Abstract
The title compound (systematic name: 9a-hydroxy-3,4a,5-trimethyl-4a,6,7,8a,9,9a-hexahydro-4H,5H-naphtho[2,3-b]furan-2,8-dione), C15H20O4, is a sesquiterpene lactone showing the typical eremophilanolide skeleton, which has been isolated from the plant Senecio candidans collected in the Chilean Magallanes region. The present study confirms the atomic connectivity assigned on the basis of 1H and 13C NMR spectroscopy, as well as the relative stereochemistry of the 4α-methyl,5α-methyl,8β-hydroxy,10β-H unit. The crystal structure is stabilized by intermolecular O—H⋯O hydrogen bonds involving the hydroxy group as donor and the oxo group as acceptor, giving chains along the a axis. The absolute structure was not determined because of the lack of suitable anomalous scatters.
Related literature
For the biological activity of metabolites isolated from plants of the Senecio species, see: Ulubelen et al. (1971 ▶); Burgueño-Tapia et al. (2007 ▶); Domínguez et al. (2008 ▶); Reina,González-Coloma, Domínguez-Díaz et al. (2006 ▶); Reina, González-Coloma, Gutiérrez et al. (2006 ▶).
Experimental
Crystal data
C15H20O4
M r = 264.31
Monoclinic,
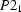
a = 7.432 (4) Å
b = 13.010 (6) Å
c = 8.161 (6) Å
β = 115.47 (4)°
V = 712.4 (8) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.45 × 0.35 × 0.25 mm
Data collection
Enraf–Nonius KappaCCD diffractometer
Absorption correction: none
4410 measured reflections
1678 independent reflections
1485 reflections with I > 2σ(I)
R int = 0.018
Refinement
R[F 2 > 2σ(F 2)] = 0.038
wR(F 2) = 0.095
S = 1.04
1678 reflections
186 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.20 e Å−3
Δρmin = −0.12 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: SCALEPACK and DENZO (Otwinowski & Minor, 1997 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶ ); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: Win GX (Farrugia,1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809033418/hg2542sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809033418/hg2542Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O4i | 0.90 (4) | 1.87 (4) | 2.750 (3) | 164 (4) |
Symmetry code: (i) 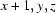 .
.
supplementary crystallographic information
Comment
The Senecio (Asteraceae) genus are widely distributed by the worldwide and the most studied constituents are sesquiterpenes with eremophilanolides and furanoeremophilanes skeleton, together with pyrrolizidine alkaloids. The chemical study of plants of the Senecio species has increased in the last few years because of the biological activity that shown the metabolites isolated from this natural sources: Reina, González-Coloma, Domínguez-Díaz et al. (2006); Reina, González-Coloma, Gutiérrez et al. (2006); Burgueño-Tapia et al. (2007); Domínguez et al. (2008).
As part of our ongoing study of Senecio genus from the chilean shouthern and Magallanes Region, in this work we report the isolation and the molecular and crystal structure determination of the title compound, which it is described for the first time as a metabolite for the Senecio genus. An Istanbulin A with optical rotation [α]D25 = + 81.5° was reported some years ago and its structure and stereochemistry determined by spectroscopic methods, but no X-ray analysys was performed (Ulubelen et al.,1971). The lack of suitable anomalous scatters did not allow us to reliably determine the absolute structure and that shown: 1-oxo-8β-hydroxy-10βH-eremophil-7(11)-en-12,8β-olide was chosen to be, on the basis of the negative optical rotation value: [α]D25 = - 68.7°, the opposite to that the previously reported compound.
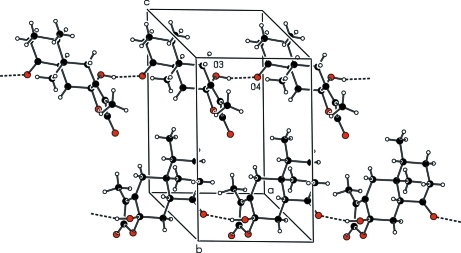
The crystal structure is stabilized by intermolecular O—H···O hydrogen bonds in which are involved the hydroxyl group at C8 acting as donor and the oxo group at C1 as acceptor, giving chains along the a axis, through a C(7) graph-set motif.
