Abstract
In the title compound, C17H15N3O5S, the thiazine ring adopts a distorted half-chair conformation. The molecular structure is stabilized by intramolecular N—H⋯O, N—H⋯N and O—H⋯O hydrogen bonding. Pairs of molecules are bound together as centrosymmetric dimers through N—H⋯O hydrogen bonds.
Related literature
For the synthesis of related molecules, see: Braun (1923 ▶); Ahmad et al. (2008 ▶); Zia-ur-Rehman et al. (2005 ▶, 2009 ▶). For the biological activity of 1,2-benzothiazine 1,1-dioxides, see: Bihovsky et al. (2004 ▶); Turck et al. (1996 ▶); Zia-ur-Rehman et al. (2006 ▶). For similar molecules, see: Kojić-Prodić & Rużić-Toroš (1982 ▶); Siddiqui et al. (2009 ▶); Weast et al. (1984 ▶); Zia-ur-Rehman et al. (2007 ▶).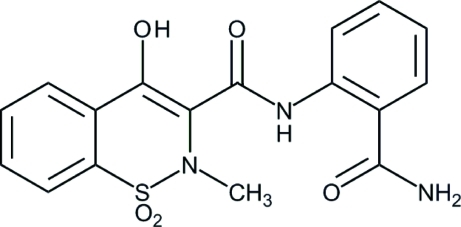
Experimental
Crystal data
C17H15N3O5S
M r = 373.38
Monoclinic,

a = 8.1377 (6) Å
b = 7.0515 (6) Å
c = 29.069 (2) Å
β = 96.502 (3)°
V = 1657.3 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 296 K
0.39 × 0.25 × 0.11 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2007 ▶) T min = 0.915, T max = 0.975
16109 measured reflections
3753 independent reflections
3011 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.087
wR(F 2) = 0.213
S = 1.09
3753 reflections
237 parameters
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.40 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809038951/bt5073sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809038951/bt5073Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O4 | 0.82 | 1.85 | 2.569 (5) | 145 |
| N2—H2⋯O5 | 0.86 | 1.92 | 2.607 (5) | 136 |
| N2—H2⋯N1 | 0.86 | 2.28 | 2.728 (5) | 113 |
| N3—H3A⋯O5i | 0.86 | 2.22 | 2.941 (6) | 141 |
Symmetry code: (i)  .
.
Acknowledgments
The authors are grateful to the PCSIR Laboratories Complex, Lahore, for the provision of facilities necessary to complete this work.
supplementary crystallographic information
Comment
Owing to the verstaile applications of 1,2-benzothiazine 1,1-dioxides, considerable attention has been given to their synthesis since their very first synthesis (Braun, 1923). Among these, Piroxicam (Zia-ur-Rehman et al., 2005), and Meloxicam (Turck et al., 1996) are familiar for their analgesic action and are being used world wide as non-steroidal anti-inflammatory drugs (NSAIDs). Some of the 3,4-dihydro-1,2-benzothiazine-3-carboxylate 1,1-dioxide α-ketomide and P(2)—P(3) peptide mimetic aldehyde compounds act as potent calpain I inhibitors (Bihovsky et al., 2004) while 1,2-benzothiazin-3-yl-quinazolin-4(3H)-ones possess anti-bacterial properties (Zia-ur-Rehman et al., 2006).
In continuation of our work on the synthesis (Zia-ur-Rehman et al., 2006, biological activity (Zia-ur-Rehman et al., 2009) and crystal structures (Zia-ur-Rehman et al., 2007; Ahmad et al., 2008, Siddiqui et al., 2009) of various 1,2-benzothiazine-1,1-dioxides, we herein report the crystal structure of the title compound (I) (Scheme and figure 1). Like its already reported dimethylsulfoxide solvate analogue (Zia-ur-Rehman et al., 2007), thiazine ring involving two double bonds, exhibits sofa conformation; with S1/C1/C2/C7 relatively planar and N1 showing significant departure from plane due to its pyramidal geometry. The enolic hydrogen on O1 is involved in intramolecular hydrogen bonding [O1—H1···O4] with the carbonyl oxygen at C9 giving rise to a six-membered hydrogen bond ring (Table 1). Atom H2 forms hydrogen bonds with both N1 and O5 giving rise to five and six-membered hydrogen bond rings respectively. The C1—S1 [1.755 Å] bond is shorter than a normal C—S single bond (1.81–2.55 Å) (Weast et al., 1984) due to partial double bond character and is in agreement with similar molecules (Kojić-Prodić & Rużić-Toroš, 1982). Each molecule is centrosymmetrically linked to its adjacent one forming a dimer through intermolecular [N—H3B···O5] hydrogen bonds (Fig. 2).
