 µl of ice-cold MBS containing protease inhibitors, probe sonicated for 3 to 4 s to create a suspension. Five µl of (G)PIPLC (B. cereus, 0.5–1 Units) was added to release GPI anchored proteins by incubation at 37°C for 1 h. The released GPI anchored proteins including Plb1 (supernatant), were collected by ultracentrifugation and are representative of the membrane-associated Plb1 fraction (Siafakas et al., 2007). The Plb1 content of all fractions was analyzed by western blotting with anti Plb1 peptide antibodies or by radiometric enzyme assay, where 1 unit=amount of enzyme hydrolyzing 1 nmol substrate per min, as previously described (Chen et al., 1997).
Cellular chitin and chitosan content assays
The chitin and chitosan content of cells was measured as described previously (Baker et al., 2007). Samples were divided into two aliquots. One aliquot was treated with acetic anhydride to measure chitin plus chitosan, and the second aliquot remained untreated to measure chitin. The difference between the two measurements was an estimate of the amount of chitosan.
Supplementary Material
Suppl. Fig. 1. Alkali-soluble cell wall -1,6-glucan dot blot analysis of H99 and deletion strains. Cells were grown at 30°C to mid-log phase and alkali-soluble polysaccharides were isolated and spotted in 1:2 serial dilutions onto nitrocellulose membrane. Membranes were probed with polyclonal anti-β-1,6-glucan antiserum.
Suppl. Fig. 2. Morphology of C. neoformans H99 and deletion strains. Cells were grown on YPD medium at 30°C for 72h and stained with Pontamine FastScarlet. All images are at 1000X magnification.
Suppl. Fig. 3. Sensitivity of mutants to temperature and cell wall inhibitors. Strains were grown overnight in YPD medium then diluted to an OD650 of 1.0. Tenfold serial dilutions were made in PBS and 5 µl of each was spotted on medium containing the indicated inhibitors. Plates were incubated at the indicated temperatures for 2 days.
ACKNOWLEDGEMENTS
We would like to thank Piet de Groot for supplying the polyclonal anti-β-1,6-glucan antiserum and Christina Hull for communication regarding serotype D microarrays. We also thank Leona Campbell and Woei Lam for helpful suggestions. This work was supported by National Institute of Health grants RO1AI072195 and RO1AI50184 to JKL and an American Heart Association predoctoral fellowship to NMG.
REFERENCES
- Aimanianda V, Clavaud C, Simenel C, Fontaine T, Delepierre M, Latge JP. Cell Wall {beta}-(1,6)-Glucan of Saccharomyces cerevisiae: Structural characterization and in situ synthesis. J Biol Chem. 2009;284:13401–13412. doi: 10.1074/jbc.M807667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Ito E. A pathway of chitosan formation in Mucor rouxii. Enzymatic deacetylation of chitin. Eur J Biochem. 1975;55:71–78. doi: 10.1111/j.1432-1033.1975.tb02139.x. [DOI] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-Garcia S. Chitosomes: past, present and future. FEMS Yeast Res. 2006;6:957–965. doi: 10.1111/j.1567-1364.2006.00158.x. [DOI] [PubMed] [Google Scholar]
- Boisramé A, Gaillardin C. Heterologous expression and characterization of a beta-1,6-glucanase from Aspergillus fumigatus. Appl Microbiol Biotechnol. 2009;82:663–669. doi: 10.1007/s00253-008-1780-z. [DOI] [PubMed] [Google Scholar]
- Bulik DA, Olczak M, Lucero HA, Osmond BC, Robbins PW, Specht CA. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot Cell. 2003;2:886–900. doi: 10.1128/EC.2.5.886-900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DA, Horisberger M, Horman I, Wursch P. The Wall Structure of Schizosaccharomyces pombe. J Gen Microbiol. 1974;81:199–206. doi: 10.1099/00221287-81-1-199. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Rosas AL, Nosanchuk JD. Melanin and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 2000;3:354–358. doi: 10.1016/s1369-5274(00)00103-x. [DOI] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Muller M, Zhou JZ, Wright LC, Sorrell TC. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J Infec Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- Coe JGS, Wilson CF, Sorrell TC, Latouche NG, Wright LC. Cloning of CnLYSO1, a novel extracellular lysophospholipase of the pathogenic fungus Cryptococcus neoformans. Gene. 2003;316:67–78. doi: 10.1016/s0378-1119(03)00740-6. [DOI] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Pintor-Toro JA, Benitez T, Llobell A. Purification and characterization of an endo-beta-1,6-glucanase from Trichoderma harzianum that is related to its mycoparasitism. J. Bacteriol. 1995;177:1864–1871. doi: 10.1128/jb.177.7.1864-1871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology (Reading, Engl) 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Djordjevic JT, Del Poeta M, Sorrell TC, Turner KM, Wright LC. Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem J. 2005;389:803–812. doi: 10.1042/BJ20050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenheer RA, Lee YJ, Blumwald E, Phinney BS, Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 2007;7:499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- Gates MA, Thorkildson P, Kozel TR. Molecular architecture of the Cryptococcus neoformans capsule. Mol Microbiol. 2004;52:13–24. doi: 10.1111/j.1365-2958.2003.03957.x. [DOI] [PubMed] [Google Scholar]