Experimental
Senecio candidans DC. (Asteraceae) was collected from the south of Chile, at the Santa Maria River, Punta Arenas(XII Region), in april 2002 and authenticated by Professor E. Pisano. A voucher specimen (n° 3483) was deposited in the herbarium of Instituto de la Patagonia, Universidad de Magallanes, Punta Arenas (Chile).
Dried aerial parts of S.candidans (3.5 kg) was extracted in methanol at room temperature during a week to give a crude methanolic extract (224 g), 6.4% yield of dry plant weight. A portion of these crude methanolic extract (124 g) was chromatographed on a silica gel vacuum-liquid chromatography column (VLC), using a hexane-ethyl acetate-methanol gradient. The portion of the extract collected at the (hexane-ethyl acetate, 75:25) solvent polarity (2.7 g) was further purified by passage over Sephadex LH-20 (hexane-methylene chloride-methanol, 3:1:1), followed by different chromatographic techniques to give (-)-Istanbuline A: 1-oxo-8β-hydroxy-10βH-eremophil-7(11)-en-12,8β-olide (6.4 mg). The molecular formula C15H20O4 was deduced from its high resolution MS spectrum which shows a molecular ion at m/z=264.1357. Complete and unambiguous assignment of de all protons and carbon were established by analysis of the mono and bidimensional NMR experiments and comparison with the previously spectroscopic data reported for Istanbulin A (Ulubelen et al.,1971).
The absolute structure was not determined because of the lack of suitable anomalous scatters. However, the value of the measured optical rotation: [α]D25 = -68.7° allowed to us to choose that shown, as the opposite to that a previously described (+)-istanbulin: [α]D25 = +81.5°.
Refinement
All H-atoms were located on sucessive difference-Fourier maps. The H-atom of the hydroxyl group was freely refined. and all other H atoms were constrained refined, with idealized geometries: C—H = 0.96(CH3), 0.97(CH2), 0.98(CH)Å. The lack of suitable anomalous scatters did not allow us to reliably determine the absolute structure and, therefore, the Friedel pairs were merged prior to the final refinement.
Figures
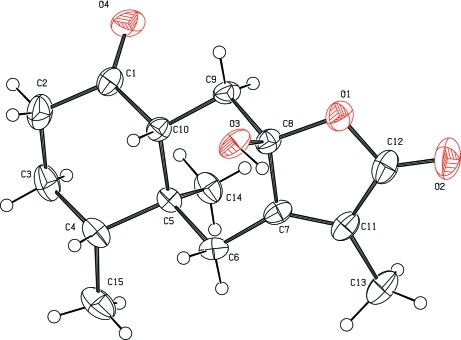
Fig. 1.
Molecular structure of the title compund showing displacement ellipsoids at the 50% probability level.
Fig. 2.
A view of the hydrogen-bonding pattern. Hydrogen atoms not involved in the O—H···O interactions have been omitted.
