Abstract
In the title compound, C14H15NO6, the ethoxycarbonyl groups adopt extended conformations. In the crystal, molecules are linked into centrosymmetric dimers via pairs of C—H⋯O hydrogen bonds with a R 2 2(20) motif.
Related literature
For biological activity of nitrogen-containing building blocks derived from α-methylene-β-hydroxy esters, see: Singh & Batra (2008 ▶); Masson et al. (2007 ▶); Basavaiah et al. (2003 ▶); Youngme et al. (2007 ▶); Ma et al. (2005 ▶); Soldatov et al. (2003 ▶); Hinckley (1969 ▶).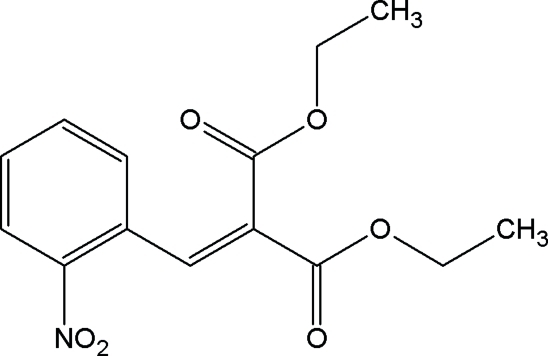
Experimental
Crystal data
C14H15NO6
M r = 293.27
Triclinic,

a = 7.8410 (2) Å
b = 8.5571 (2) Å
c = 12.3533 (4) Å
α = 80.866 (2)°
β = 75.037 (1)°
γ = 64.402 (1)°
V = 721.10 (3) Å3
Z = 2
Mo Kα radiation
μ = 0.11 mm−1
T = 293 K
0.21 × 0.19 × 0.17 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.978, T max = 0.982
19570 measured reflections
4226 independent reflections
3259 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.145
S = 1.06
4226 reflections
192 parameters
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.28 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809032668/bt5022sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809032668/bt5022Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O3i | 0.93 | 2.47 | 3.1683 (18) | 132 |
Symmetry code: (i) 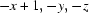 .
.
Acknowledgments
ST and ASP thank Dr Babu Varghese, SAIF, IIT, Chennai, India, for the X-ray data collection.
supplementary crystallographic information
Comment
Nitrogen-containing building blocks derived from a-methylene-β-hydroxy esters (Morita-Baylis-Hillman adducts) have been widely employed in modern organic chemistry for the synthesis of natural products and heterocycles of biological relevance (Singh & Batra, 2008; Masson et al., 2007; Basavaiah et al., 2003). β-Diketone as an excellent chelating group has been widely used in supramolecular chemistry. It can form a variety of complexes with various transition-metals (e.g. Cu, Co, Ni, Mn, Pd) or rare-earth metals (e.g. Eu, Sm, La, Gd) (Youngme et al., 2007; Ma et al., 2005). These metal complexes have significant applications in material science or act as chemical shift reagents (Soldatov et al., 2003; Hinckley, 1969). In view of this importance, the crystal structure determination of the title compound (Fig.1) has been carried out.
A perspective view of the title compound with the atom-numbering scheme is shown in Fig. 1. The deviations of the atoms N, O1 and O2 from the least-squares plane of the phenyl rings are -0.080 (1), 0.296 (2) and -0.527 (1) Å. The ethoxycarbonyl groups adopt extended conformation as can be seen from the torsion angles C8- C9- O6- C10 [175.6 (1)°], C9- O6- C10- C11 [-79.9 (2)°], C8- C12- O5- C13[178.6 (1)°] and C12- O5- C13- C14 [178.5 (1)°]. The C2–H2···O3 hydrogen bonds form a cyclic centrosymmetric dimer [R22(20)] shown in Fig.2.
Experimental
A mixture of 2-nitrobenzaldehyde (5 g, 33.11 mmol) in dry xylene (50 ml), ethylene diamine di acetate (1.2 g, 6.66 mmol) and diethyl malonate (6.36 g, 39.7 mmol) were added. The reaction was then refluxed for 12 h, it was then poured over ice - water (100 ml), extracted with CHCl3 (60 ml) and dried (Na2 SO4). The removal of solvent followed by recrystallization from methanol. Single crystals of the title compound suitable for X-ray diffraction were obtained by slow evaporation of a solution in methanol.
