Abstract
An intramolecular Claisen-like cyclization of ethyl 2-acetoxy-4,4-dimethyl-1-(3-methylbut-2-enyl)cyclohex-2-enecarboxylate followed by dialkylation yielded the bicyclic title compound, C23H26O4. In both of the fused six-membered rings exist fragments of four atoms which are planar, whereas the remaining two atoms deviate by up to 0.682 (3) Å on one side of the plane of the ring containing an O atom and by up to 0.415 (3) Å on opposite sides of the other ring. The dihedral anglebetween the planar fragments of the six-membered rings is 41.76 (10)°
Related literature
For literature related to the synthesis, see: Ciochina & Grossman (2006 ▶).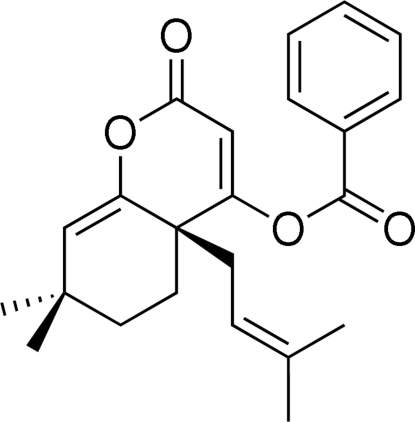
Experimental
Crystal data
C23H26O4
M r = 366.44
Triclinic,

a = 9.559 (4) Å
b = 10.201 (5) Å
c = 11.087 (5) Å
α = 69.23 (2)°
β = 83.649 (17)°
γ = 74.823 (17)°
V = 975.4 (8) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 173 K
0.45 × 0.40 × 0.20 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: none
12665 measured reflections
3547 independent reflections
1691 reflections with I > 2σ(I)
R int = 0.065
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.067
S = 0.81
3547 reflections
248 parameters
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.14 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL-Plus (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809030918/hg2544sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809030918/hg2544Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Experimental
Diethylether was dryed over sodium. All other solvents and reagents were commercially available and used as received. Flash-chromatography was done on silicagel 60 (230–400 mesh) using head pressure by means of compressed air. Infrared spectra (IR) were recorded as a thin film between KBr-plates. The instrument used was a Bruker IFS 66 F T—IR spectrophotometer. GC—MS spectra were recorded on a Finnigan Polaris GCQ spectrometer. Proton (1H NMR, 500 MHz) and carbon (13C NMR, 125 MHz) nuclear magnetic resonance spectra were recorded in chloroform(d-1) and referenced to the solvent signal. The instrument used was a Bruker DRX 500. The multiplicities of the signals are given as s (singlet), d (doublet), t (triplet), and m (multiplet).
HMDS (468 µL, 2.25 mmol) was dissolved in diethylether (3 ml) at 273 K. Butyllithium (1.3 ml, 2.03 mmol, 1.6 M in hexane) was added at that temperature and the mixture was stirred for 15 minutes. After cooling to 195 K a suspension of CuI (216 mg, 1.13 mmol) and ethyl 2-acetoxy-4,4-dimethyl-1-(3-methylbut-2-enyl)cyclohex-2-enecarboxylate (348 mg, 1.13 mmol) in diethylether (3 ml) was added. The mixture was stirred for 2 h. Benzoyl chloride (88 µl, 2.25 mmol) was added dropwise and the mixture was stirred for 5 days at room temperature. An aqueous Seignette salt-solution was added. Phase were separated and the aqueous layer was extracted with Et2O (3 x 10 ml). The combined organic layers were dried over Na2SO4 and concentrated in vacuum (Ciochina & Grossman, 2006). The crude product was purified via column chromatography (10:1 i-hexane/ethyl acetate). The product was obtained in 12% yield (50 mg, 0.136 mmol). The purified product was crystallized by slow evaporation of a mixture of diethylether and i-hexane.