Crystal data
| C15H20O4 | F(000) = 284 |
| Mr = 264.31 | Dx = 1.232 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 297 reflections |
| a = 7.432 (4) Å | θ = 4.3–23.7° |
| b = 13.010 (6) Å | µ = 0.09 mm−1 |
| c = 8.161 (6) Å | T = 293 K |
| β = 115.47 (4)° | Block, colourless |
| V = 712.4 (8) Å3 | 0.45 × 0.35 × 0.25 mm |
| Z = 2 |
Data collection
| Enraf–Nonius KappaCCD diffractometer | 1485 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.018 |
| graphite | θmax = 27.5°, θmin = 6.4° |
| Detector resolution: 9 pixels mm-1 | h = −9→9 |
| φ and ω scans | k = −16→15 |
| 4410 measured reflections | l = −10→10 |
| 1678 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.038 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.095 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0482P)2 + 0.0898P] where P = (Fo2 + 2Fc2)/3 |
| 1678 reflections | (Δ/σ)max = 0.001 |
| 186 parameters | Δρmax = 0.20 e Å−3 |
| 1 restraint | Δρmin = −0.11 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.4955 (3) | 0.50383 (14) | 0.8319 (2) | 0.0568 (5) | |
| O2 | 0.5421 (4) | 0.5731 (2) | 0.6018 (3) | 0.0869 (7) | |
| O3 | 0.6082 (2) | 0.37119 (15) | 1.04064 (19) | 0.0521 (4) | |
| H3 | 0.713 (5) | 0.354 (3) | 1.018 (4) | 0.079 (9)* | |
| O4 | −0.0560 (2) | 0.35876 (16) | 0.9801 (3) | 0.0635 (5) | |
| C1 | 0.0324 (3) | 0.28290 (19) | 0.9675 (3) | 0.0456 (5) | |
| C2 | −0.0216 (4) | 0.1763 (2) | 0.9971 (4) | 0.0635 (7) | |
| H2A | −0.1422 | 0.1778 | 1.0153 | 0.079 (9)* | |
| H2B | 0.0844 | 0.1476 | 1.1053 | 0.073 (9)* | |
| C3 | −0.0542 (4) | 0.1087 (2) | 0.8333 (4) | 0.0643 (7) | |
| H3A | −0.1759 | 0.1299 | 0.7312 | 0.074 (9)* | |
| H3B | −0.0713 | 0.0379 | 0.8615 | 0.059 (8)* | |
| C4 | 0.1189 (4) | 0.11441 (18) | 0.7792 (3) | 0.0518 (6) | |
| H4 | 0.2377 | 0.0891 | 0.8827 | 0.059 (7)* | |
| C5 | 0.1632 (3) | 0.22659 (16) | 0.7435 (3) | 0.0375 (4) | |
| C6 | 0.3567 (3) | 0.22904 (18) | 0.7137 (3) | 0.0448 (5) | |
| H6A | 0.4602 | 0.1896 | 0.8084 | 0.057 (7)* | |
| H6B | 0.3309 | 0.1977 | 0.5979 | 0.050 (7)* | |
| C7 | 0.4259 (3) | 0.33673 (18) | 0.7168 (3) | 0.0422 (5) | |
| C8 | 0.4541 (3) | 0.40177 (17) | 0.8797 (3) | 0.0412 (5) | |
| C9 | 0.2648 (3) | 0.40183 (17) | 0.9078 (3) | 0.0407 (5) | |
| H9A | 0.1589 | 0.4368 | 0.8076 | 0.036 (5)* | |
| H9B | 0.2875 | 0.4377 | 1.0192 | 0.043 (6)* | |
| C10 | 0.2059 (3) | 0.29068 (16) | 0.9181 (3) | 0.0372 (4) | |
| H10 | 0.3200 | 0.2583 | 1.0170 | 0.044 (6)* | |
| C11 | 0.4554 (3) | 0.3931 (2) | 0.5961 (3) | 0.0488 (6) | |
| C12 | 0.5040 (4) | 0.4988 (3) | 0.6676 (4) | 0.0588 (6) | |
| C13 | 0.4395 (4) | 0.3664 (3) | 0.4119 (3) | 0.0668 (8) | |