Refinement
All H atoms were fixed geometrically and allowed to ride on their parent C atoms, with C–H distances fixed in the range 0.93–0.97 Å with Uiso(H) = 1.5Ueq(C) for methyl H 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
View of the title molecule with the atom labeling scheme. The displacement ellipsoids are drawn at the 30% probability level while the H atoms are shown as small spheres of arbitrary radii.
Fig. 2.
The crystal structure showing the centrosymmetric hydrogen bond motif R22(20). For the sake of clarity, the H atoms not involved in the motif have been omitted. The atoms marked with an asterisk (*) are at the symmetry position (1 - x, -y, -z). The dashed lines indicate the hydrogen bonds.
Crystal data
| C14H15NO6 | Z = 2 |
| Mr = 293.27 | F(000) = 308 |
| Triclinic, P1 | Dx = 1.351 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.8410 (2) Å | Cell parameters from 4226 reflections |
| b = 8.5571 (2) Å | θ = 1.7–30.7° |
| c = 12.3533 (4) Å | µ = 0.11 mm−1 |
| α = 80.866 (2)° | T = 293 K |
| β = 75.037 (1)° | Block, colourless |
| γ = 64.402 (1)° | 0.21 × 0.19 × 0.17 mm |
| V = 721.10 (3) Å3 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 4226 independent reflections |
| Radiation source: fine-focus sealed tube | 3259 reflections with I > 2σ(I) |
| graphite | Rint = 0.025 |
| ω scans | θmax = 30.2°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −6→11 |
| Tmin = 0.978, Tmax = 0.982 | k = −11→12 |
| 19570 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.145 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0732P)2 + 0.1321P] where P = (Fo2 + 2Fc2)/3 |
| 4226 reflections | (Δ/σ)max < 0.001 |
| 192 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.32734 (17) | 0.26054 (16) | 0.00483 (10) | 0.0324 (3) | |
| C2 | 0.4208 (2) | 0.18112 (18) | −0.09512 (11) | 0.0393 (3) | |
| H2 | 0.3530 | 0.1563 | −0.1367 | 0.047* | |
| C3 | 0.6156 (2) | 0.1390 (2) | −0.13275 (12) | 0.0445 (3) | |
| H3 | 0.6808 | 0.0854 | −0.2002 | 0.053* | |
| C4 | 0.7138 (2) | 0.1765 (2) | −0.07003 (12) | 0.0444 (3) | |
| H4 | 0.8458 | 0.1469 | −0.0951 | 0.053* | |
| C5 | 0.61807 (19) | 0.25790 (18) | 0.02979 (11) | 0.0380 (3) | |
| H5 | 0.6867 | 0.2834 | 0.0704 | 0.046* | |
| C6 | 0.42104 (17) | 0.30233 (16) | 0.07069 (10) | 0.0307 (2) | |
| C7 | 0.32275 (18) | 0.40052 (17) | 0.17243 (11) | 0.0335 (3) | |
| H7 | 0.2096 | 0.4996 | 0.1694 | 0.040* | |
| C8 | 0.38277 (18) | 0.35895 (16) | 0.26840 (10) | 0.0322 (3) | |
| C9 | 0.54181 (19) | 0.18778 (16) | 0.28991 (10) | 0.0339 (3) | |
