Abstract
Two enantiomeric counterparts (l- and d-asparginium cations related by glide planes) are present in the structure of the title compound, C4H9N2O3 +·ClO4 −, with a 1:1 cation–anion ratio. The structure is built up from asparginium cations and perchlorate anions. In the crystal, molecules assemble in double layers parallel to (100) through N—H⋯O, O—H⋯O and C—H⋯O hydrogen bonds. In the asparginium layers, hydrogen bonds generate alternating R 2 2(8) and R 4 3(18) graph-set motifs. Further hydrogen bonds involving the anions and cations result in the formation of a three-dimensional network.
Related literature
For the use of dl-asparagine in growth-media for bacteria, see: Gerhardt & Wilson (1948 ▶); Palleroni et al. (1973 ▶); van Wagtendonk et al. (1963 ▶). For related structures, see: Aarthy et al. (2005 ▶); Anitha et al. (2005 ▶); Bendjeddou et al. (2009 ▶); Verbist et al. (1972 ▶); Wang et al. (1985 ▶); Yamada et al. (2007 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).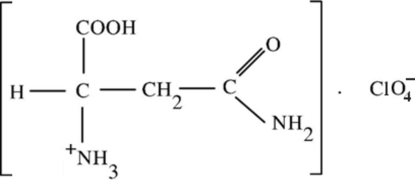
Experimental
Crystal data
C4H9N2O3 +·ClO4 −
M r = 232.58
Orthorhombic,
a = 9.861 (5) Å
b = 10.289 (4) Å
c = 16.700 (5) Å
V = 1694.4 (12) Å3
Z = 8
Mo Kα radiation
μ = 0.47 mm−1
T = 100 K
0.09 × 0.04 × 0.02 mm
Data collection
Oxford Diffraction Xcalibur Saphire2 CCD diffractometer
Absorption correction: none
45509 measured reflections
2818 independent reflections
2205 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.034
wR(F 2) = 0.100
S = 1.12
2818 reflections
127 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.68 e Å−3
Δρmin = −0.38 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2008 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2008 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SIR92 (Altomare et al., 1993 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶), PARST97 (Nardelli, 1995 ▶) and Mercury (Macrae et al., 2006 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809033534/at2865sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809033534/at2865Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O3i | 0.82 | 1.76 | 2.5485 (19) | 161 |
| N1—H1A⋯O4ii | 0.89 | 2.02 | 2.837 (2) | 152 |
| N1—H1B⋯O5iii | 0.89 | 2.03 | 2.910 (2) | 171 |
| N1—H1C⋯O3 | 0.89 | 2.30 | 2.886 (2) | 123 |
| N1—H1C⋯O5 | 0.89 | 2.16 | 2.907 (2) | 142 |
| N2—H4N⋯O2iv | 0.84 | 2.54 | 3.341 (2) | 159 |
| N2—H5N⋯O2v | 0.84 | 2.57 | 3.362 (2) | 157 |
| N2—H5N⋯O5v | 0.84 | 2.55 | 3.089 (2) | 123 |
| C2—H2⋯O7vi | 0.98 | 2.44 | 3.201 (2) | 134 |
| C3—H3A⋯O4ii | 0.97 | 2.58 | 3.326 (2) | 134 |
| C3—H3B⋯O2v | 0.97 | 2.41 | 3.253 (2) | 145 |
Symmetry codes: (i) 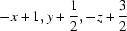 ; (ii)
; (ii) 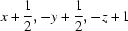 ; (iii)
; (iii) 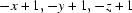 ; (iv)
; (iv) 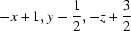 ; (v)
; (v) 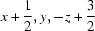 ; (vi)
; (vi) 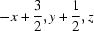 .
.
Acknowledgments
Technical support (X-ray measurements at SCDRX) from Université Henry Poincaré, Nancy 1 is gratefully acknowledged.
supplementary crystallographic information
Comment
The asparagine is one of twenty natural amino acids the most common land. DL-asparagine has been used in growth-media for bacteria-growth such as Brucellae (Gerhardt & Wilson, 1948), Pseudomonas fluorescens (Palleroni et al., 1973) and lambda particles (Wagtendonk et al., 1963).