Rf: 0.22 (i-hexane/ethyl acetate 10:1), mp: 387 K, 1H NMR (400 MHz, CDCl3): δ (p.p.m.) = 8.08–7.50 (m, 5H, CH), 6.33 (s, 1H, CH), 5.28 (s, 1H, CH), 5.21 (m, 1H, CH), 2.62 (dd, J = 13.9, 7.7 Hz, 1H, CH2), 2.43 (dd, J = 13.9, 8.5 Hz, 1H, CH2), 2.06 (dt, J = 13.4, 3.5 Hz, 1H, CH2), 1.72 (dt, J = 13.9, 3.0 Hz, 1H, CH2), 1.65 (s, 3H, CH3), 1.59 (s, 3H, CH3), 1.57–1.52 (m, 1H, CH2), 1.10 (s, 3H, CH3), 1.03 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3): δ (p.p.m.) = 168.7, 162.9, 162.4, 147.8, 136.8, 134.5, 130.4, 129.0, 120.3, 118.3, 105.1, 42.3, 36.7, 32.8, 31.9, 31.2, 26.5, 26.1. IR (film): ν (cm-1) = 2966 (m), 2945 (m), 2919 (m), 2863 (m), 1759 (s), 1736 (s), 1673 (m), 1636 (m), 1452 (m), 1366 (m), 1234 (s), 1145 (s), 1077 (s), 1065 (s), 1020 (m). MS (EI, 70 eV): m / z (%) = 366 (10) [M+], 351 (1) [C22H23O4+], 311 (1) [C19H19O4+], 261 (100) [C16H21O3+], 245 (5) [C16H21O2+], 219 (2) [C13H15O3+], 121 (1) [C7H5O2+], 105 (99) [C7H5O+], 77 (26) [C6H5+]. LRMS (FAB+LR,C23H26O4) calc. [(M+H)+]: 367.18; found: 367.02.
Refinement
H atoms were placed in calculated positions, with C—H = 0.95–0.99 Å and were refined as riding, with Uiso= 1.5Ueq for methyl and 1.2Ueq for others; the methyl were allowed to rotate but not to tip.
Figures
Fig. 1.
: The asymmetric unit of the title compound showing the labelling of all non-H atoms. Displacement ellipsoids are shown at the 30% probability level. Of the two disordered positions C35 and C35' only one is shown.
Crystal data
| C23H26O4 | Z = 2 |
| Mr = 366.44 | F(000) = 392 |
| Triclinic, P1 | Dx = 1.248 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.559 (4) Å | Cell parameters from 12656 reflections |
| b = 10.201 (5) Å | θ = 2.9–25.3° |
| c = 11.087 (5) Å | µ = 0.08 mm−1 |
| α = 69.23 (2)° | T = 173 K |
| β = 83.649 (17)° | Block, colourless |
| γ = 74.823 (17)° | 0.45 × 0.40 × 0.20 mm |
| V = 975.4 (8) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 1691 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.065 |
| graphite | θmax = 25.3°, θmin = 2.9° |
| Detector resolution: 19 vertical, 18 horizontal pixels mm-1 | h = −11→11 |
| 239 frames via ω–rotation (Δω=2°) and two times 20 s per frame (four sets at different κ–angles) scans | k = −11→12 |
| 12665 measured reflections | l = −13→13 |
