Abstract
The title compound, C6H7N3, has an ethylene group connecting an imidazole ring and a –CN group. These groups are in a staggered conformation. The shortest intermolecular contact is found between the imidazole N atom and a –CH2– group of a neighboring molecule.
Related literature
For background and applications of ionic liquids, see: Hayashi et al. (2006 ▶); Kozlova et al. (2009a
▶,b
▶); Lombardo et al. (2007 ▶); Macaev et al. (2007 ▶); Sawa & Okamura (1969 ▶); Scheers et al. (2008 ▶); Visser et al. (2001 ▶); Wang et al. (2003 ▶); Wasserscheid & Keim (2000 ▶); Xu et al. (2007 ▶); Yamauchi & Masui (1976 ▶); Yang et al. (2006 ▶).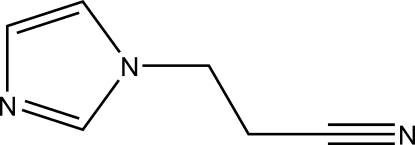
Experimental
Crystal data
C6H7N3
M r = 121.15
Monoclinic,

a = 7.2712 (3) Å
b = 5.5917 (2) Å
c = 15.4625 (5) Å
β = 100.979 (1)°
V = 617.17 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 173 K
0.45 × 0.40 × 0.30 mm
Data collection
Bruker–Nonius X8 APEX diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2007 ▶) T min = 0.946, T max = 0.975
11604 measured reflections
1542 independent reflections
1456 reflections with I > 2σ(I)
R int = 0.018
Refinement
R[F 2 > 2σ(F 2)] = 0.032
wR(F 2) = 0.086
S = 1.04
1542 reflections
111 parameters
All H-atom parameters refined
Δρmax = 0.27 e Å−3
Δρmin = −0.16 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: ct.exe (Köckerling, 1996 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809035685/om2269sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809035685/om2269Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5A⋯N2i | 0.97 (1) | 2.66 (1) | 3.366 (1) | 135.7 (9) |
Symmetry code: (i)  .
.
Acknowledgments
We thank Professor Dr Helmut Reinke (University of Rostock/Germany, Institute of Chemistry) for maintaining the X-ray equipment. Support from the DFG (priority program SPP 1191-Ionic Liquids, KO-1616/4-1 and 1616/4-2) is gratefully acknowledged.
supplementary crystallographic information
Comment
Ionic Liquids (ILs) are more and more attracting remarkable attention as new solvents and reaction media for organic and inorganic synthesis and catalysis in order to replace volatile organic solvents (Wasserscheid & Keim, 2000). Imidazolium based ILs have emerged as leading candidates since they have very low vapor pressures, are moisture and air stable, and are highly solvating for both molecular and ionic species. Furthermore, ILs are finding use in separation processes (Visser et al., 2001), in battery applications (Scheers et al., 2008), as electrolytes in solar cells (Wang et al., 2003), or as part of metal catalysts and magnetic liquids (Lombardo et al., 2007; Hayashi et al., 2006; Kozlova et al., 2009a; Kozlova et al., 2009b). Ionic Liquids and imidazole compounds incorporating nitrile functionalities have shown applications in heterocyclic and terpene chemistry (Macaev et al., 2007; Yamauchi & Masui, 1976), in Michael addition or aza-Michael reactions (Xu et al., 2007; Yang et al., 2006).
In this context we present the molecular and single-crystal structure of 3-(1H-imidazol-1-yl)propanenitrile, an important precursor of nitrile functionalized ILs. Although all other sources report this compound to be a liquid at room temperature, there is one single indication in the literature that it is a solid melting at 33–35 °C (Sawa & Okamura, 1969). A freshly distilled sample crystallizes spontaneously within days, forming big block-shaped colourless crystals, which are barely deliquescent and melting at 37 °C.
All the bond lengths are within the expected ranges. The molecular structure of the title compound is shown in Figure 1. The C6–N3 bond length of 1.141 (1) Å indicates the triple-bond nitrile character. This nitrile group is connected through an ethylene group with the planar imidazolyl ring. The ethylene group has a staggered conformation with an N1–C4–C5–C6 torsion angle of -65.43 (9)°.
The shortest intermolecular contacts are found between one of the H atoms bonded at C5 and the N2 atom of the symmetry equivalent neighboring molecule. The H5A···N2# distance measures 2.66 (1) Å, the corresponding C5···N2# distance 3.366 (1) Å, and the C5–H5a···N2# angle 135.6° (symmetry code #: x, 1/2-y, z+1/2). Therefore the contact can be considered as a weak hydrogen bond. Figure 2 shows the orientation of two molecules, which are attached through this weak hydrogen bond.
In the crystal the molecules are arranged in rows, such that this weak hydrogen bond is oriented approximately parallel to the crystallographic c direction. Due to the c glide plane, every second row, stacked along a is shifted by c/2. The packing diagram, Figure 3, shows this assembly.