Experimental
N-[2-(Aminocarbonyl)phenyl]-4-hydroxy-2-methyl-2H-1,2-benzothiazine- 3-carboxamide 1,1-dioxide was synthesized according to a literature method (Zia-ur-Rehman et al., 2006). Suitable crystals were obtained by dissoving the compound in chloroform followed by slow evaporation at room temperature.The compound was dissolved in a mixture of methanol and DMSO (80:20 v/v) at room temperature. Crystals were obtained by slow evaporation and dried under high vacuum.
Refinement
All hydrogen atoms were identified in the difference map. They were refined using a riding model with O—H = 0.84 Å, N—H = 0.86 Å, Cmethyl—H 0.98 Å and Caromatic—H = 0.95 Å. and U(H) set to 1.2Ueq of the parent atoms or set to 1.5Ueq(Cmethyl).
Figures
Fig. 1.
The molecular structure of the title compound with displacement ellipsoids at the 50% probability level.
Fig. 2.
Perspective view of the three-dimensional crystal packing showing hydrogen-bonded interactions (dashed lines). H atoms not involved in hydrogen bonding have been omitted for clarity.
Crystal data
| C17H15N3O5S | F(000) = 776 |
| Mr = 373.38 | Dx = 1.496 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6164 reflections |
| a = 8.1377 (6) Å | θ = 2.5–27.1° |
| b = 7.0515 (6) Å | µ = 0.23 mm−1 |
| c = 29.069 (2) Å | T = 296 K |
| β = 96.502 (3)° | Needles, colourless |
| V = 1657.3 (2) Å3 | 0.39 × 0.25 × 0.11 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 3753 independent reflections |
| Radiation source: fine-focus sealed tube | 3011 reflections with I > 2σ(I) |
| graphite | Rint = 0.039 |
| φ and ω scans | θmax = 27.5°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2007) | h = −10→10 |
| Tmin = 0.915, Tmax = 0.975 | k = −9→9 |
| 16109 measured reflections | l = −32→37 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.087 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.213 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0335P)2 + 9.427P] where P = (Fo2 + 2Fc2)/3 |
| 3753 reflections | (Δ/σ)max < 0.001 |
| 237 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.40 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.35684 (14) | 0.5031 (2) | 0.33289 (4) | 0.0343 (3) | |
| O2 | 0.2977 (5) | 0.6850 (5) | 0.34450 (12) | 0.0427 (9) | |
| O3 | 0.5306 (4) | 0.4668 (7) | 0.33922 (13) | 0.0538 (11) | |
| O4 | −0.1641 (4) | 0.3152 (6) | 0.38428 (12) | 0.0480 (10) | |
| O5 | 0.3670 (4) | 0.3198 (6) | 0.47838 (12) | 0.0484 (10) | |
| N1 | 0.2640 (5) | 0.3427 (6) | 0.36214 (13) | 0.0310 (9) | |
| N2 | 0.0743 (5) | 0.2848 (6) | 0.43247 (12) | 0.0306 (8) | |