| 3547 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.067 | H-atom parameters constrained |
| S = 0.81 | w = 1/[σ2(Fo2)] |
| 3547 reflections | (Δ/σ)max < 0.001 |
| 248 parameters | Δρmax = 0.17 e Å−3 |
| 0 restraints | Δρmin = −0.14 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.57097 (14) | 0.80118 (14) | −0.03317 (12) | 0.0520 (4) | |
| O2 | 0.46540 (13) | 0.63064 (14) | 0.09192 (11) | 0.0425 (4) | |
| O3 | 0.14946 (14) | 0.93092 (13) | 0.21014 (11) | 0.0436 (4) | |
| O4 | 0.08624 (14) | 1.10136 (14) | 0.01484 (13) | 0.0592 (4) | |
| C1 | 0.2779 (2) | 0.6848 (2) | 0.25269 (17) | 0.0372 (5) | |
| C2 | 0.2597 (2) | 0.8390 (2) | 0.16647 (17) | 0.0386 (5) | |
| C3 | 0.3498 (2) | 0.8812 (2) | 0.06716 (17) | 0.0420 (5) | |
| H3A | 0.3364 | 0.9805 | 0.0163 | 0.050* | |
| C4 | 0.4696 (2) | 0.7729 (2) | 0.03743 (18) | 0.0424 (6) | |
| C5 | 0.3483 (2) | 0.5906 (2) | 0.17409 (17) | 0.0374 (5) | |
| C6 | 0.3123 (2) | 0.4741 (2) | 0.17565 (16) | 0.0403 (5) | |
| H6A | 0.3664 | 0.4240 | 0.1214 | 0.048* | |
| C7 | 0.1911 (2) | 0.4141 (2) | 0.25740 (17) | 0.0427 (5) | |
| C8 | 0.1416 (2) | 0.48837 (19) | 0.35812 (17) | 0.0438 (5) | |
| H8A | 0.0457 | 0.4717 | 0.3940 | 0.053* | |
| H8B | 0.2110 | 0.4442 | 0.4297 | 0.053* | |
| C9 | 0.1305 (2) | 0.65023 (19) | 0.30311 (17) | 0.0423 (5) | |
| H9A | 0.0926 | 0.6932 | 0.3710 | 0.051* | |
| H9B | 0.0608 | 0.6948 | 0.2317 | 0.051* | |
| C10 | 0.0654 (2) | 0.4414 (2) | 0.16994 (17) | 0.0564 (6) | |
| H10A | 0.1009 | 0.4002 | 0.1016 | 0.085* | |
| H10B | 0.0254 | 0.5456 | 0.1311 | 0.085* | |
| H10C | −0.0104 | 0.3960 | 0.2212 | 0.085* | |
| C11 | 0.2449 (2) | 0.25092 (19) | 0.32494 (17) | 0.0554 (6) | |
| H11A | 0.2777 | 0.2040 | 0.2601 | 0.083* | |
| H11B | 0.1657 | 0.2123 | 0.3771 | 0.083* | |
| H11C | 0.3256 | 0.2321 | 0.3808 | 0.083* | |
| C12 | 0.3724 (2) | 0.6598 (2) | 0.36940 (16) | 0.0435 (5) | |
| H12A | 0.3274 | 0.7340 | 0.4095 | 0.052* | |
| H12B | 0.3699 | 0.5644 | 0.4347 | 0.052* | |
| C13 | 0.5899 (2) | 0.7717 (2) | 0.32049 (17) | 0.0432 (5) | |
| C14 | 0.5284 (2) | 0.6650 (2) | 0.33670 (16) | 0.0437 (5) | |
| H14A | 0.5908 | 0.5802 | 0.3264 | 0.052* | |
| C15 | 0.7482 (2) | 0.7596 (2) | 0.28583 (18) | 0.0613 (7) | |
| H15A | 0.7934 | 0.6623 | 0.2839 | 0.092* | |