Experimental
1H-Imidazole (50.0 g, 0.7 mol) and acrylonitrile (117.0 g, 2.2 mol) were refluxed in 150 ml of ethanol overnight. Excess acrylonitrile and the solvent were evaporated and the residue was distilled in vacuum, yielding a colourless supercooled liquid, which crystallized at room temperature after several days. Yield: 75.0 g (84%), mp 37 °C.
Refinement
The positions of the hydrogen atoms were located from a difference Fourier map and refined isotropically.
Figures
Fig. 1.
Molecular structure of 3-(1H-imidazol-1-yl)propanenitrile with atom labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
Two neighboring C6H7N3 molecules with the short N2···H(C5) contact.
Fig. 3.
Packing diagram of 3-(1H-imidazol-1-yl)propanenitrile in a view down the crystallographic b direction.
Crystal data
| C6H7N3 | Dx = 1.304 Mg m−3 |
| Mr = 121.15 | Melting point: 310 K |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.2712 (3) Å | Cell parameters from 8906 reflections |
| b = 5.5917 (2) Å | θ = 2.7–28.3° |
| c = 15.4625 (5) Å | µ = 0.09 mm−1 |
| β = 100.979 (1)° | T = 173 K |
| V = 617.17 (4) Å3 | Block, colourless |
| Z = 4 | 0.45 × 0.40 × 0.30 mm |
| F(000) = 256 |
Data collection
| Bruker–Nonius X8 Apex diffractometer | 1542 independent reflections |
| Radiation source: fine-focus sealed tube | 1456 reflections with I > 2σ(I) |
| graphite | Rint = 0.018 |
| φ and ω scans | θmax = 28.3°, θmin = 3.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2007) | h = −9→9 |
| Tmin = 0.946, Tmax = 0.975 | k = −7→7 |
| 11604 measured reflections | l = −20→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.032 | All H-atom parameters refined |
| wR(F2) = 0.086 | w = 1/[σ2(Fo2) + (0.0486P)2 + 0.1206P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 1542 reflections | Δρmax = 0.27 e Å−3 |
| 111 parameters | Δρmin = −0.16 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.064 (15) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.76561 (9) | 0.2518 (1) | 0.30688 (4) | 0.0207 (2) | |
| C1 | 0.6541 (1) | 0.1964 (2) | 0.22892 (5) | 0.0254 (2) | |
| H1 | 0.579 (2) | 0.053 (2) | 0.2217 (7) | 0.033 (3)* | |
| N2 | 0.6623 (1) | 0.3580 (1) | 0.16800 (4) | 0.0302 (2) | |
| C2 | 0.7866 (1) | 0.5266 (2) | 0.20919 (6) | 0.0281 (2) | |
| H2 | 0.817 (2) | 0.670 (2) | 0.1771 (8) | 0.037 (3)* | |
| C3 | 0.8519 (1) | 0.4648 (1) | 0.29465 (5) | 0.0249 (2) | |
| H3 | 0.944 (2) | 0.535 (2) | 0.3408 (8) | 0.035 (3)* | |
| C4 | 0.7876 (1) | 0.1115 (1) | 0.38757 (5) | 0.0239 (2) | |
| H4A | 0.919 (2) | 0.109 (2) | 0.4147 (7) | 0.030 (3)* | |
| H4B | 0.744 (2) | −0.051 (2) | 0.3700 (7) | 0.031 (3)* | |
| C5 | 0.6708 (1) | 0.2069 (2) | 0.45232 (5) | 0.0254 (2) | |
| H5A | 0.683 (2) | 0.100 (2) | 0.5022 (8) | 0.037 (3)* | |
| H5B | 0.538 (2) | 0.217 (2) | 0.4243 (7) | 0.035 (3)* | |
| C6 | 0.7298 (1) | 0.4436 (2) | 0.48692 (5) | 0.0244 (2) | |
| N3 | 0.7779 (1) | 0.6275 (1) | 0.51425 (5) | 0.0333 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0242 (3) | 0.0212 (3) | 0.0176 (3) | 0.0012 (2) | 0.0061 (2) | 0.0007 (2) |
| C1 | 0.0293 (4) | 0.0279 (4) | 0.0194 (4) | 0.0001 (3) | 0.0057 (3) | −0.0026 (3) |
| N2 | 0.0345 (4) | 0.0371 (4) | 0.0196 (3) | 0.0033 (3) | 0.0064 (3) | 0.0029 (3) |
| C2 | 0.0290 (4) | 0.0307 (4) | 0.0271 (4) | 0.0027 (3) | 0.0113 (3) | 0.0075 (3) |
| C3 | 0.0246 (4) | 0.0249 (4) | 0.0257 (4) | −0.0015 (3) | 0.0065 (3) | 0.0023 (3) |