| H13A | 0.5521 | 0.3940 | 0.3982 | 0.090* | |
| H13B | 0.3190 | 0.3950 | 0.3207 | 0.090* | |
| H13C | 0.4374 | 0.2930 | 0.3988 | 0.090* | |
| C14 | −0.0102 (3) | 0.27228 (19) | 0.5782 (3) | 0.0512 (5) | |
| H14A | 0.0173 | 0.3431 | 0.5648 | 0.090* | |
| H14B | −0.1303 | 0.2676 | 0.5947 | 0.090* | |
| H14C | −0.0266 | 0.2348 | 0.4713 | 0.090* | |
| C15 | 0.0800 (5) | 0.0415 (2) | 0.6210 (5) | 0.0771 (9) | |
| H15A | 0.1886 | 0.0454 | 0.5872 | 0.090* | |
| H15B | −0.0415 | 0.0607 | 0.5195 | 0.090* | |
| H15C | 0.0685 | −0.0276 | 0.6569 | 0.090* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0612 (10) | 0.0542 (10) | 0.0633 (10) | −0.0224 (8) | 0.0348 (8) | −0.0122 (8) |
| O2 | 0.0968 (16) | 0.0837 (16) | 0.0944 (16) | −0.0249 (13) | 0.0548 (14) | 0.0156 (13) |
| O3 | 0.0339 (7) | 0.0817 (12) | 0.0434 (7) | −0.0078 (8) | 0.0194 (6) | −0.0091 (8) |
| O4 | 0.0465 (9) | 0.0680 (12) | 0.0928 (12) | −0.0034 (9) | 0.0460 (9) | −0.0125 (10) |
| C1 | 0.0355 (10) | 0.0562 (14) | 0.0480 (11) | −0.0048 (10) | 0.0208 (9) | −0.0015 (11) |
| C2 | 0.0559 (14) | 0.0680 (18) | 0.0766 (17) | −0.0052 (13) | 0.0378 (13) | 0.0159 (15) |
| C3 | 0.0604 (15) | 0.0452 (14) | 0.0848 (18) | −0.0128 (12) | 0.0289 (14) | 0.0092 (14) |
| C4 | 0.0517 (12) | 0.0354 (11) | 0.0592 (13) | 0.0019 (10) | 0.0153 (11) | 0.0043 (11) |
| C5 | 0.0343 (9) | 0.0339 (10) | 0.0398 (9) | 0.0004 (8) | 0.0118 (8) | −0.0005 (9) |
| C6 | 0.0463 (11) | 0.0476 (12) | 0.0440 (10) | 0.0054 (10) | 0.0228 (9) | −0.0063 (10) |
| C7 | 0.0325 (9) | 0.0558 (13) | 0.0408 (10) | −0.0020 (9) | 0.0183 (8) | −0.0071 (10) |
| C8 | 0.0384 (10) | 0.0472 (12) | 0.0423 (10) | −0.0099 (9) | 0.0213 (8) | −0.0077 (9) |
| C9 | 0.0367 (9) | 0.0430 (12) | 0.0488 (11) | −0.0050 (8) | 0.0244 (9) | −0.0109 (9) |
| C10 | 0.0282 (8) | 0.0438 (11) | 0.0407 (10) | 0.0015 (8) | 0.0161 (7) | 0.0002 (9) |
| C11 | 0.0378 (10) | 0.0703 (16) | 0.0414 (10) | −0.0015 (10) | 0.0201 (8) | 0.0010 (11) |
| C12 | 0.0495 (13) | 0.0745 (18) | 0.0584 (13) | −0.0133 (12) | 0.0290 (11) | 0.0039 (13) |
| C13 | 0.0641 (14) | 0.099 (2) | 0.0423 (11) | −0.0002 (16) | 0.0278 (11) | 0.0043 (14) |
| C14 | 0.0465 (12) | 0.0464 (13) | 0.0465 (11) | 0.0016 (10) | 0.0066 (9) | 0.0057 (11) |
| C15 | 0.089 (2) | 0.0412 (14) | 0.093 (2) | −0.0098 (14) | 0.0316 (18) | −0.0157 (14) |
Geometric parameters (Å, °)
| O1—C12 | 1.371 (3) | C6—H6A | 0.9700 |
| O1—C8 | 1.454 (3) | C6—H6B | 0.9700 |
| O2—C12 | 1.198 (4) | C7—C11 | 1.320 (3) |
| O3—C8 | 1.379 (3) | C7—C8 | 1.512 (3) |
| O3—H3 | 0.90 (4) | C8—C9 | 1.519 (3) |
| O4—C1 | 1.214 (3) | C9—C10 | 1.524 (3) |