The crystal structure of L-asparagine (Yamada et al., 2007), L-asparagine monohydrate, (Verbist et al., 1972), L-asparagine-L-aspartic acid monohydrate (Wang et al., 1985), L-asparaginium nitrate (Aarthy et al., 2005) and L-asparaginium picrate (Anitha et al., 2005) have been solved. In this paper, the crystal structure information of DL-asparaginium perchlorate at 100 K was undertaken.
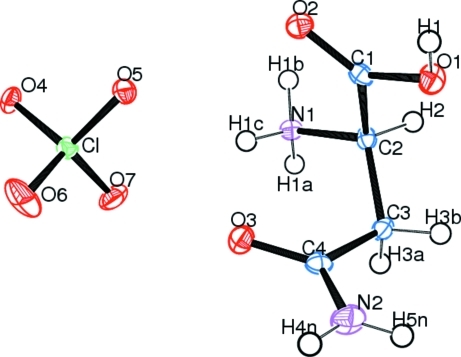
The asymmetric unit of (I) (Fig. 1) is formed by a monoprotonated asparaginium cation and a perchlorate anion. A proton transfer from the perchloric acid to atom N(1) of aspargine resulted in the formation of salts. This protonation lead to the different C—O bond distances [1.2179 (18)Å and 1.3086 (18) Å] and bond angle [126.42 (13)°] of the carboxyl group. This type of protonation is observed in various aspargine acid complexes (Anitha et al., 2005; Aarthy, et al., 2005).
The average Cl—O bond distances and O—Cl—O bond angles of the perchlorate anion are 1.4434%A and 109.47 °, respectively, confirming a tetrahedral configuration, similar to other perchlorate studied at low temperature (Bendjeddou et al., 2009).
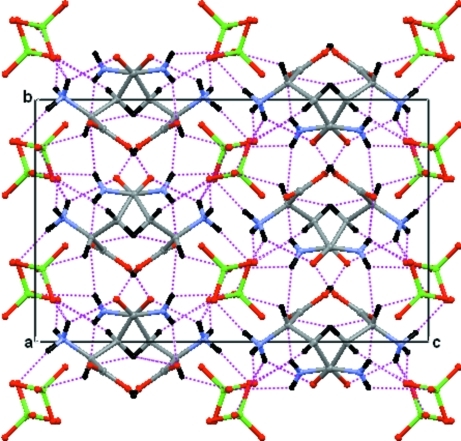
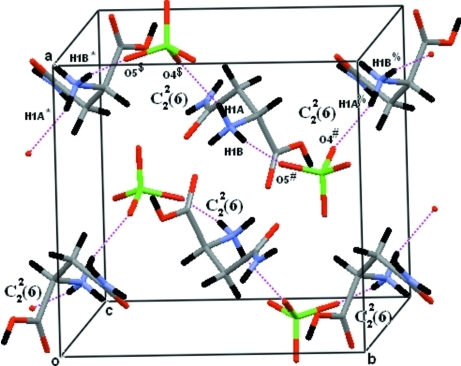
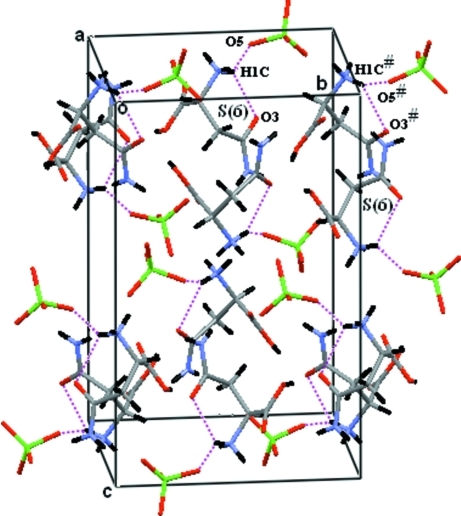
In (I), the ions are connected via N—H···O, O—H···O and C—H···O hydrogen bonds (Table 1) into three-dimensional hydrogen bonded double layers which run parallel to the (100) plane (Fig. 2). All ammonium H atoms are involved in hydrogen bonds, with three different perchlorate ions, while two anions accepts one hydrogen bond. These Two interactions link the anions and cations in to zigzag infinite chains along the [010] direction, which can be described by the graph-set motif C22(6) (Bernstein et al., 1995) (Fig. 3). The third anion participate in two centred hydrogen bonds with O(5) atom to form a finite chaine D(4). An intramolecular hydrogen bond is also observed between the α -amino group and the γ-carbonyl group with the graph-set motif S(6) (Fig. 4).
