Abstract
In the title compound, C13H11NO3S, the two aromatic rings are oriented at an angle of 88.18 (8)°. Intramolecular N—H⋯O and C—H⋯O hydrogen bonds are observed, each of which generates an S(6) ring motif. In the crystal, molecules are linked into C(7) chains along [010] by intermolecular C—H⋯O hydrogen bonds. The structure is further stabilized by intermolecular C—H⋯π interactions involving the sulfonyl-bound phenyl ring.
Related literature
For the biological activity of sulfonamides, see: Zareef et al. (2007 ▶); Chohan et al. (2007 ▶); Brown (1971 ▶); Pomarnacka & Kozlarska-Kedra (2003 ▶); Sethu Sankar et al. (2002 ▶). For related structures, see: Bassindale (1984 ▶); Cotton & Stokley (1970 ▶); Usha et al. (2005 ▶); Zhu et al. (2008 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).
Experimental
Crystal data
C13H11NO3S
M r = 261.29
Triclinic,
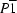
a = 7.7656 (2) Å
b = 9.0080 (2) Å
c = 9.5855 (2) Å
α = 86.293 (1)°
β = 77.912 (1)°
γ = 68.826 (1)°
V = 611.35 (2) Å3
Z = 2
Mo Kα radiation
μ = 0.26 mm−1
T = 293 K
0.21 × 0.19 × 0.17 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.768, T max = 0.956
15490 measured reflections
3960 independent reflections
3228 reflections with I > 2σ(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.119
S = 1.02
3960 reflections
167 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.31 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809032681/ci2869sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809032681/ci2869Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O3 | 0.80 (2) | 1.99 (2) | 2.6751 (19) | 144 (2) |
| C2—H2⋯O1 | 0.93 | 2.46 | 3.0879 (18) | 125 |
| C3—H3⋯O2i | 0.93 | 2.56 | 3.2691 (19) | 133 |
| C5—H5⋯Cg1ii | 0.93 | 2.80 | 3.700 (2) | 162 |
Symmetry codes: (i)  ; (ii)
; (ii)  . Cg1 is the centroid of the C8–C13 ring.
. Cg1 is the centroid of the C8–C13 ring.
Acknowledgments
ST and AS thank Dr Babu Varghese, SAIF, IIT-Madras, Chennai, India, for his help with the data collection.
supplementary crystallographic information
Comment
Sulfonamides have been recognized for their wide variety of pharmacological activities such as antibacterial, antitumor, anti-carbonic anhydrase, diuretic, hypoglycaemic, antithyroid and protease inhibitory activities. Sulfonamides particularly sulfadiazine and sulfadoxine have been used clinically as antimalarial agents (Zareef et al., 2007). Due to their significant pharmacological applications and widespread use in medicine, these compounds have also gained attention in bioinorganic and metal-based drug chemistry (Chohan et al., 2007). Sulfonamide derivates are well known drugs and are used to control diseases caused by bacterial infections. The antibacterial action of this group of drugs is exerted by the complete inhibition of dihydropteroate synthase enzyme towards the p-amino benzonate (Brown, 1971). Benzene sulfonamide derivatives are known to exhibit anticancer and HIV activities (Pomarnacka & Kozlarska-Kedra, 2003). The sulfonamides inhibit the growth of bacterial organism and are also useful for treating urinary and gastrointestinal infections (Sethu Sankar et al., 2002). In view of this medicinal importance, the crystal structure determination of the title compound (Fig.1) was carried out and the results are presented here.
Atom S1 has a distorted tetrahedral configuration. The widening of angle O1—S1—O2 [119.76 (7)°] and narrowing of angle C8—S1—N1 [106.08 (6)°] from the ideal tetrahedral value are attributed to the Thorpe-Ingold effect (Bassindale, 1984). The two aromatic rings are oriented at an angle of 88.18 (8)°. The angles around atom S1 deviate significantly from the regular tetrahedral value, with the largest deviation of 119.7 (7)° for O1—S1—O2 angle. This may be due to non-bonding interactions between S—O bonds (Cotton & Stokley, 1970). The aldyhyde group is coplanar with the benzene ring, as evidenced by the torsion angle C1—C6—C7—O3 of -2.2 (3)°. The geometrical parameters agree well with those reported for related sulfonamide structures (Usha et al., 2005; Zhu et al., 2008). Intramolecular N1—H1···O3 and C2—H2···O1 hydrogen bonds are observed, each of which generates an S(6) ring motif (Bernstein et al., 1995).