| C10 | 0.8427 (2) | 0.0493 (2) | 0.34618 (14) | 0.0517 (4) | |
| H10A | 0.9576 | 0.0709 | 0.3360 | 0.062* | |
| H10B | 0.8725 | −0.0413 | 0.2972 | 0.062* | |
| C11 | 0.7894 (3) | −0.0099 (2) | 0.46438 (15) | 0.0570 (4) | |
| H11A | 0.7533 | 0.0818 | 0.5126 | 0.086* | |
| H11B | 0.8980 | −0.1092 | 0.4840 | 0.086* | |
| H11C | 0.6824 | −0.0408 | 0.4730 | 0.086* | |
| C12 | 0.27995 (18) | 0.48151 (17) | 0.36030 (11) | 0.0350 (3) | |
| C13 | 0.2334 (2) | 0.51216 (19) | 0.55376 (11) | 0.0432 (3) | |
| H13A | 0.0935 | 0.5618 | 0.5646 | 0.052* | |
| H13B | 0.2752 | 0.6061 | 0.5419 | 0.052* | |
| C14 | 0.2947 (3) | 0.4002 (2) | 0.65347 (13) | 0.0581 (4) | |
| H14A | 0.2440 | 0.3131 | 0.6678 | 0.087* | |
| H14B | 0.2462 | 0.4697 | 0.7174 | 0.087* | |
| H14C | 0.4334 | 0.3452 | 0.6397 | 0.087* | |
| N | 0.12284 (17) | 0.29396 (16) | 0.04462 (12) | 0.0446 (3) | |
| O1 | 0.03150 (19) | 0.3042 (2) | −0.02475 (13) | 0.0753 (4) | |
| O2 | 0.05294 (17) | 0.30788 (19) | 0.14427 (11) | 0.0651 (4) | |
| O3 | 0.53708 (18) | 0.05173 (13) | 0.28408 (10) | 0.0536 (3) | |
| O4 | 0.17045 (18) | 0.63004 (14) | 0.34780 (9) | 0.0564 (3) | |
| O5 | 0.32173 (14) | 0.40462 (12) | 0.45770 (8) | 0.0394 (2) | |
| O6 | 0.68593 (13) | 0.20702 (12) | 0.31542 (9) | 0.0417 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0360 (6) | 0.0334 (6) | 0.0324 (6) | −0.0158 (5) | −0.0140 (5) | 0.0017 (5) |
| C2 | 0.0546 (8) | 0.0390 (7) | 0.0318 (6) | −0.0213 (6) | −0.0185 (6) | −0.0007 (5) |
| C3 | 0.0560 (8) | 0.0465 (8) | 0.0301 (7) | −0.0220 (7) | −0.0034 (6) | −0.0068 (6) |
| C4 | 0.0409 (7) | 0.0516 (8) | 0.0402 (7) | −0.0221 (6) | 0.0000 (6) | −0.0065 (6) |
| C5 | 0.0384 (6) | 0.0467 (7) | 0.0357 (7) | −0.0220 (6) | −0.0098 (5) | −0.0037 (6) |
| C6 | 0.0364 (6) | 0.0320 (6) | 0.0264 (6) | −0.0150 (5) | −0.0105 (4) | 0.0001 (4) |
| C7 | 0.0345 (6) | 0.0339 (6) | 0.0331 (6) | −0.0130 (5) | −0.0095 (5) | −0.0037 (5) |
| C8 | 0.0374 (6) | 0.0303 (6) | 0.0299 (6) | −0.0130 (5) | −0.0083 (5) | −0.0052 (5) |
| C9 | 0.0445 (6) | 0.0313 (6) | 0.0249 (6) | −0.0119 (5) | −0.0099 (5) | −0.0057 (4) |
| C10 | 0.0354 (7) | 0.0476 (8) | 0.0584 (10) | −0.0017 (6) | −0.0121 (6) | −0.0072 (7) |
| C11 | 0.0533 (9) | 0.0496 (9) | 0.0595 (10) | −0.0091 (7) | −0.0232 (7) | 0.0035 (8) |
| C12 | 0.0382 (6) | 0.0330 (6) | 0.0333 (6) | −0.0120 (5) | −0.0089 (5) | −0.0059 (5) |
| C13 | 0.0523 (8) | 0.0386 (7) | 0.0336 (7) | −0.0118 (6) | −0.0064 (6) | −0.0129 (6) |
| C14 | 0.0844 (12) | 0.0509 (9) | 0.0335 (8) | −0.0210 (8) | −0.0129 (8) | −0.0067 (7) |