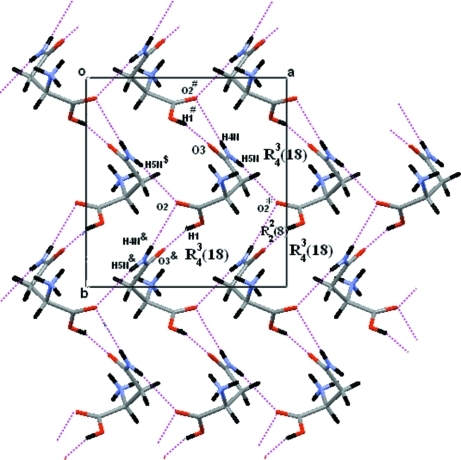
The carboxylic acid H and carbonyl O atoms participates respectively with a neighbouring cation through an O—H···O and N—H···O hydrogen bond. The combination of these hydrogen bonds generates an alternating noncentrosymmetric rings in two-dimensional network which can be described by the graph-set motif R22(8) and R43(18) (Fig. 5).
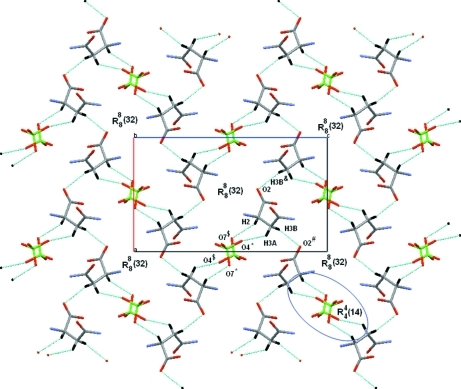
The junction between the cationic entities is consolidated by three weaks independent C—H···O hydrogen bonds via the perchlorate anions, forming an R88(32) and R44(14) centrosymmetric Rings in two-dimensional network (Fig. 6).
Experimental
The monocrystals of the compound DL-asparaginium perchlorate are obtained by slow evaporation at room temperature of an aqueous solution containing DL-asparagine monohydrate and the perchloric acid in a 1:1 stochiometric ratio. The solution was maintained in 293 K under agitation during twenty minutes.
Refinement
H atoms were positioned geometrically and refined in the riding-model approximation, with C—H = 0.98 Å (methine) or 0.97 Å (methylene), N—H = 0.89 Å (ammonium) or 0.84 Å (amine), O—H = 0.82 Å, and with Uiso(H) = 1.2Ueq(C, N) or 1.5Ueq(O).
Figures
Fig. 1.
The asymmetric unit of of DL-asparaginium perchlorate, showing the crystallographic numbering scheme. Displacement ellipsoids are drawn at the 50% probability level and H atoms are shown as spheres of arbitrary radii.
Fig. 2.
A packing diagram for the title compound, viewed down the a axis, showing the double layers. Dashed lines indicate N—H···O, O—H···O and C—H···O hydrogen bonds
Fig. 3.
Part of the crystal structure, showing the aggregation of C22(6) motif via N—H···O hydrogen bonds. Atoms marked with a hash symbol (#), dollar sign ($), a percent sign (%), or a star (*) are at the symmetry positions (1 - x, 1 - y, 1 - z), (1/2 + x,1/2 - y, 1 - z), (1.5 - x, 1/2 + y, z), (1.5 - x, -1/2 + y, z) respectively.
Fig. 4.
Part of the crystal structure, showing the formation of a finite chaine D(4) and S(6) rings. Atoms marked with an hash symbol (#), are at the symmetry position (x, 1.5 - y,-1/2 + z).