Intermolecular C—H···O hydrogen bonds involving atoms C3 and O2 link molecules into C(7) chains running along the [010] (Fig. 2). The crystal packing is further stabilized by C—H···π interactions involving the sulfonyl-bound phenyl ring.
Experimental
N-[2-(Hydroxymethyl)phenyl]benzenesulfonamide (2 g, 7.6 mmol) was added to a solution of pyridinium chlorochromate (3.25 g, 15.11 mmol) in dry dichloromethane (20 ml) at room temperature and the reaction mixture was stirred for 4 h. The solvent was removed to obtain a crude aldehyde as a white solid. Single crystals of the title compound suitable for X-ray diffraction were obtained by slow evaporation of a methanol solution.
Refinement
Atom H1 was located in a difference map and refined freely. All other H atoms were positioned geometrically and constrained to ride on their parent atoms, with C-H = 0.93 Å and Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% probability displacement ellipsoids and the atomic numbering scheme. Hydrogen bonds are shown as dashed lines.
Fig. 2.
Part of a C(7) chain in the title compound.
Crystal data
| C13H11NO3S | Z = 2 |
| Mr = 261.29 | F(000) = 272 |
| Triclinic, P1 | Dx = 1.419 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.7656 (2) Å | Cell parameters from 3960 reflections |
| b = 9.0080 (2) Å | θ = 2.2–31.3° |
| c = 9.5855 (2) Å | µ = 0.26 mm−1 |
| α = 86.293 (1)° | T = 293 K |
| β = 77.912 (1)° | Block, colourless |
| γ = 68.826 (1)° | 0.21 × 0.19 × 0.17 mm |
| V = 611.35 (2) Å3 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 3960 independent reflections |
| Radiation source: fine-focus sealed tube | 3228 reflections with I > 2σ(I) |
| graphite | Rint = 0.022 |
| ω scans | θmax = 31.3°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −11→11 |
| Tmin = 0.768, Tmax = 0.956 | k = −13→13 |
| 15490 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.119 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0582P)2 + 0.1353P] where P = (Fo2 + 2Fc2)/3 |
| 3960 reflections | (Δ/σ)max = 0.001 |
| 167 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.19429 (17) | 0.98956 (14) | 0.61966 (13) | 0.0409 (2) | |
| C2 | 0.1595 (2) | 1.10035 (15) | 0.72614 (15) | 0.0470 (3) | |
| H2 | 0.1207 | 1.0779 | 0.8210 | 0.056* | |
| C3 | 0.1827 (2) | 1.24378 (17) | 0.69056 (18) | 0.0563 (3) | |
| H3 | 0.1595 | 1.3170 | 0.7626 | 0.068* | |
| C4 | 0.2392 (3) | 1.2813 (2) | 0.5513 (2) | 0.0695 (5) | |
| H4 | 0.2547 | 1.3783 | 0.5292 | 0.083* | |