| N | 0.0378 (6) | 0.0457 (7) | 0.0561 (8) | −0.0183 (5) | −0.0164 (5) | −0.0047 (6) |
| O1 | 0.0558 (7) | 0.1031 (11) | 0.0868 (10) | −0.0357 (7) | −0.0377 (7) | −0.0131 (8) |
| O2 | 0.0455 (6) | 0.0896 (10) | 0.0628 (8) | −0.0328 (6) | 0.0022 (5) | −0.0185 (7) |
| O3 | 0.0800 (8) | 0.0325 (5) | 0.0580 (7) | −0.0194 (5) | −0.0361 (6) | −0.0042 (5) |
| O4 | 0.0665 (7) | 0.0376 (6) | 0.0441 (6) | 0.0029 (5) | −0.0168 (5) | −0.0086 (5) |
| O5 | 0.0511 (5) | 0.0328 (5) | 0.0292 (5) | −0.0096 (4) | −0.0096 (4) | −0.0086 (4) |
| O6 | 0.0364 (5) | 0.0370 (5) | 0.0497 (6) | −0.0103 (4) | −0.0137 (4) | −0.0033 (4) |
Geometric parameters (Å, °)
| C1—C2 | 1.3756 (19) | C10—O6 | 1.4566 (17) |
| C1—C6 | 1.3991 (16) | C10—C11 | 1.482 (2) |
| C1—N | 1.4615 (17) | C10—H10A | 0.9700 |
| C2—C3 | 1.375 (2) | C10—H10B | 0.9700 |
| C2—H2 | 0.9300 | C11—H11A | 0.9600 |
| C3—C4 | 1.377 (2) | C11—H11B | 0.9600 |
| C3—H3 | 0.9300 | C11—H11C | 0.9600 |
| C4—C5 | 1.383 (2) | C12—O4 | 1.1998 (16) |
| C4—H4 | 0.9300 | C12—O5 | 1.3244 (16) |
| C5—C6 | 1.3917 (17) | C13—O5 | 1.4512 (16) |
| C5—H5 | 0.9300 | C13—C14 | 1.483 (2) |
| C6—C7 | 1.4680 (17) | C13—H13A | 0.9700 |
| C7—C8 | 1.3278 (17) | C13—H13B | 0.9700 |
| C7—H7 | 0.9300 | C14—H14A | 0.9600 |
| C8—C12 | 1.4878 (17) | C14—H14B | 0.9600 |
| C8—C9 | 1.4967 (17) | C14—H14C | 0.9600 |
| C9—O3 | 1.1947 (16) | N—O2 | 1.2132 (18) |
| C9—O6 | 1.3283 (16) | N—O1 | 1.2222 (17) |
| C2—C1—C6 | 123.07 (12) | O6—C10—H10B | 109.4 |
| C2—C1—N | 117.21 (11) | C11—C10—H10B | 109.4 |
| C6—C1—N | 119.66 (12) | H10A—C10—H10B | 108.0 |
| C3—C2—C1 | 119.15 (12) | C10—C11—H11A | 109.5 |
| C3—C2—H2 | 120.4 | C10—C11—H11B | 109.5 |
| C1—C2—H2 | 120.4 | H11A—C11—H11B | 109.5 |
| C4—C3—C2 | 119.66 (13) | C10—C11—H11C | 109.5 |
| C4—C3—H3 | 120.2 | H11A—C11—H11C | 109.5 |
| C2—C3—H3 | 120.2 | H11B—C11—H11C | 109.5 |
| C3—C4—C5 | 120.72 (13) | O4—C12—O5 | 124.51 (12) |
| C3—C4—H4 | 119.6 | O4—C12—C8 | 124.15 (12) |
| C5—C4—H4 | 119.6 | O5—C12—C8 | 111.33 (10) |
| C4—C5—C6 | 121.29 (12) | O5—C13—C14 | 107.61 (12) |
| C4—C5—H5 | 119.4 | O5—C13—H13A | 110.2 |
| C6—C5—H5 | 119.4 | C14—C13—H13A | 110.2 |
| C5—C6—C1 | 116.11 (11) | O5—C13—H13B | 110.2 |
| C5—C6—C7 | 119.28 (11) | C14—C13—H13B | 110.2 |
| C1—C6—C7 | 124.41 (11) | H13A—C13—H13B | 108.5 |
| C8—C7—C6 | 125.13 (11) | C13—C14—H14A | 109.5 |
| C8—C7—H7 | 117.4 | C13—C14—H14B | 109.5 |
| C6—C7—H7 | 117.4 | H14A—C14—H14B | 109.5 |
| C7—C8—C12 | 118.91 (11) | C13—C14—H14C | 109.5 |
| C7—C8—C9 | 122.12 (11) | H14A—C14—H14C | 109.5 |