| H15B | 0.7957 | 0.7777 | 0.3503 | 0.092* | |
| H15C | 0.7589 | 0.8308 | 0.2007 | 0.092* | |
| C16 | 0.5118 (2) | 0.9153 (2) | 0.33373 (17) | 0.0561 (6) | |
| H16A | 0.4091 | 0.9170 | 0.3540 | 0.084* | |
| H16B | 0.5209 | 0.9931 | 0.2525 | 0.084* | |
| H16C | 0.5548 | 0.9291 | 0.4033 | 0.084* | |
| C21 | −0.0634 (2) | 1.1136 (2) | 0.19952 (18) | 0.0364 (5) | |
| C22 | −0.0832 (2) | 1.0524 (2) | 0.33214 (17) | 0.0460 (6) | |
| H22A | −0.0149 | 0.9691 | 0.3809 | 0.055* | |
| C23 | −0.2037 (2) | 1.1143 (2) | 0.39196 (19) | 0.0520 (6) | |
| H23A | −0.2180 | 1.0729 | 0.4824 | 0.062* | |
| C24 | −0.3031 (2) | 1.2347 (2) | 0.3223 (2) | 0.0518 (6) | |
| H24A | −0.3854 | 1.2758 | 0.3649 | 0.062* | |
| C25 | −0.2839 (2) | 1.2966 (2) | 0.19050 (19) | 0.0496 (6) | |
| H25A | −0.3526 | 1.3798 | 0.1421 | 0.059* | |
| C26 | −0.1632 (2) | 1.2356 (2) | 0.13033 (18) | 0.0445 (5) | |
| H26A | −0.1487 | 1.2781 | 0.0400 | 0.053* | |
| C27 | 0.0632 (2) | 1.0537 (2) | 0.1279 (2) | 0.0422 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0492 (10) | 0.0545 (10) | 0.0489 (9) | −0.0150 (8) | 0.0184 (8) | −0.0175 (8) |
| O2 | 0.0430 (9) | 0.0443 (9) | 0.0372 (8) | −0.0111 (8) | 0.0074 (7) | −0.0122 (7) |
| O3 | 0.0457 (9) | 0.0418 (9) | 0.0353 (8) | 0.0002 (7) | 0.0020 (7) | −0.0118 (7) |
| O4 | 0.0581 (11) | 0.0648 (11) | 0.0345 (9) | −0.0001 (8) | 0.0044 (8) | −0.0049 (8) |
| C1 | 0.0352 (13) | 0.0413 (13) | 0.0327 (12) | −0.0075 (11) | 0.0046 (10) | −0.0124 (10) |
| C2 | 0.0388 (14) | 0.0386 (14) | 0.0359 (12) | −0.0029 (11) | −0.0013 (11) | −0.0141 (11) |
| C3 | 0.0446 (14) | 0.0410 (13) | 0.0367 (12) | −0.0068 (11) | 0.0022 (11) | −0.0120 (10) |
| C4 | 0.0491 (16) | 0.0439 (15) | 0.0321 (13) | −0.0131 (13) | 0.0013 (11) | −0.0097 (11) |
| C5 | 0.0363 (14) | 0.0414 (14) | 0.0293 (12) | −0.0099 (11) | 0.0023 (10) | −0.0062 (10) |
| C6 | 0.0432 (14) | 0.0431 (14) | 0.0326 (12) | −0.0082 (12) | 0.0032 (10) | −0.0131 (10) |
| C7 | 0.0450 (14) | 0.0433 (14) | 0.0379 (12) | −0.0109 (12) | 0.0019 (11) | −0.0124 (11) |
| C8 | 0.0441 (14) | 0.0475 (14) | 0.0387 (12) | −0.0137 (11) | 0.0062 (10) | −0.0133 (11) |
| C9 | 0.0411 (14) | 0.0486 (14) | 0.0352 (12) | −0.0078 (11) | 0.0043 (10) | −0.0154 (10) |
| C10 | 0.0586 (16) | 0.0652 (16) | 0.0495 (14) | −0.0228 (13) | 0.0018 (13) | −0.0197 (12) |