- Goins CL, Gerik KJ, Lodge JK. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol. 2006;43:531–544. doi: 10.1016/j.fgb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- de Groot PWJ, Yin QY, Weig M, Sosinska GJ, Klis FM, de Koster CG. Mass spectrometric identification of covalently bound cell wall proteins from the fission yeast Schizosaccharomyces pombe. Yeast. 2007;24:267–278. doi: 10.1002/yea.1443. [DOI] [PubMed] [Google Scholar]
- Hartland RP, Vermeulen CA, Klis FM, Sietsma JH, Wessels JG. The linkage of (1–3)-beta-glucan to chitin during cell wall assembly in Saccharomyces cerevisiae. Yeast. 1994;10:1591–1599. doi: 10.1002/yea.320101208. [DOI] [PubMed] [Google Scholar]
- Herrero AB, Magnelli P, Mansour MK, Levitz SM, Bussey H, Abeijon C. KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot Cell. 2004;3:1423–1432. doi: 10.1128/EC.3.6.1423-1432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch HC, Galvani CD, Szarowski DH, Turner JN. Two new fluorescent dye applicable for visualization of fungal cell walls. Mycologia. 2005;97(no. 3):580–588. doi: 10.3852/mycologia.97.3.580. [DOI] [PubMed] [Google Scholar]
- Hochstenbach F, Klis FM, van den Ende H, van Donselaar E, Peters PJ, Klausner RD. Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc Nat Acad Sci USA. 1998;95:9161–9166. doi: 10.1073/pnas.95.16.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyer JD, Lodge JK. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin Diagn Lab Immunol. 2000;7:125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. Cell-wall glucans of Cryptococcus neoformans Cap 67. Carbohydr Res. 1990;198:23–38. doi: 10.1016/0008-6215(90)84273-w. [DOI] [PubMed] [Google Scholar]
- Kapteyn JC, Montijn RC, Vink E, de la Cruz J, Llobell A, Douwes JE, et al. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiesterlinked beta-1,3-/beta-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- Kapteyn JC, Ram AF, Groos EM, Kollar R, Montijn RC, Van Den Ende H, et al. Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. JBacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Someya K, Hata M, Nakajima R, Takemura M. Discovery of a Small-Molecule Inhibitor of β-1,6-Glucan Synthesis. Antimicrob Agen Chemo. 2009;53:670–677. doi: 10.1128/AAC.00844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM, De Groot P, Hellingwerf K. Molecular organization of the cell wall of Candida albicans. Med Mycol. 2001;39 Suppl 1:1–8. [PubMed] [Google Scholar]
- Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Lesage G, Shapiro J, Specht CA, Sdicu A, Ménard P, Hussein S, et al. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Specht CA. The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 2006;6:513–524. doi: 10.1111/j.1567-1364.2006.00071.x. [DOI] [PubMed] [Google Scholar]
- López-Sáez JF, Mingo R, González-Fernández A. ATP level and caffeine efficiency on cytokinesis inhibition in plants. Eur J Cell Biol. 1982;27:185–190. [PubMed] [Google Scholar]
- Maneesri J, Azuma M, Sakai Y, Igarashi K, Matsumoto T, Fukuda H, et al. Deletion of MCD 4 involved in glycosylphosphatidylinositol (GPI) anchor synthesis leads to an increase in beta-1,6-glucan level and a decrease in GPI-anchored protein and mannan levels in the cell wall of Saccharomyces cerevisiae. J Biosci Bioeng. 2005;99:354–360. doi: 10.1263/jbb.99.354. [DOI] [PubMed] [Google Scholar]
- McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Meaden P, Hill K, Wagne rJ, Slipetz D, Sommer SS, Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1----6)-beta- D-glucan synthesis and normal cell growth. Mol Cell Biol. 1990;10:3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missal TA, Moran JM, Corbett JA, Lodge JK. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot Cell. 2005;4:202–208. doi: 10.1128/EC.4.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RT, Huaa J, Pryor BA, Lodge JK. Identification of Virulence Mutants of the Fungal Pathogen Cryptococcus neoformans Using Signature-Tagged Mutagenesis. Genetics. 2001;157:935–947. doi: 10.1093/genetics/157.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RT, Pryor BA, Lodge JK. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet Bio. 2003;38:1–9. doi: 10.1016/s1087-1845(02)00510-8. [DOI] [PubMed] [Google Scholar]