| C1—C2 | 1.493 (4) | C9—H9A | 0.9700 |
| C1—C10 | 1.511 (3) | C9—H9B | 0.9700 |
| C2—C3 | 1.530 (4) | C10—H10 | 0.9800 |
| C2—H2A | 0.9700 | C11—C12 | 1.477 (4) |
| C2—H2B | 0.9700 | C11—C13 | 1.497 (3) |
| C3—C4 | 1.532 (4) | C13—H13A | 0.9600 |
| C3—H3A | 0.9700 | C13—H13B | 0.9600 |
| C3—H3B | 0.9700 | C13—H13C | 0.9600 |
| C4—C15 | 1.526 (4) | C14—H14A | 0.9600 |
| C4—C5 | 1.551 (3) | C14—H14B | 0.9600 |
| C4—H4 | 0.9800 | C14—H14C | 0.9600 |
| C5—C14 | 1.529 (3) | C15—H15A | 0.9600 |
| C5—C6 | 1.558 (3) | C15—H15B | 0.9600 |
| C5—C10 | 1.561 (3) | C15—H15C | 0.9600 |
| C6—C7 | 1.489 (3) | ||
| C12—O1—C8 | 108.86 (19) | O1—C8—C7 | 103.91 (17) |
| C8—O3—H3 | 108 (2) | O3—C8—C9 | 107.47 (17) |
| O4—C1—C2 | 123.30 (19) | O1—C8—C9 | 110.88 (18) |
| O4—C1—C10 | 121.5 (2) | C7—C8—C9 | 110.20 (16) |
| C2—C1—C10 | 115.2 (2) | C8—C9—C10 | 108.33 (16) |
| C1—C2—C3 | 110.2 (2) | C8—C9—H9A | 110.0 |
| C1—C2—H2A | 109.6 | C10—C9—H9A | 110.0 |
| C3—C2—H2A | 109.6 | C8—C9—H9B | 110.0 |
| C1—C2—H2B | 109.6 | C10—C9—H9B | 110.0 |
| C3—C2—H2B | 109.6 | H9A—C9—H9B | 108.4 |
| H2A—C2—H2B | 108.1 | C1—C10—C9 | 112.13 (17) |
| C2—C3—C4 | 112.8 (2) | C1—C10—C5 | 110.24 (17) |
| C2—C3—H3A | 109.0 | C9—C10—C5 | 114.02 (16) |
| C4—C3—H3A | 109.0 | C1—C10—H10 | 106.7 |
| C2—C3—H3B | 109.0 | C9—C10—H10 | 106.7 |
| C4—C3—H3B | 109.0 | C5—C10—H10 | 106.7 |
| H3A—C3—H3B | 107.8 | C7—C11—C12 | 108.2 (2) |
| C15—C4—C3 | 109.7 (2) | C7—C11—C13 | 130.8 (3) |
| C15—C4—C5 | 113.8 (2) | C12—C11—C13 | 120.9 (2) |
| C3—C4—C5 | 111.89 (19) | O2—C12—O1 | 121.3 (3) |
| C15—C4—H4 | 107.0 | O2—C12—C11 | 129.7 (2) |
| C3—C4—H4 | 107.0 | O1—C12—C11 | 108.9 (2) |
| C5—C4—H4 | 107.0 | C11—C13—H13A | 109.5 |
| C14—C5—C4 | 111.40 (18) | C11—C13—H13B | 109.5 |
| C14—C5—C6 | 109.80 (18) | H13A—C13—H13B | 109.5 |
| C4—C5—C6 | 109.48 (18) | C11—C13—H13C | 109.5 |
| C14—C5—C10 | 111.15 (18) | H13A—C13—H13C | 109.5 |
| C4—C5—C10 | 107.89 (17) | H13B—C13—H13C | 109.5 |
| C6—C5—C10 | 106.99 (16) | C5—C14—H14A | 109.5 |
| C7—C6—C5 | 110.60 (17) | C5—C14—H14B | 109.5 |
| C7—C6—H6A | 109.5 | H14A—C14—H14B | 109.5 |
| C5—C6—H6A | 109.5 | C5—C14—H14C | 109.5 |
| C7—C6—H6B | 109.5 | H14A—C14—H14C | 109.5 |
| C5—C6—H6B | 109.5 | H14B—C14—H14C | 109.5 |
| H6A—C6—H6B | 108.1 | C4—C15—H15A | 109.5 |
| C11—C7—C6 | 132.6 (2) | C4—C15—H15B | 109.5 |
| C11—C7—C8 | 109.9 (2) | H15A—C15—H15B | 109.5 |
| C6—C7—C8 | 117.24 (18) | C4—C15—H15C | 109.5 |
| O3—C8—O1 | 109.53 (17) | H15A—C15—H15C | 109.5 |
| O3—C8—C7 | 114.85 (19) | H15B—C15—H15C | 109.5 |
| O4—C1—C2—C3 | 126.2 (3) | O3—C8—C9—C10 | −72.0 (2) |
| C10—C1—C2—C3 | −53.2 (3) | O1—C8—C9—C10 | 168.34 (16) |