| C11 | 0.0642 (16) | 0.0468 (14) | 0.0504 (14) | −0.0153 (13) | 0.0086 (12) | −0.0122 (11) |
| C12 | 0.0496 (15) | 0.0458 (14) | 0.0330 (12) | −0.0120 (12) | 0.0012 (11) | −0.0110 (10) |
| C13 | 0.0446 (15) | 0.0510 (15) | 0.0308 (12) | −0.0122 (13) | −0.0014 (10) | −0.0092 (11) |
| C14 | 0.0449 (15) | 0.0474 (14) | 0.0327 (12) | −0.0048 (12) | −0.0036 (11) | −0.0099 (11) |
| C15 | 0.0491 (16) | 0.0791 (18) | 0.0491 (14) | −0.0152 (13) | −0.0003 (12) | −0.0141 (12) |
| C16 | 0.0627 (16) | 0.0566 (15) | 0.0473 (13) | −0.0165 (13) | 0.0000 (12) | −0.0143 (11) |
| C21 | 0.0355 (13) | 0.0374 (13) | 0.0352 (12) | −0.0069 (11) | 0.0019 (10) | −0.0133 (10) |
| C22 | 0.0450 (15) | 0.0465 (14) | 0.0385 (13) | −0.0026 (12) | −0.0001 (11) | −0.0110 (11) |
| C23 | 0.0543 (16) | 0.0579 (16) | 0.0393 (13) | −0.0048 (13) | 0.0042 (12) | −0.0188 (12) |
| C24 | 0.0419 (15) | 0.0588 (16) | 0.0540 (15) | −0.0029 (13) | 0.0039 (12) | −0.0264 (13) |
| C25 | 0.0428 (15) | 0.0520 (15) | 0.0480 (15) | −0.0006 (12) | −0.0073 (12) | −0.0158 (12) |
| C26 | 0.0432 (14) | 0.0456 (14) | 0.0400 (12) | −0.0055 (12) | −0.0041 (11) | −0.0115 (11) |
| C27 | 0.0396 (14) | 0.0412 (14) | 0.0410 (13) | −0.0042 (11) | −0.0052 (12) | −0.0110 (12) |
Geometric parameters (Å, °)
| O1—C4 | 1.206 (2) | C11—H11B | 0.9800 |
| O2—C4 | 1.369 (2) | C11—H11C | 0.9800 |
| O2—C5 | 1.414 (2) | C12—C14 | 1.505 (2) |
| O3—C27 | 1.377 (2) | C12—H12A | 0.9900 |
| O3—C2 | 1.3843 (19) | C12—H12B | 0.9900 |
| O4—C27 | 1.191 (2) | C13—C14 | 1.317 (2) |
| C1—C2 | 1.498 (2) | C13—C15 | 1.504 (2) |
| C1—C5 | 1.504 (2) | C13—C16 | 1.510 (2) |
| C1—C9 | 1.539 (2) | C14—H14A | 0.9500 |
| C1—C12 | 1.568 (2) | C15—H15A | 0.9800 |
| C2—C3 | 1.337 (2) | C15—H15B | 0.9800 |
| C3—C4 | 1.468 (2) | C15—H15C | 0.9800 |
| C3—H3A | 0.9500 | C16—H16A | 0.9800 |
| C5—C6 | 1.316 (2) | C16—H16B | 0.9800 |
| C6—C7 | 1.511 (2) | C16—H16C | 0.9800 |
| C6—H6A | 0.9500 | C21—C26 | 1.382 (2) |
| C7—C8 | 1.530 (2) | C21—C22 | 1.392 (2) |
| C7—C11 | 1.532 (2) | C21—C27 | 1.487 (2) |
| C7—C10 | 1.534 (2) | C22—C23 | 1.383 (2) |
| C8—C9 | 1.522 (2) | C22—H22A | 0.9500 |
| C8—H8A | 0.9900 | C23—C24 | 1.373 (2) |
| C8—H8B | 0.9900 | C23—H23A | 0.9500 |
| C9—H9A | 0.9900 | C24—C25 | 1.384 (2) |