Fig. 5.
Part of the crystal structure, showing the formation of R22(8) and R43(18) rings. Atoms marked with an ampersand (&), a hash symbol (#), dollar sign ($), or a star (*) are at the symmetry positions (1 - x, 1/2 + y, 1.5 - z), (1 - x, -1/2 + y, 1.5 - z), (-1/2 + x, y, 1.5 - z), (1/2 + x, y, 1.5 - z), respectively.
Fig. 6.
Part of the crystal structure, showing the formation of R88(32) rings. Atoms marked with a star (*), dollar sign ($), an ampersand (&), a hash symbol (#) are at the symmetry positions (1/2 + x, 1/2 - y, 1 - z), (1.5 - x, 1/2 + y, z), (-1/2 + x, y, 1.5 - z), (1/2 + x, y, 1.5 - z) respectively.
Crystal data
| C4H9N2O3+·ClO4− | F(000) = 960 |
| Mr = 232.58 | Dx = 1.823 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2818 reflections |
| a = 9.861 (5) Å | θ = 3.1–31.5° |
| b = 10.289 (4) Å | µ = 0.47 mm−1 |
| c = 16.700 (5) Å | T = 100 K |
| V = 1694.4 (12) Å3 | Needle, colourless |
| Z = 8 | 0.09 × 0.04 × 0.02 mm |
Data collection
| Oxford Diffraction Xcalibur Saphire2 CCD diffractometer | 2205 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.033 |
| graphite | θmax = 31.5°, θmin = 3.1° |
| φ and ω scans | h = −14→14 |
| 45509 measured reflections | k = −15→11 |
| 2818 independent reflections | l = −24→24 |
Refinement
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.034 | w = 1/[σ2(Fo2) + (0.0538P)2 + 0.6998P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.100 | (Δ/σ)max = 0.001 |
| S = 1.12 | Δρmax = 0.68 e Å−3 |
| 2818 reflections | Δρmin = −0.38 e Å−3 |
| 127 parameters |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.58079 (10) | 0.69351 (10) | 0.73615 (6) | 0.0162 (3) | |
| O2 | 0.45528 (9) | 0.59815 (10) | 0.63971 (6) | 0.0139 (3) | |
| O3 | 0.61640 (11) | 0.32442 (10) | 0.70705 (6) | 0.0158 (3) | |
| N1 | 0.67333 (11) | 0.48834 (11) | 0.57093 (6) | 0.0115 (3) | |
| N2 | 0.71473 (14) | 0.37342 (13) | 0.82581 (7) | 0.0204 (4) | |
| C1 | 0.56293 (14) | 0.62144 (13) | 0.67245 (8) | 0.0117 (3) | |
| C2 | 0.69857 (13) | 0.56919 (13) | 0.64328 (8) | 0.0111 (3) | |
| C3 | 0.77716 (13) | 0.49753 (13) | 0.70849 (8) | 0.0126 (3) | |
| C4 | 0.69625 (14) | 0.39103 (13) | 0.74834 (8) | 0.0127 (3) | |
| Cl | 0.47309 (3) | 0.19831 (3) | 0.51836 (2) | 0.0127 (1) | |
| O4 | 0.40552 (10) | 0.17287 (10) | 0.44286 (6) | 0.0158 (3) | |
| O5 | 0.43440 (10) | 0.32809 (10) | 0.54535 (6) | 0.0145 (3) | |
| O6 | 0.43348 (15) | 0.10450 (12) | 0.57652 (7) | 0.0315 (4) | |
| O7 | 0.61761 (11) | 0.19650 (11) | 0.50614 (7) | 0.0226 (3) | |
| H1 | 0.50733 | 0.72120 | 0.75158 | 0.0243* | |
| H1A | 0.75164 | 0.45681 | 0.55290 | 0.0172* | |