| C5 | 0.2722 (3) | 1.1734 (2) | 0.44619 (19) | 0.0675 (4) | |
| H5 | 0.3085 | 1.1987 | 0.3517 | 0.081* | |
| C6 | 0.2529 (2) | 1.02670 (18) | 0.47720 (15) | 0.0503 (3) | |
| C7 | 0.2926 (3) | 0.9189 (3) | 0.35857 (18) | 0.0692 (5) | |
| H7 | 0.3264 | 0.9554 | 0.2677 | 0.083* | |
| C8 | 0.36455 (18) | 0.70690 (14) | 0.85278 (13) | 0.0413 (3) | |
| C9 | 0.5123 (2) | 0.57064 (16) | 0.79634 (16) | 0.0519 (3) | |
| H9 | 0.4963 | 0.5055 | 0.7325 | 0.062* | |
| C10 | 0.6827 (2) | 0.5337 (2) | 0.83658 (19) | 0.0628 (4) | |
| H10 | 0.7829 | 0.4425 | 0.7999 | 0.075* | |
| C11 | 0.7062 (2) | 0.6304 (2) | 0.93052 (19) | 0.0621 (4) | |
| H11 | 0.8225 | 0.6047 | 0.9563 | 0.074* | |
| C12 | 0.5587 (2) | 0.7649 (2) | 0.98675 (18) | 0.0599 (4) | |
| H12 | 0.5758 | 0.8296 | 1.0503 | 0.072* | |
| C13 | 0.3853 (2) | 0.80416 (16) | 0.94909 (15) | 0.0487 (3) | |
| H13 | 0.2847 | 0.8941 | 0.9877 | 0.058* | |
| N1 | 0.16942 (19) | 0.84378 (14) | 0.64708 (14) | 0.0509 (3) | |
| O1 | 0.00479 (14) | 0.85915 (13) | 0.90061 (12) | 0.0576 (3) | |
| O2 | 0.13222 (17) | 0.60899 (13) | 0.76162 (14) | 0.0649 (3) | |
| O3 | 0.2861 (2) | 0.78653 (18) | 0.36620 (14) | 0.0795 (4) | |
| S1 | 0.14948 (5) | 0.75289 (4) | 0.79852 (4) | 0.04575 (11) | |
| H1 | 0.199 (3) | 0.790 (2) | 0.5767 (19) | 0.064 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0388 (6) | 0.0369 (5) | 0.0461 (6) | −0.0096 (4) | −0.0123 (5) | −0.0046 (5) |
| C2 | 0.0562 (7) | 0.0365 (6) | 0.0482 (7) | −0.0149 (5) | −0.0113 (6) | −0.0055 (5) |
| C3 | 0.0638 (9) | 0.0377 (6) | 0.0699 (9) | −0.0173 (6) | −0.0180 (7) | −0.0056 (6) |
| C4 | 0.0839 (12) | 0.0498 (8) | 0.0803 (11) | −0.0310 (8) | −0.0182 (10) | 0.0112 (8) |
| C5 | 0.0769 (11) | 0.0679 (10) | 0.0581 (9) | −0.0294 (9) | −0.0116 (8) | 0.0132 (8) |
| C6 | 0.0475 (7) | 0.0533 (8) | 0.0471 (7) | −0.0125 (6) | −0.0117 (6) | −0.0036 (6) |
| C7 | 0.0711 (10) | 0.0802 (12) | 0.0485 (8) | −0.0171 (9) | −0.0081 (7) | −0.0160 (8) |
| C8 | 0.0431 (6) | 0.0316 (5) | 0.0466 (6) | −0.0155 (4) | 0.0004 (5) | 0.0003 (4) |
| C9 | 0.0505 (7) | 0.0380 (6) | 0.0589 (8) | −0.0127 (5) | 0.0027 (6) | −0.0044 (5) |
| C10 | 0.0474 (7) | 0.0506 (8) | 0.0725 (10) | −0.0056 (6) | 0.0036 (7) | 0.0057 (7) |
| C11 | 0.0481 (7) | 0.0680 (10) | 0.0680 (9) | −0.0209 (7) | −0.0118 (7) | 0.0184 (8) |
| C12 | 0.0638 (9) | 0.0618 (9) | 0.0604 (9) | −0.0276 (7) | −0.0175 (7) | 0.0038 (7) |
| C13 | 0.0518 (7) | 0.0414 (6) | 0.0511 (7) | −0.0160 (5) | −0.0062 (6) | −0.0039 (5) |