| H2 | 0.1801 | 0.2826 | 0.4328 | 0.037* | |
| N3 | 0.3804 (6) | 0.4241 (8) | 0.55160 (15) | 0.0534 (13) | |
| H3A | 0.4817 | 0.4585 | 0.5513 | 0.064* | |
| H3B | 0.3319 | 0.4407 | 0.5761 | 0.064* | |
| O1 | −0.1513 (4) | 0.3895 (6) | 0.29823 (12) | 0.0436 (9) | |
| H1 | −0.1968 | 0.3747 | 0.3218 | 0.065* | |
| C1 | 0.2765 (6) | 0.4493 (7) | 0.27578 (16) | 0.0329 (10) | |
| C2 | 0.1069 (6) | 0.4046 (7) | 0.26901 (16) | 0.0322 (10) | |
| C3 | 0.0347 (8) | 0.3766 (8) | 0.22406 (17) | 0.0445 (13) | |
| H3 | −0.0780 | 0.3516 | 0.2183 | 0.053* | |
| C4 | 0.1301 (9) | 0.3861 (9) | 0.18781 (19) | 0.0577 (17) | |
| H4 | 0.0805 | 0.3685 | 0.1577 | 0.069* | |
| C5 | 0.2966 (9) | 0.4210 (10) | 0.19540 (19) | 0.0580 (17) | |
| H5 | 0.3594 | 0.4221 | 0.1706 | 0.070* | |
| C6 | 0.3718 (7) | 0.4546 (9) | 0.23979 (19) | 0.0470 (14) | |
| H6 | 0.4845 | 0.4802 | 0.2451 | 0.056* | |
| C7 | 0.0124 (6) | 0.3801 (7) | 0.30885 (16) | 0.0314 (10) | |
| C8 | 0.0874 (6) | 0.3479 (7) | 0.35254 (16) | 0.0308 (10) | |
| C9 | −0.0106 (6) | 0.3133 (7) | 0.39093 (15) | 0.0315 (10) | |
| C10 | 0.0150 (6) | 0.2579 (6) | 0.47583 (15) | 0.0287 (9) | |
| C11 | 0.1236 (6) | 0.2888 (7) | 0.51660 (16) | 0.0323 (10) | |
| C12 | 0.0636 (7) | 0.2643 (7) | 0.55876 (17) | 0.0402 (12) | |
| H12 | 0.1350 | 0.2810 | 0.5858 | 0.048* | |
| C13 | −0.0980 (7) | 0.2160 (8) | 0.56205 (18) | 0.0441 (13) | |
| H13 | −0.1358 | 0.2046 | 0.5909 | 0.053* | |
| C14 | −0.2039 (7) | 0.1845 (8) | 0.5223 (2) | 0.0448 (13) | |
| H14 | −0.3134 | 0.1514 | 0.5245 | 0.054* | |
| C15 | −0.1485 (6) | 0.2019 (7) | 0.47945 (17) | 0.0374 (11) | |
| H15 | −0.2197 | 0.1764 | 0.4528 | 0.045* | |
| C16 | 0.2986 (6) | 0.3449 (8) | 0.51412 (16) | 0.0368 (11) | |
| C17 | 0.3393 (7) | 0.1508 (9) | 0.3632 (2) | 0.0521 (15) | |
| H17A | 0.3025 | 0.0790 | 0.3882 | 0.078* | |
| H17B | 0.4576 | 0.1619 | 0.3678 | 0.078* | |
| H17C | 0.3065 | 0.0873 | 0.3344 | 0.078* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0273 (5) | 0.0450 (7) | 0.0309 (6) | −0.0003 (5) | 0.0047 (4) | 0.0006 (5) |
| O2 | 0.048 (2) | 0.041 (2) | 0.0391 (19) | −0.0070 (17) | 0.0065 (16) | −0.0037 (16) |
| O3 | 0.0284 (18) | 0.084 (3) | 0.049 (2) | 0.000 (2) | 0.0067 (16) | 0.010 (2) |
| O4 | 0.0289 (18) | 0.079 (3) | 0.0368 (19) | 0.0027 (19) | 0.0076 (14) | 0.0050 (19) |
| O5 | 0.0350 (19) | 0.075 (3) | 0.0354 (19) | −0.0012 (19) | 0.0067 (15) | −0.0072 (19) |
| N1 | 0.0278 (19) | 0.038 (2) | 0.0276 (19) | 0.0065 (17) | 0.0039 (15) | 0.0006 (16) |
| N2 | 0.0260 (19) | 0.039 (2) | 0.0280 (19) | 0.0001 (17) | 0.0063 (15) | 0.0053 (17) |