| C12—C8—C9 | 118.84 (10) | H14B—C14—H14C | 109.5 |
| O3—C9—O6 | 125.05 (12) | O2—N—O1 | 123.34 (13) |
| O3—C9—C8 | 123.16 (12) | O2—N—C1 | 118.78 (11) |
| O6—C9—C8 | 111.79 (11) | O1—N—C1 | 117.87 (13) |
| O6—C10—C11 | 111.16 (12) | C12—O5—C13 | 116.56 (10) |
| O6—C10—H10A | 109.4 | C9—O6—C10 | 116.65 (11) |
| C11—C10—H10A | 109.4 | ||
| C6—C1—C2—C3 | 0.5 (2) | C7—C8—C9—O6 | 123.02 (13) |
| N—C1—C2—C3 | −176.61 (12) | C12—C8—C9—O6 | −61.15 (15) |
| C1—C2—C3—C4 | 0.1 (2) | C7—C8—C12—O4 | −15.0 (2) |
| C2—C3—C4—C5 | −0.7 (2) | C9—C8—C12—O4 | 169.01 (14) |
| C3—C4—C5—C6 | 0.8 (2) | C7—C8—C12—O5 | 163.66 (12) |
| C4—C5—C6—C1 | −0.28 (19) | C9—C8—C12—O5 | −12.30 (16) |
| C4—C5—C6—C7 | −175.29 (12) | C2—C1—N—O2 | 155.65 (14) |
| C2—C1—C6—C5 | −0.36 (19) | C6—C1—N—O2 | −21.53 (19) |
| N—C1—C6—C5 | 176.64 (11) | C2—C1—N—O1 | −23.42 (19) |
| C2—C1—C6—C7 | 174.37 (12) | C6—C1—N—O1 | 159.40 (14) |
| N—C1—C6—C7 | −8.63 (18) | O4—C12—O5—C13 | −2.7 (2) |
| C5—C6—C7—C8 | −50.59 (19) | C8—C12—O5—C13 | 178.64 (11) |
| C1—C6—C7—C8 | 134.84 (14) | C14—C13—O5—C12 | 178.49 (13) |
| C6—C7—C8—C12 | 173.32 (11) | O3—C9—O6—C10 | −4.6 (2) |
| C6—C7—C8—C9 | −10.8 (2) | C8—C9—O6—C10 | 175.61 (11) |
| C7—C8—C9—O3 | −56.78 (19) | C11—C10—O6—C9 | −79.96 (17) |
| C12—C8—C9—O3 | 119.05 (15) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O3i | 0.93 | 2.47 | 3.1683 (18) | 132 |
Symmetry codes: (i) −x+1, −y, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5022).
References
- Basavaiah, D., Rao, A. J. & Satyanarayana, T. (2003). Chem. Rev.103, 811–891. [DOI] [PubMed]
- Bruker (2004). SAINT and SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hinckley, C. C. (1969). J. Am. Chem. Soc.91, 5160–5162. [DOI] [PubMed]
- Ma, D.-Z., Wu, Y.-Q. & Zuo, X. (2005). Mater. Lett.59, 3678–3681.
- Masson, G., Housseman, C. & Zhu, J. (2007). Angew. Chem. Int. Ed.46, 4614–4628. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Gottingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Singh, V. & Batra, S. (2008). Tetrahedron, 64, 4511–4574.
- Soldatov, D. V., Tinnemans, P., Enright, G. D., Ratcliff, C. I., Diamente, P. R. & Ripmeester, J. A. (2003). Chem. Mater.15, 3826–3840.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Youngme, S., Chotkhun, T., Chaichit, N., van Albada, G. A. & Reedijk, J. (2007). Inorg. Chem. Commun.10, 843–848.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809032668/bt5022sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809032668/bt5022Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