| N1 | 0.0662 (7) | 0.0391 (5) | 0.0508 (6) | −0.0199 (5) | −0.0134 (5) | −0.0108 (5) |
| O1 | 0.0440 (5) | 0.0534 (6) | 0.0689 (7) | −0.0164 (4) | 0.0051 (5) | −0.0129 (5) |
| O2 | 0.0657 (7) | 0.0436 (5) | 0.0938 (8) | −0.0315 (5) | −0.0089 (6) | −0.0106 (5) |
| O3 | 0.0860 (9) | 0.0802 (9) | 0.0672 (8) | −0.0192 (7) | −0.0129 (7) | −0.0333 (7) |
| S1 | 0.04451 (18) | 0.03489 (16) | 0.0591 (2) | −0.01869 (12) | −0.00239 (14) | −0.00769 (12) |
Geometric parameters (Å, °)
| C1—C2 | 1.3900 (17) | C8—C9 | 1.3871 (18) |
| C1—N1 | 1.3957 (17) | C8—S1 | 1.7515 (14) |
| C1—C6 | 1.4052 (19) | C9—C10 | 1.375 (2) |
| C2—C3 | 1.3797 (19) | C9—H9 | 0.93 |
| C2—H2 | 0.93 | C10—C11 | 1.374 (3) |
| C3—C4 | 1.375 (2) | C10—H10 | 0.93 |
| C3—H3 | 0.93 | C11—C12 | 1.376 (2) |
| C4—C5 | 1.365 (3) | C11—H11 | 0.93 |
| C4—H4 | 0.93 | C12—C13 | 1.383 (2) |
| C5—C6 | 1.390 (2) | C12—H12 | 0.93 |
| C5—H5 | 0.93 | C13—H13 | 0.93 |
| C6—C7 | 1.453 (2) | N1—S1 | 1.6265 (13) |
| C7—O3 | 1.208 (2) | N1—H1 | 0.798 (18) |
| C7—H7 | 0.93 | O1—S1 | 1.4212 (10) |
| C8—C13 | 1.3838 (18) | O2—S1 | 1.4241 (10) |
| C2—C1—N1 | 123.02 (12) | C10—C9—C8 | 118.75 (15) |
| C2—C1—C6 | 118.86 (12) | C10—C9—H9 | 120.6 |
| N1—C1—C6 | 118.10 (12) | C8—C9—H9 | 120.6 |
| C3—C2—C1 | 119.78 (14) | C11—C10—C9 | 120.54 (15) |
| C3—C2—H2 | 120.1 | C11—C10—H10 | 119.7 |
| C1—C2—H2 | 120.1 | C9—C10—H10 | 119.7 |
| C4—C3—C2 | 121.69 (15) | C10—C11—C12 | 120.40 (15) |
| C4—C3—H3 | 119.2 | C10—C11—H11 | 119.8 |
| C2—C3—H3 | 119.2 | C12—C11—H11 | 119.8 |
| C5—C4—C3 | 118.78 (15) | C11—C12—C13 | 120.29 (16) |
| C5—C4—H4 | 120.6 | C11—C12—H12 | 119.9 |
| C3—C4—H4 | 120.6 | C13—C12—H12 | 119.9 |
| C4—C5—C6 | 121.51 (16) | C12—C13—C8 | 118.66 (14) |
| C4—C5—H5 | 119.2 | C12—C13—H13 | 120.7 |
| C6—C5—H5 | 119.2 | C8—C13—H13 | 120.7 |
| C5—C6—C1 | 119.37 (14) | C1—N1—S1 | 128.49 (9) |
| C5—C6—C7 | 117.69 (16) | C1—N1—H1 | 112.3 (14) |
| C1—C6—C7 | 122.94 (15) | S1—N1—H1 | 116.5 (13) |
| O3—C7—C6 | 126.49 (17) | O1—S1—O2 | 119.76 (7) |
| O3—C7—H7 | 116.8 | O1—S1—N1 | 109.04 (7) |
| C6—C7—H7 | 116.8 | O2—S1—N1 | 103.70 (7) |
| C13—C8—C9 | 121.35 (13) | O1—S1—C8 | 108.57 (7) |
| C13—C8—S1 | 120.66 (10) | O2—S1—C8 | 108.83 (7) |
| C9—C8—S1 | 117.98 (11) | N1—S1—C8 | 106.08 (6) |
| N1—C1—C2—C3 | −178.37 (13) | C9—C10—C11—C12 | 0.6 (2) |
| C6—C1—C2—C3 | 0.1 (2) | C10—C11—C12—C13 | −0.1 (2) |
| C1—C2—C3—C4 | 0.2 (2) | C11—C12—C13—C8 | −0.9 (2) |