| N3 | 0.042 (3) | 0.082 (4) | 0.036 (2) | −0.006 (3) | 0.0011 (19) | −0.009 (2) |
| O1 | 0.0307 (18) | 0.065 (3) | 0.0335 (18) | −0.0013 (18) | −0.0011 (14) | 0.0024 (18) |
| C1 | 0.040 (3) | 0.032 (2) | 0.028 (2) | 0.001 (2) | 0.0079 (19) | 0.0016 (18) |
| C2 | 0.037 (2) | 0.031 (2) | 0.029 (2) | −0.001 (2) | 0.0064 (19) | 0.0005 (19) |
| C3 | 0.063 (4) | 0.042 (3) | 0.029 (2) | −0.005 (3) | 0.005 (2) | −0.002 (2) |
| C4 | 0.087 (5) | 0.056 (4) | 0.029 (3) | −0.013 (3) | 0.004 (3) | −0.005 (3) |
| C5 | 0.076 (4) | 0.071 (4) | 0.031 (3) | −0.005 (4) | 0.024 (3) | −0.004 (3) |
| C6 | 0.046 (3) | 0.057 (4) | 0.041 (3) | −0.003 (3) | 0.016 (2) | −0.001 (3) |
| C7 | 0.030 (2) | 0.034 (2) | 0.030 (2) | 0.0008 (19) | 0.0067 (18) | −0.0015 (19) |
| C8 | 0.028 (2) | 0.034 (2) | 0.030 (2) | 0.0032 (19) | 0.0029 (17) | 0.0000 (19) |
| C9 | 0.034 (2) | 0.032 (2) | 0.029 (2) | 0.0000 (19) | 0.0034 (18) | −0.0003 (19) |
| C10 | 0.032 (2) | 0.026 (2) | 0.028 (2) | 0.0039 (18) | 0.0082 (18) | 0.0025 (18) |
| C11 | 0.039 (3) | 0.028 (2) | 0.031 (2) | 0.004 (2) | 0.0072 (19) | 0.0029 (19) |
| C12 | 0.058 (3) | 0.035 (3) | 0.029 (2) | 0.002 (2) | 0.011 (2) | 0.001 (2) |
| C13 | 0.063 (4) | 0.039 (3) | 0.035 (3) | 0.000 (3) | 0.023 (2) | 0.003 (2) |
| C14 | 0.045 (3) | 0.039 (3) | 0.054 (3) | −0.002 (2) | 0.021 (3) | 0.005 (3) |
| C15 | 0.036 (3) | 0.039 (3) | 0.039 (3) | 0.000 (2) | 0.012 (2) | 0.007 (2) |
| C16 | 0.034 (3) | 0.044 (3) | 0.031 (2) | 0.006 (2) | −0.0011 (19) | 0.003 (2) |
| C17 | 0.052 (3) | 0.049 (3) | 0.058 (4) | 0.020 (3) | 0.020 (3) | 0.014 (3) |
Geometric parameters (Å, °)
| S1—O2 | 1.424 (4) | C4—C5 | 1.371 (9) |
| S1—O3 | 1.428 (4) | C4—H4 | 0.9300 |
| S1—N1 | 1.648 (4) | C5—C6 | 1.384 (8) |
| S1—C1 | 1.755 (5) | C5—H5 | 0.9300 |
| O4—C9 | 1.242 (6) | C6—H6 | 0.9300 |
| O5—C16 | 1.246 (6) | C7—C8 | 1.364 (6) |
| N1—C8 | 1.433 (6) | C8—C9 | 1.464 (6) |
| N1—C17 | 1.484 (7) | C10—C15 | 1.403 (6) |
| N2—C9 | 1.337 (6) | C10—C11 | 1.413 (6) |
| N2—C10 | 1.412 (5) | C11—C12 | 1.380 (6) |
| N2—H2 | 0.8600 | C11—C16 | 1.488 (7) |
| N3—C16 | 1.333 (6) | C12—C13 | 1.372 (8) |
| N3—H3A | 0.8600 | C12—H12 | 0.9300 |
| N3—H3B | 0.8600 | C13—C14 | 1.378 (8) |
| O1—C7 | 1.335 (5) | C13—H13 | 0.9300 |
| O1—H1 | 0.8200 | C14—C15 | 1.379 (7) |
| C1—C6 | 1.372 (7) | C14—H14 | 0.9300 |
| C1—C2 | 1.407 (7) | C15—H15 | 0.9300 |
| C2—C3 | 1.385 (7) | C17—H17A | 0.9600 |
| C2—C7 | 1.471 (6) | C17—H17B | 0.9600 |
| C3—C4 | 1.379 (8) | C17—H17C | 0.9600 |
| C3—H3 | 0.9300 | ||
| O2—S1—O3 | 119.2 (3) | O1—C7—C2 | 114.2 (4) |