| H1B | 0.63499 | 0.53694 | 0.53310 | 0.0172* | |
| H1C | 0.61828 | 0.42291 | 0.58350 | 0.0172* | |
| H2 | 0.75352 | 0.64382 | 0.62663 | 0.0133* | |
| H3A | 0.85826 | 0.46000 | 0.68513 | 0.0151* | |
| H3B | 0.80548 | 0.55964 | 0.74887 | 0.0151* | |
| H4N | 0.67437 | 0.31063 | 0.84737 | 0.0246* | |
| H5N | 0.77523 | 0.41807 | 0.84813 | 0.0246* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0144 (5) | 0.0187 (5) | 0.0156 (5) | 0.0014 (4) | 0.0022 (4) | −0.0059 (4) |
| O2 | 0.0116 (4) | 0.0155 (5) | 0.0146 (5) | 0.0008 (3) | 0.0014 (3) | 0.0001 (4) |
| O3 | 0.0162 (5) | 0.0177 (5) | 0.0135 (4) | −0.0048 (4) | −0.0012 (4) | 0.0035 (4) |
| N1 | 0.0113 (5) | 0.0127 (5) | 0.0104 (5) | 0.0015 (4) | 0.0010 (4) | 0.0002 (4) |
| N2 | 0.0253 (7) | 0.0250 (7) | 0.0110 (5) | 0.0020 (5) | −0.0013 (4) | 0.0023 (5) |
| C1 | 0.0134 (6) | 0.0092 (5) | 0.0125 (6) | −0.0007 (4) | 0.0028 (4) | 0.0024 (5) |
| C2 | 0.0104 (5) | 0.0108 (5) | 0.0120 (5) | −0.0010 (4) | 0.0013 (4) | −0.0014 (4) |
| C3 | 0.0102 (5) | 0.0141 (6) | 0.0135 (6) | −0.0005 (4) | −0.0017 (4) | −0.0010 (5) |
| C4 | 0.0128 (5) | 0.0133 (6) | 0.0119 (6) | 0.0045 (5) | 0.0010 (4) | −0.0002 (5) |
| Cl | 0.0164 (2) | 0.0106 (2) | 0.0112 (2) | −0.0003 (1) | −0.0021 (1) | 0.0004 (1) |
| O4 | 0.0155 (5) | 0.0178 (5) | 0.0142 (5) | −0.0034 (4) | −0.0043 (4) | −0.0027 (4) |
| O5 | 0.0165 (5) | 0.0125 (5) | 0.0145 (5) | 0.0006 (3) | −0.0008 (4) | −0.0023 (4) |
| O6 | 0.0588 (9) | 0.0177 (5) | 0.0179 (6) | −0.0056 (5) | 0.0043 (5) | 0.0082 (4) |
| O7 | 0.0145 (5) | 0.0267 (6) | 0.0265 (6) | 0.0061 (4) | −0.0080 (4) | −0.0079 (4) |
Geometric parameters (Å, °)
| Cl—O5 | 1.4601 (13) | N1—H1C | 0.8900 |
| Cl—O6 | 1.4239 (15) | N1—H1B | 0.8900 |
| Cl—O4 | 1.4499 (13) | N2—H4N | 0.8400 |
| Cl—O7 | 1.4398 (13) | N2—H5N | 0.8400 |
| O1—C1 | 1.3086 (18) | C1—C2 | 1.522 (2) |
| O2—C1 | 1.2179 (18) | C2—C3 | 1.527 (2) |
| O3—C4 | 1.2511 (18) | C3—C4 | 1.510 (2) |
| O1—H1 | 0.8200 | C2—H2 | 0.9800 |
| N1—C2 | 1.4879 (19) | C3—H3A | 0.9700 |
| N2—C4 | 1.3190 (19) | C3—H3B | 0.9700 |
| N1—H1A | 0.8900 | ||
| O5—Cl—O6 | 109.74 (7) | O1—C1—C2 | 110.01 (11) |
| O5—Cl—O7 | 108.32 (6) | O1—C1—O2 | 126.42 (13) |
| O6—Cl—O7 | 111.07 (8) | N1—C2—C3 | 113.22 (11) |
| O4—Cl—O5 | 108.27 (6) | N1—C2—C1 | 108.09 (10) |
| O4—Cl—O6 | 110.17 (7) | C1—C2—C3 | 112.86 (11) |
| O4—Cl—O7 | 109.23 (6) | C2—C3—C4 | 113.37 (11) |
| C1—O1—H1 | 109.00 | N2—C4—C3 | 117.32 (12) |
| C2—N1—H1A | 109.00 | O3—C4—N2 | 123.53 (13) |
| C2—N1—H1B | 109.00 | O3—C4—C3 | 119.16 (12) |
| H1B—N1—H1C | 109.00 | N1—C2—H2 | 107.00 |
| H1A—N1—H1C | 109.00 | C1—C2—H2 | 107.00 |