| C4 | 0.0318 (4) | 0.0213 (4) | 0.0188 (3) | 0.0026 (3) | 0.0058 (3) | 0.0025 (3) |
| C5 | 0.0321 (4) | 0.0265 (4) | 0.0190 (4) | −0.0056 (3) | 0.0084 (3) | −0.0011 (3) |
| C6 | 0.0265 (4) | 0.0288 (4) | 0.0186 (3) | 0.0011 (3) | 0.0060 (3) | −0.0002 (3) |
| N3 | 0.0386 (4) | 0.0303 (4) | 0.0313 (4) | −0.0002 (3) | 0.0078 (3) | −0.0047 (3) |
Geometric parameters (Å, °)
| N1—C1 | 1.354 (1) | C6—N3 | 1.141 (1) |
| N1—C3 | 1.376 (1) | C1—H1 | 0.97 (1) |
| N1—C4 | 1.4565 (9) | C2—H2 | 0.99 (1) |
| C1—N2 | 1.315 (1) | C3—H3 | 0.97 (1) |
| N2—C2 | 1.376 (1) | C4—H4A | 0.97 (1) |
| C2—C3 | 1.361 (1) | C4—H4B | 0.99 (1) |
| C4—C5 | 1.527 (1) | C5—H5A | 0.97 (1) |
| C5—C6 | 1.461 (1) | C5—H5B | 0.98 (1) |
| C1—N1—C3 | 106.68 (6) | H2—C2—N2 | 120.9 (7) |
| C1—N1—C4 | 126.06 (7) | H3—C3—C2 | 132.8 (7) |
| C3—N1—C4 | 127.27 (7) | H3—C3—N1 | 121.4 (7) |
| N2—C1—N1 | 112.30 (7) | H4A—C4—H4B | 110.4 (9) |
| C1—N2—C2 | 104.76 (7) | H4A—C4—N1 | 108.5 (7) |
| C3—C2—N2 | 110.64 (7) | H4B—C4—N1 | 106.5 (7) |
| C2—C3—N1 | 105.63 (7) | H4A—C4—C5 | 110.2 (7) |
| N1—C4—C5 | 112.92 (6) | H4B—C4—C5 | 108.2 (7) |
| C6—C5—C4 | 113.28 (7) | H5A—C5—H5B | 109 (1) |
| N3—C6—C5 | 179.24 (9) | H5A—C5—C6 | 107.0 (7) |
| H1—C1—N2 | 126.2 (7) | H5B—C5—C6 | 107.9 (7) |
| H1—C1—N1 | 121.5 (7) | H5A—C5—C4 | 109.2 (7) |
| H2—C2—C3 | 128.4 (7) | H5B—C5—C4 | 110.7 (7) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5A···N2i | 0.97 (1) | 2.66 (1) | 3.366 (1) | 135.7 (9) |
Symmetry codes: (i) x, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: OM2269).
References
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2007). SAINT, SMART and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Hayashi, S., Saha, S. & Hamaguchi, H. (2006). IEEE Trans. Magn.42, 12–14.
- Köckerling, M. (1996). ct.exe. Universität Rostock, Germany.
- Kozlova, S. A., Verevkin, S. P., Heintz, A., Peppel, T. & Köckerling, M. (2009a). J. Chem. Eng. Data, 54, 1524–1528.
- Kozlova, S. A., Verevkin, S. P., Heintz, A., Peppel, T. & Köckerling, M. (2009b). J. Chem. Thermodyn 41, 330–333.
- Lombardo, M., Pasi, F., Trombini, C., Seddon, K. R. & Pittner, W. R. (2007). Green Chem.9, 321–322.
- Macaev, F., Gavrilov, K., Muntyanu, V., Styngach, E., Vlad, L., Bets, L., Pogrebnoi, S. & Barba, A. (2007). Chem. Nat. Compd.43, 136–139.
- Sawa, N. & Okamura, S. (1969). Nippon Kagaku Zasshi, 90, 704–707.
- Scheers, J., Johansson, P. & Jacobsson, P. (2008). J. Electrochem. Soc.155, A628–A634.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Visser, A. E., Swatloski, R. P., Reichert, W. M., Mayton, R., Sheff, S., Wierzbicki, A., Davis, J. H. Jr & Rogers, R. D. (2001). Chem. Commun. pp. 135–136.
- Wang, P., Zakeeruddin, S. M., Comte, P., Exnar, I. & Grätzel, M. (2003). J. Am. Chem. Soc.125, 1166–1167. [DOI] [PubMed]
- Wasserscheid, P. & Keim, W. (2000). Angew. Chem. Int. Ed.39, 3772–3789. [DOI] [PubMed]
- Xu, J.-M., Qian, C., Liu, B.-K., Wu, Q. & Lin, X.-F. (2007). Tetrahedron, 63, 986–990.
- Yamauchi, M. & Masui, M. (1976). Chem. Pharm. Bull.24, 1480–1484.
- Yang, L., Xu, L.-W., Zhou, W., Li, L. & Xia, C.-G. (2006). Tetrahedron Lett.47, 7723–7726.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809035685/om2269sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809035685/om2269Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