| C2—C3—C4—C5 | 0.3 (3) | C9—C8—C13—C12 | 1.3 (2) |
| C3—C4—C5—C6 | −1.1 (3) | S1—C8—C13—C12 | −178.76 (11) |
| C4—C5—C6—C1 | 1.4 (3) | C2—C1—N1—S1 | −16.6 (2) |
| C4—C5—C6—C7 | −179.47 (18) | C6—C1—N1—S1 | 164.92 (11) |
| C2—C1—C6—C5 | −0.8 (2) | C1—N1—S1—O1 | 52.78 (14) |
| N1—C1—C6—C5 | 177.67 (14) | C1—N1—S1—O2 | −178.56 (12) |
| C2—C1—C6—C7 | −179.97 (14) | C1—N1—S1—C8 | −63.97 (14) |
| N1—C1—C6—C7 | −1.4 (2) | C13—C8—S1—O1 | −19.79 (13) |
| C5—C6—C7—O3 | 178.68 (18) | C9—C8—S1—O1 | 160.20 (10) |
| C1—C6—C7—O3 | −2.2 (3) | C13—C8—S1—O2 | −151.70 (11) |
| C13—C8—C9—C10 | −0.7 (2) | C9—C8—S1—O2 | 28.29 (13) |
| S1—C8—C9—C10 | 179.30 (11) | C13—C8—S1—N1 | 97.27 (11) |
| C8—C9—C10—C11 | −0.2 (2) | C9—C8—S1—N1 | −82.73 (11) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O3 | 0.80 (2) | 1.99 (2) | 2.6751 (19) | 144 (2) |
| C2—H2···O1 | 0.93 | 2.46 | 3.0879 (18) | 125 |
| C3—H3···O2i | 0.93 | 2.56 | 3.2691 (19) | 133 |
| C5—H5···Cg1ii | 0.93 | 2.80 | 3.700 (2) | 162 |
Symmetry codes: (i) x, y+1, z; (ii) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI2869).
References
- Bassindale, A. (1984). The Third Dimension in Organic Chemistry, ch. 1, p. 11. New York: John Wiley and Sons.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Brown, G. M. (1971). Adv. Biochem.35, 35–40.
- Bruker (2004). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chohan, Z. H. & Shad, H. A. (2007). J. Enz. Inhib. Med. Chem.23, 369–379. [DOI] [PubMed]
- Cotton, F. A. & Stokley, P. F. (1970). J. Chem. Soc.92, 294–302.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Pomarnacka, E. & Kozlarska-Kedra, I. (2003). Farmaco, 58, 423–429. [DOI] [PubMed]
- Sethu Sankar, K., Kannadasan, S., Velmurugan, D., Srinivasan, P. C., Shanmuga Sundara Raj, S. & Fun, H.-K. (2002). Acta Cryst. C58, o277–o279. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Gottingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Usha, G., Selvanayagam, S., Velmurugan, D., Ravikumar, K., Jaisankar, P. & Srinivasan, P. C. (2005). Acta Cryst. E61, o1916–o1918.
- Zareef, M., Iqbal, R., De Dominguez, N. G., Rodrigues, J., Zaidi, J. H., Arfan, M. & Supuran, C. T. (2007). J. Enz. Inhib. Med. Chem.22, 301–308. [DOI] [PubMed]
- Zhu, H.-Y., Wu, Z. & Jiang, S. (2008). Acta Cryst. E64, o596. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809032681/ci2869sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809032681/ci2869Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