| C9—H9B | 0.9900 | C24—H24A | 0.9500 |
| C10—H10A | 0.9800 | C25—C26 | 1.383 (2) |
| C10—H10B | 0.9800 | C25—H25A | 0.9500 |
| C10—H10C | 0.9800 | C26—H26A | 0.9500 |
| C11—H11A | 0.9800 | ||
| C4—O2—C5 | 120.47 (15) | H11A—C11—H11B | 109.5 |
| C27—O3—C2 | 122.64 (14) | C7—C11—H11C | 109.5 |
| C2—C1—C5 | 107.80 (15) | H11A—C11—H11C | 109.5 |
| C2—C1—C9 | 111.32 (16) | H11B—C11—H11C | 109.5 |
| C5—C1—C9 | 107.82 (16) | C14—C12—C1 | 115.38 (15) |
| C2—C1—C12 | 107.95 (16) | C14—C12—H12A | 108.4 |
| C5—C1—C12 | 112.42 (15) | C1—C12—H12A | 108.4 |
| C9—C1—C12 | 109.54 (14) | C14—C12—H12B | 108.4 |
| C3—C2—O3 | 125.00 (17) | C1—C12—H12B | 108.4 |
| C3—C2—C1 | 122.81 (17) | H12A—C12—H12B | 107.5 |
| O3—C2—C1 | 111.89 (16) | C14—C13—C15 | 121.7 (2) |
| C2—C3—C4 | 119.40 (18) | C14—C13—C16 | 124.62 (19) |
| C2—C3—H3A | 120.3 | C15—C13—C16 | 113.69 (19) |
| C4—C3—H3A | 120.3 | C13—C14—C12 | 128.56 (19) |
| O1—C4—O2 | 117.76 (19) | C13—C14—H14A | 115.7 |
| O1—C4—C3 | 124.16 (19) | C12—C14—H14A | 115.7 |
| O2—C4—C3 | 118.08 (18) | C13—C15—H15A | 109.5 |
| C6—C5—O2 | 117.13 (17) | C13—C15—H15B | 109.5 |
| C6—C5—C1 | 126.20 (18) | H15A—C15—H15B | 109.5 |
| O2—C5—C1 | 116.66 (17) | C13—C15—H15C | 109.5 |
| C5—C6—C7 | 124.84 (18) | H15A—C15—H15C | 109.5 |
| C5—C6—H6A | 117.6 | H15B—C15—H15C | 109.5 |
| C7—C6—H6A | 117.6 | C13—C16—H16A | 109.5 |
| C6—C7—C8 | 109.01 (16) | C13—C16—H16B | 109.5 |
| C6—C7—C11 | 109.75 (16) | H16A—C16—H16B | 109.5 |
| C8—C7—C11 | 109.79 (16) | C13—C16—H16C | 109.5 |
| C6—C7—C10 | 108.99 (15) | H16A—C16—H16C | 109.5 |
| C8—C7—C10 | 110.54 (16) | H16B—C16—H16C | 109.5 |
| C11—C7—C10 | 108.74 (17) | C26—C21—C22 | 119.58 (18) |
| C9—C8—C7 | 112.82 (15) | C26—C21—C27 | 117.90 (17) |
| C9—C8—H8A | 109.0 | C22—C21—C27 | 122.51 (18) |
| C7—C8—H8A | 109.0 | C23—C22—C21 | 119.15 (19) |
| C9—C8—H8B | 109.0 | C23—C22—H22A | 120.4 |
| C7—C8—H8B | 109.0 | C21—C22—H22A | 120.4 |
| H8A—C8—H8B | 107.8 | C24—C23—C22 | 120.90 (19) |
| C8—C9—C1 | 112.10 (15) | C24—C23—H23A | 119.6 |
| C8—C9—H9A | 109.2 | C22—C23—H23A | 119.6 |
| C1—C9—H9A | 109.2 | C23—C24—C25 | 120.33 (19) |
| C8—C9—H9B | 109.2 | C23—C24—H24A | 119.8 |
| C1—C9—H9B | 109.2 | C25—C24—H24A | 119.8 |
| H9A—C9—H9B | 107.9 | C26—C25—C24 | 119.00 (19) |
| C7—C10—H10A | 109.5 | C26—C25—H25A | 120.5 |