| O2—S1—N1 | 108.0 (2) | C8—C7—C2 | 122.3 (4) |
| O3—S1—N1 | 108.4 (2) | C7—C8—N1 | 121.2 (4) |
| O2—S1—C1 | 108.6 (2) | C7—C8—C9 | 120.8 (4) |
| O3—S1—C1 | 109.9 (2) | N1—C8—C9 | 117.9 (4) |
| N1—S1—C1 | 101.3 (2) | O4—C9—N2 | 123.4 (4) |
| C8—N1—C17 | 115.4 (4) | O4—C9—C8 | 120.3 (4) |
| C8—N1—S1 | 113.0 (3) | N2—C9—C8 | 116.3 (4) |
| C17—N1—S1 | 115.1 (3) | C15—C10—N2 | 121.8 (4) |
| C9—N2—C10 | 129.2 (4) | C15—C10—C11 | 119.3 (4) |
| C9—N2—H2 | 115.4 | N2—C10—C11 | 118.9 (4) |
| C10—N2—H2 | 115.4 | C12—C11—C10 | 118.4 (5) |
| C16—N3—H3A | 120.0 | C12—C11—C16 | 120.9 (5) |
| C16—N3—H3B | 120.0 | C10—C11—C16 | 120.8 (4) |
| H3A—N3—H3B | 120.0 | C13—C12—C11 | 122.1 (5) |
| C7—O1—H1 | 109.5 | C13—C12—H12 | 119.0 |
| C6—C1—C2 | 122.0 (5) | C11—C12—H12 | 119.0 |
| C6—C1—S1 | 122.2 (4) | C12—C13—C14 | 119.7 (5) |
| C2—C1—S1 | 115.8 (3) | C12—C13—H13 | 120.2 |
| C3—C2—C1 | 118.0 (5) | C14—C13—H13 | 120.2 |
| C3—C2—C7 | 121.5 (5) | C13—C14—C15 | 120.4 (5) |
| C1—C2—C7 | 120.5 (4) | C13—C14—H14 | 119.8 |
| C4—C3—C2 | 119.9 (6) | C15—C14—H14 | 119.8 |
| C4—C3—H3 | 120.1 | C14—C15—C10 | 120.2 (5) |
| C2—C3—H3 | 120.1 | C14—C15—H15 | 119.9 |
| C5—C4—C3 | 121.2 (5) | C10—C15—H15 | 119.9 |
| C5—C4—H4 | 119.4 | O5—C16—N3 | 120.8 (5) |
| C3—C4—H4 | 119.4 | O5—C16—C11 | 121.6 (4) |
| C4—C5—C6 | 120.4 (5) | N3—C16—C11 | 117.6 (4) |
| C4—C5—H5 | 119.8 | N1—C17—H17A | 109.5 |
| C6—C5—H5 | 119.8 | N1—C17—H17B | 109.5 |
| C1—C6—C5 | 118.5 (5) | H17A—C17—H17B | 109.5 |
| C1—C6—H6 | 120.7 | N1—C17—H17C | 109.5 |
| C5—C6—H6 | 120.7 | H17A—C17—H17C | 109.5 |
| O1—C7—C8 | 123.6 (4) | H17B—C17—H17C | 109.5 |
| O2—S1—N1—C8 | 58.6 (4) | O1—C7—C8—C9 | 2.6 (8) |
| O3—S1—N1—C8 | −171.0 (3) | C2—C7—C8—C9 | −176.4 (4) |
| C1—S1—N1—C8 | −55.4 (4) | C17—N1—C8—C7 | −96.6 (6) |
| O2—S1—N1—C17 | −165.8 (4) | S1—N1—C8—C7 | 38.8 (6) |
| O3—S1—N1—C17 | −35.4 (4) | C17—N1—C8—C9 | 82.0 (5) |
| C1—S1—N1—C17 | 80.2 (4) | S1—N1—C8—C9 | −142.6 (4) |
| O2—S1—C1—C6 | 106.1 (5) | C10—N2—C9—O4 | −2.0 (8) |
| O3—S1—C1—C6 | −25.9 (5) | C10—N2—C9—C8 | 176.5 (4) |
| N1—S1—C1—C6 | −140.4 (5) | C7—C8—C9—O4 | −0.9 (8) |
| O2—S1—C1—C2 | −72.5 (4) | N1—C8—C9—O4 | −179.6 (5) |
| O3—S1—C1—C2 | 155.5 (4) | C7—C8—C9—N2 | −179.5 (5) |
| N1—S1—C1—C2 | 41.0 (4) | N1—C8—C9—N2 | 1.9 (7) |
| C6—C1—C2—C3 | −3.9 (8) | C9—N2—C10—C15 | 19.3 (8) |
| S1—C1—C2—C3 | 174.7 (4) | C9—N2—C10—C11 | −160.5 (5) |
| C6—C1—C2—C7 | 173.4 (5) | C15—C10—C11—C12 | −0.6 (7) |
| S1—C1—C2—C7 | −8.0 (6) | N2—C10—C11—C12 | 179.2 (4) |
| C1—C2—C3—C4 | 2.4 (8) | C15—C10—C11—C16 | 178.9 (4) |
| C7—C2—C3—C4 | −174.9 (5) | N2—C10—C11—C16 | −1.3 (7) |