| C2—N1—H1C | 109.00 | C3—C2—H2 | 107.00 |
| H1A—N1—H1B | 109.00 | H3A—C3—H3B | 108.00 |
| C4—N2—H5N | 117.00 | C2—C3—H3A | 109.00 |
| H4N—N2—H5N | 125.00 | C2—C3—H3B | 109.00 |
| C4—N2—H4N | 117.00 | C4—C3—H3A | 109.00 |
| O2—C1—C2 | 123.57 (12) | C4—C3—H3B | 109.00 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O3i | 0.8200 | 1.7600 | 2.5485 (19) | 161.00 |
| N1—H1A···O4ii | 0.8900 | 2.0200 | 2.837 (2) | 152.00 |
| N1—H1B···O5iii | 0.8900 | 2.0300 | 2.910 (2) | 171.00 |
| N1—H1C···O3 | 0.8900 | 2.3000 | 2.886 (2) | 123.00 |
| N1—H1C···O5 | 0.8900 | 2.1600 | 2.907 (2) | 142.00 |
| N2—H4N···O2iv | 0.8400 | 2.5400 | 3.341 (2) | 159.00 |
| N2—H5N···O2v | 0.8400 | 2.5700 | 3.362 (2) | 157.00 |
| N2—H5N···O5v | 0.8400 | 2.5500 | 3.089 (2) | 123.00 |
| C2—H2···O7vi | 0.9800 | 2.4400 | 3.201 (2) | 134.00 |
| C3—H3A···O4ii | 0.9700 | 2.5800 | 3.326 (2) | 134.00 |
| C3—H3B···O2v | 0.9700 | 2.4100 | 3.253 (2) | 145.00 |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2; (ii) x+1/2, −y+1/2, −z+1; (iii) −x+1, −y+1, −z+1; (iv) −x+1, y−1/2, −z+3/2; (v) x+1/2, y, −z+3/2; (vi) −x+3/2, y+1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AT2865).
References
- Aarthy, A., Anitha, K., Athimoolam, S., Bahadur, S. A. & Rajaram, R. K. (2005). Acta Cryst. E61, o2042–o2044.
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst.26, 343–350.
- Anitha, K., Athimoolam, S. & Rajaram, R. K. (2005). Acta Cryst. E61, o1463–o1465.
- Bendjeddou, L., Cherouana, A., Hadjadj, N., Dahaoui, S. & Lecomte, C. (2009). Acta Cryst. E65, o1770–o1771. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Gerhardt, P. & Wilson, J. B. (1948). J. Bacteriol.56, 17–24. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Nardelli, M. (1995). J. Appl. Cryst.28, 659.
- Oxford Diffraction (2008). CrysAlis CCD and CrysAlis RED Oxford Diffraction, Wrocław, Poland.
- Palleroni, N. J., Kunisawa, R., Contopoulou, R. & Doudoroff, M. (1973). Int. J. Syst. Bacteriol.23, 333–339.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Verbist, J. J., Lehmann, M. S., Koetzle, T. F. & Hamilton, W. C. (1972). Acta Cryst. B28, 3006–3013.
- Wagtendonk, W. J. van, Clark, J. A. D. & Godoy, G. A. (1963). Proc. Natl. Acad. Sci. USA, 50, 835–838.
- Wang, J. L., Berkovitch-Yellin, Z. & Leiserowitz, L. (1985). Acta Cryst. B41, 341–348.
- Yamada, K., Hashizume, D., Shimizu, T. & Yokoyama, S. (2007). Acta Cryst. E63, o3802–o3803.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809033534/at2865sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809033534/at2865Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report