| C1—C2—C3—C4 | 51.1 (3) | C7—C8—C9—C10 | 53.8 (2) |
| C2—C3—C4—C15 | 177.4 (2) | O4—C1—C10—C9 | 6.0 (3) |
| C2—C3—C4—C5 | −55.3 (3) | C2—C1—C10—C9 | −174.5 (2) |
| C15—C4—C5—C14 | 59.4 (3) | O4—C1—C10—C5 | −122.1 (2) |
| C3—C4—C5—C14 | −65.6 (3) | C2—C1—C10—C5 | 57.3 (2) |
| C15—C4—C5—C6 | −62.2 (3) | C8—C9—C10—C1 | 173.39 (16) |
| C3—C4—C5—C6 | 172.73 (19) | C8—C9—C10—C5 | −60.4 (2) |
| C15—C4—C5—C10 | −178.3 (2) | C14—C5—C10—C1 | 66.0 (2) |
| C3—C4—C5—C10 | 56.6 (2) | C4—C5—C10—C1 | −56.4 (2) |
| C14—C5—C6—C7 | 69.0 (2) | C6—C5—C10—C1 | −174.11 (18) |
| C4—C5—C6—C7 | −168.42 (18) | C14—C5—C10—C9 | −61.1 (2) |
| C10—C5—C6—C7 | −51.8 (2) | C4—C5—C10—C9 | 176.45 (16) |
| C5—C6—C7—C11 | −120.6 (2) | C6—C5—C10—C9 | 58.7 (2) |
| C5—C6—C7—C8 | 53.2 (2) | C6—C7—C11—C12 | 173.9 (2) |
| C12—O1—C8—O3 | 119.6 (2) | C8—C7—C11—C12 | −0.3 (2) |
| C12—O1—C8—C7 | −3.5 (2) | C6—C7—C11—C13 | −3.7 (4) |
| C12—O1—C8—C9 | −121.9 (2) | C8—C7—C11—C13 | −177.9 (2) |
| C11—C7—C8—O3 | −117.3 (2) | C8—O1—C12—O2 | −177.8 (2) |
| C6—C7—C8—O3 | 67.5 (2) | C8—O1—C12—C11 | 3.5 (3) |
| C11—C7—C8—O1 | 2.3 (2) | C7—C11—C12—O2 | 179.5 (3) |
| C6—C7—C8—O1 | −172.85 (17) | C13—C11—C12—O2 | −2.6 (4) |
| C11—C7—C8—C9 | 121.1 (2) | C7—C11—C12—O1 | −2.0 (3) |
| C6—C7—C8—C9 | −54.0 (2) | C13—C11—C12—O1 | 175.8 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O4i | 0.90 (4) | 1.87 (4) | 2.750 (3) | 164 (4) |
Symmetry codes: (i) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2542).
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst.32, 115–119.
- Burgueño-Tapia, E., González-Coloma, A., Martín-Benito, D. & Joseph-Nathan, P. Z. (2007). Z. Naturforsch. Teil C, 62,362–366. [DOI] [PubMed]
- Domínguez, D. M., Reina, M., Villarroel, L., Fajardo, V. & González-Coloma, A. (2008). Z. Naturforsch. Teil C, 63, 837–842. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Nonius (2000). KappaCCD Server Software, Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Reina, M., González-Coloma, A., Domínguez-Díaz, D. M., Cabrera, R., Giménez-Mariño, C., López-Rodríguez, M. & Villarroel, L. (2006). J. Nat. Prod.64, 6–11. [DOI] [PubMed]
- Reina, M., González-Coloma, A., Gutiérrez, C., Cabrera, R., López-Rodríguez, M., Fajardo, V. & Villarroel, L. (2006). Nat. Prod. Res.20, 13–19. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Ulubelen, A., Öksüs, S., Samek, Z. & Holub, M. (1971). Tetrahedron Lett.12, 4455–4456.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809033418/hg2542sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809033418/hg2542Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report