| C2—C3—C4—C5 | 0.7 (10) | C10—C11—C12—C13 | −1.7 (8) |
| C3—C4—C5—C6 | −2.4 (11) | C16—C11—C12—C13 | 178.8 (5) |
| C2—C1—C6—C5 | 2.2 (9) | C11—C12—C13—C14 | 2.2 (8) |
| S1—C1—C6—C5 | −176.3 (5) | C12—C13—C14—C15 | −0.2 (8) |
| C4—C5—C6—C1 | 0.9 (10) | C13—C14—C15—C10 | −2.1 (8) |
| C3—C2—C7—O1 | −20.1 (7) | N2—C10—C15—C14 | −177.3 (5) |
| C1—C2—C7—O1 | 162.7 (5) | C11—C10—C15—C14 | 2.5 (7) |
| C3—C2—C7—C8 | 159.0 (5) | C12—C11—C16—O5 | 160.7 (5) |
| C1—C2—C7—C8 | −18.2 (7) | C10—C11—C16—O5 | −18.7 (8) |
| O1—C7—C8—N1 | −178.8 (4) | C12—C11—C16—N3 | −19.5 (7) |
| C2—C7—C8—N1 | 2.1 (7) | C10—C11—C16—N3 | 161.1 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O4 | 0.82 | 1.85 | 2.569 (5) | 145 |
| N2—H2···O5 | 0.86 | 1.92 | 2.607 (5) | 136 |
| N2—H2···N1 | 0.86 | 2.28 | 2.728 (5) | 113 |
| N3—H3A···O5i | 0.86 | 2.22 | 2.941 (6) | 141 |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5073).
References
- Ahmad, M., Siddiqui, H. L., Zia-ur-Rehman, M., Ashiq, M. I. & Tizzard, G. J. (2008). Acta Cryst. E64, o788. [DOI] [PMC free article] [PubMed]
- Bihovsky, R., Tao, M., Mallamo, J. P. & Wells, G. J. (2004). Bioorg. Med. Chem. Lett 14, 1035–1038. [DOI] [PubMed]
- Braun, J. (1923). Chem. Ber 56, 2332–2343.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Kojić-Prodić, B. & Rużić-Toroš, Ž. (1982). Acta Cryst. B38, 2948–2951.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Sheldrick, G. M. (2007). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siddiqui, W. A., Ali, M., Zia-ur-Rehman, M., Sharif, S. & Tizzard, G. J. (2009). Acta Cryst. E65, o900–o901. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Turck, D., Busch, U., Heinzel, G., Narjes, H. & Nehmiz, G. (1996). J. Clin. Pharmacol 36, 79–84. [DOI] [PubMed]
- Weast, R. C., Astle, M. J. & Beyer, W. H. (1984). Handbook of Chemistry and Physics, 65th ed. Boca Raton, Florida: CRC Press.
- Zia-ur-Rehman, M., Choudary, J. A. & Ahmad, S. (2005). Bull. Korean Chem. Soc 26,1771–1775.
- Zia-ur-Rehman, M., Choudary, J. A., Ahmad, S. & Siddiqui, H. L. (2006). Chem. Pharm. Bull 54, 1175–1178. [DOI] [PubMed]
- Zia-ur-Rehman, M., Choudary, J. A., Elsegood, M. R. J., Siddiqui, H. L. & Ahmad, S. (2007). Acta Cryst. E63, o900–o901.
- Zia-ur-Rehman, M., Choudary, J. A., Elsegood, M. R. J., Siddiqui, H. L. & Khan, K. M. (2009). Eur. J. Med. Chem 44, 1311–1316. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809038951/bt5073sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809038951/bt5073Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