- Pagé N, Gérard-Vincent M, Ménard P, Beaulieu M, Azuma M, Dijkgraaf GJP, et al. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics. 2003;163:875–894. doi: 10.1093/genetics/163.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck I, Hearing VJ, Kwon-Chung KJ. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. J Bacteriol. 1982;150:1212–1220. doi: 10.1128/jb.150.3.1212-1220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AJ, Doering TL. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 2003;50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- Reese AJ, Yoneda A, Breger JA, Beauvais A, Liu H, Griffith CL, et al. Loss of cell wall alpha(1–3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol Microbiol. 2007;63:1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Alvarez M, Fonseca FL, Casadevall A. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell. 2008;7:602–609. doi: 10.1128/EC.00307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci U.S.A. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Delaney S, Bussey H. SKN1 and KRE6 define a pair of functional homologs encoding putative membrane proteins involved in beta-glucan synthesis. Mol Cell Biol. 1993;13:4039–4048. doi: 10.1128/mcb.13.7.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Paravicini G, Payton MA, Bussey H. Characterization of the yeast (1-->6)-beta-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C, Durán A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herreraa J, Ortiz-Castellanosa L, Martínezb AI, León-Ramíreza C, Sentandreub R. Analysis of the proteins involved in the structure and synthesis of the cell wall of Ustilago maydis. Fungal Genet Biol. 2008;45:S71–S76. doi: 10.1016/j.fgb.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shahinian S, Bussey H. beta-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:477–489. doi: 10.1046/j.1365-2958.2000.01713.x. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Durán A, et al. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shematek EM, Braatz JA, Cabib E. Biosynthesis of the yeast cell wall. I. Preparation and properties of beta-(1 leads to 3)glucan synthetase. J Biol Chem. 1980;255:888–894. [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, et al. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem. 2007;282:37508–37514. doi: 10.1074/jbc.M707913200. [DOI] [PubMed] [Google Scholar]
- Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sietsma JH, Wessels JG. The occurrence of glucosaminoglycan in the wall of Schizosaccharomyces pombe. J Gen Microbiol. 1990;136:2261–2265. doi: 10.1099/00221287-136-11-2261. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J of Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LC, Payne J, Santangelo RT, Simpanya MF, Chen SCA, Widmer F, et al. Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii. Biochem J. 2004;384:377–384. doi: 10.1042/BJ20041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LC, Santangelo RM, Ganendren R, Payne J, Djordjevic JT, Sorrell TC. Cryptococcal lipid metabolism: phospholipase B1 is implicated in transcellular metabolism of macrophage-derived lipids. Eukaryot Cell. 2007;6:37–47. doi: 10.1128/EC.00262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QY, de Groot PWJ, Dekker HL, de Jong L, Klis FM, de Koster CG. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkalisensitive linkages. J Biol Chem. 2005;280:20894–20901. doi: 10.1074/jbc.M500334200. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Schekman RW. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol Biol Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1. Alkali-soluble cell wall -1,6-glucan dot blot analysis of H99 and deletion strains. Cells were grown at 30°C to mid-log phase and alkali-soluble polysaccharides were isolated and spotted in 1:2 serial dilutions onto nitrocellulose membrane. Membranes were probed with polyclonal anti-β-1,6-glucan antiserum.
Suppl. Fig. 2. Morphology of C. neoformans H99 and deletion strains. Cells were grown on YPD medium at 30°C for 72h and stained with Pontamine FastScarlet. All images are at 1000X magnification.
Suppl. Fig. 3. Sensitivity of mutants to temperature and cell wall inhibitors. Strains were grown overnight in YPD medium then diluted to an OD650 of 1.0. Tenfold serial dilutions were made in PBS and 5 µl of each was spotted on medium containing the indicated inhibitors. Plates were incubated at the indicated temperatures for 2 days.