| C7—C10—H10B | 109.5 | C24—C25—H25A | 120.5 |
| H10A—C10—H10B | 109.5 | C21—C26—C25 | 121.03 (18) |
| C7—C10—H10C | 109.5 | C21—C26—H26A | 119.5 |
| H10A—C10—H10C | 109.5 | C25—C26—H26A | 119.5 |
| H10B—C10—H10C | 109.5 | O4—C27—O3 | 123.95 (18) |
| C7—C11—H11A | 109.5 | O4—C27—C21 | 125.59 (19) |
| C7—C11—H11B | 109.5 | O3—C27—C21 | 110.43 (17) |
| C27—O3—C2—C3 | 39.7 (3) | C6—C7—C8—C9 | 42.2 (2) |
| C27—O3—C2—C1 | −146.56 (16) | C11—C7—C8—C9 | 162.48 (16) |
| C5—C1—C2—C3 | −29.9 (3) | C10—C7—C8—C9 | −77.6 (2) |
| C9—C1—C2—C3 | −147.98 (18) | C7—C8—C9—C1 | −62.2 (2) |
| C12—C1—C2—C3 | 91.8 (2) | C2—C1—C9—C8 | 163.77 (15) |
| C5—C1—C2—O3 | 156.15 (15) | C5—C1—C9—C8 | 45.7 (2) |
| C9—C1—C2—O3 | 38.1 (2) | C12—C1—C9—C8 | −76.91 (19) |
| C12—C1—C2—O3 | −82.17 (19) | C2—C1—C12—C14 | −68.5 (2) |
| O3—C2—C3—C4 | 175.08 (16) | C5—C1—C12—C14 | 50.2 (2) |
| C1—C2—C3—C4 | 2.0 (3) | C9—C1—C12—C14 | 170.09 (16) |
| C5—O2—C4—O1 | −179.08 (16) | C15—C13—C14—C12 | −179.07 (16) |
| C5—O2—C4—C3 | 0.1 (2) | C16—C13—C14—C12 | 0.7 (3) |
| C2—C3—C4—O1 | −165.68 (19) | C1—C12—C14—C13 | 100.6 (2) |
| C2—C3—C4—O2 | 15.2 (3) | C26—C21—C22—C23 | 0.6 (3) |
| C4—O2—C5—C6 | 149.39 (17) | C27—C21—C22—C23 | 180.00 (18) |
| C4—O2—C5—C1 | −31.3 (2) | C21—C22—C23—C24 | −0.1 (3) |
| C2—C1—C5—C6 | −137.1 (2) | C22—C23—C24—C25 | −0.2 (3) |
| C9—C1—C5—C6 | −16.8 (3) | C23—C24—C25—C26 | −0.1 (3) |
| C12—C1—C5—C6 | 104.0 (2) | C22—C21—C26—C25 | −1.0 (3) |
| C2—C1—C5—O2 | 43.7 (2) | C27—C21—C26—C25 | 179.64 (17) |
| C9—C1—C5—O2 | 163.97 (14) | C24—C25—C26—C21 | 0.7 (3) |
| C12—C1—C5—O2 | −75.2 (2) | C2—O3—C27—O4 | −9.8 (3) |
| O2—C5—C6—C7 | 179.69 (15) | C2—O3—C27—C21 | 168.29 (15) |
| C1—C5—C6—C7 | 0.5 (3) | C26—C21—C27—O4 | −0.4 (3) |
| C5—C6—C7—C8 | −12.7 (3) | C22—C21—C27—O4 | −179.8 (2) |
| C5—C6—C7—C11 | −133.0 (2) | C26—C21—C27—O3 | −178.48 (17) |
| C5—C6—C7—C10 | 108.0 (2) | C22—C21—C27—O3 | 2.2 (3) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2544).
References
- Ciochina, R. & Grossman, R. B. (2006). Chem. Rev.106, 3963–3986. [DOI] [PubMed]
- Nonius (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809030918/hg2544sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809030918/hg2544Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



