Abstract
In this paper we propose a novel approach for the mapping of 3D surfaces. With the Reeb graph of Laplace-Beltrami eigenmaps, our method automatically detects stable landmark features intrinsic to the surface geometry and use them as boundary conditions to compute harmonic maps to the unit sphere. The resulting maps are diffeomorphic, robust to natural pose variations, and establish correspondences for geometric features shared across population. In the experiments, we demonstrate our method on three subcortical structures: the hippocampus, putamen, and caudate nucleus. A group study is also performed to generate a statistically significant map of local volume losses in the hippocampus of patients with secondary progressive multiple sclerosis.
1 Introduction
In many imaging studies, surface mapping plays an important role as it can provide localized information complementary to volume measurements [1]. The task of mapping general 3D surface models, however, remains challenging because it is usually difficult to define homologous points across population. In this paper, we propose a novel method of computing maps for a class of subcortical structures by using Laplace-Beltrami eigenmaps to capture their salient geometry and harmonic maps to establish diffeomorphic correspondences.
Various approaches have been proposed in previous work for the automated construction of surface maps. The first approach computes surface maps based on a canonical parameterization such as the spherical parameterization [2–5]. Using techniques from image registration, a map between two surfaces can be established by warping the image space surrounding the anatomical structure of interest [6–8]. The medial model, or skeleton, is a popular tool to represent shapes and it can also be used to construct geometrically intuitive maps between surfaces [9]. Because the skeleton is sensitive to noise, a simplified topology was usually assumed for robustness and consistency [10, 11].
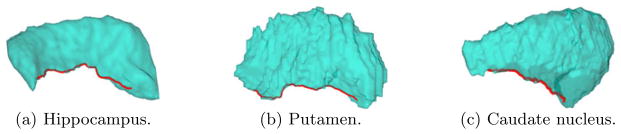
In this work we propose a novel approach for the automated mapping of three subcortical structures: the hippocampus, putamen and caudate nucleus. As shown in Fig. 1, these shapes share a similar profile globally. For each shape, we can consider its thinner part on the left as the “tail”, and the thicker part on the right as the “head”. This kind of division can also be useful in anatomical and functional studies. Not only globally, these shapes also have locally similar ridge features as highlighted by the red curve in each picture. By using Laplace-Beltrami eigenmaps, we describe in section 2 an automated approach to capture these salient features. After that, the landmarks are used to guide harmonic maps to the sphere in section 3 and generate feature-aligned, one-to-one correspondences across shapes. Experimental results will be presented to demonstrate our algorithm in section 4. Finally, conclusions are made in section 5.
Fig. 1.
Examples of subcortical structures.
2 Laplace-Beltrami Eigenmaps of Surfaces
Given a surface  , we compute a function f:
, we compute a function f:  → ℝ that maps
→ ℝ that maps  smoothly into ℝ by solving
smoothly into ℝ by solving
| (1) |
where 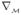 is the intrinsic gradient operator on
is the intrinsic gradient operator on  . Using Stokes’ theorem, we have
. Using Stokes’ theorem, we have
| (2) |
where 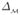 is the Laplace-Beltrami operator on
is the Laplace-Beltrami operator on  . For a manifold with boundary, the above equation still holds if we choose the Neumann boundary condition. Based on (2), we find the optimal map f by considering the spectrum of the operator
. For a manifold with boundary, the above equation still holds if we choose the Neumann boundary condition. Based on (2), we find the optimal map f by considering the spectrum of the operator 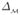 , which is discrete for a compact manifold. Let us denote the eigenvalues of
, which is discrete for a compact manifold. Let us denote the eigenvalues of 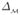 as λ0 ≤ λ1 ≤ λ2 ≤ ··· and the corresponding eigenfunctions as f0, f1, f2, ···. In previous work on shape analysis, the set of eigenvalues was used for classification [12] and the eigenfunctions were used for denoising [13]. Here we use the spectrum to characterize the salient geometry of
as λ0 ≤ λ1 ≤ λ2 ≤ ··· and the corresponding eigenfunctions as f0, f1, f2, ···. In previous work on shape analysis, the set of eigenvalues was used for classification [12] and the eigenfunctions were used for denoising [13]. Here we use the spectrum to characterize the salient geometry of  . For λ0 = 0, the eigenfunction f0 is constant. So the smoothest and non-trivial map from
. For λ0 = 0, the eigenfunction f0 is constant. So the smoothest and non-trivial map from  to ℝ is f1 because it achieves the minimal energy λ1 =
to ℝ is f1 because it achieves the minimal energy λ1 = 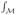 ||
|| 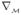 f1||2 d
f1||2 d .
.
For compact manifolds, a generic property of the Laplace-Beltrami operator is that its eigenfunctions are Morse functions [14]. This motivates us to use the Reeb graph [15] of f1 to capture the global property of the surface intrinsically. For the eigenmap f1:  → ℝ, its Reeb graph is defined as the quotient space of
→ ℝ, its Reeb graph is defined as the quotient space of  × ℝ with the equivalent relation (x1, f1(x1)) ≃ (x2, f1(x2)) for x1, x2 ∈
× ℝ with the equivalent relation (x1, f1(x1)) ≃ (x2, f1(x2)) for x1, x2 ∈  . The structure of the Reeb graph is closely related to the global characteristics, such as topology, of the manifold
. The structure of the Reeb graph is closely related to the global characteristics, such as topology, of the manifold  . For example, the number of loops in the Reeb graph of a Morse function on
. For example, the number of loops in the Reeb graph of a Morse function on  equals its number of genus. Besides that, we can see below it reveals richer structural similarities between different shapes, such as the hippocampus and the sphere. To build the Reeb graph numerically, we assume the surface
equals its number of genus. Besides that, we can see below it reveals richer structural similarities between different shapes, such as the hippocampus and the sphere. To build the Reeb graph numerically, we assume the surface  is represented as a triangular mesh and compute its Laplace-Beltrami spectrum with the finite element method [12, 13]. As a result, the eigenmap f1 is defined on each vertex of
is represented as a triangular mesh and compute its Laplace-Beltrami spectrum with the finite element method [12, 13]. As a result, the eigenmap f1 is defined on each vertex of  . Let f1 ∈ [fmin, fmax]. We trace the level sets of f1 on
. Let f1 ∈ [fmin, fmax]. We trace the level sets of f1 on  at K values (α1, α2, ···, αK) such that fmin = α0 < α1 < α2 < ··· < αK < fmax = αK+1 and
at K values (α1, α2, ···, αK) such that fmin = α0 < α1 < α2 < ··· < αK < fmax = αK+1 and
| (3) |
where 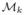 = {x ∈
= {x ∈  |αk < f1(x) ≤ αk+1}. This generates a set of level contours distributing evenly spaced on the surface. Note that each level set can be composed of multiple contours for arbitrary surfaces. With each contour as a node in the Reeb graph, two neighboring level contours are connected with an edge if they can deform into each other without crossing other contours.
|αk < f1(x) ≤ αk+1}. This generates a set of level contours distributing evenly spaced on the surface. Note that each level set can be composed of multiple contours for arbitrary surfaces. With each contour as a node in the Reeb graph, two neighboring level contours are connected with an edge if they can deform into each other without crossing other contours.
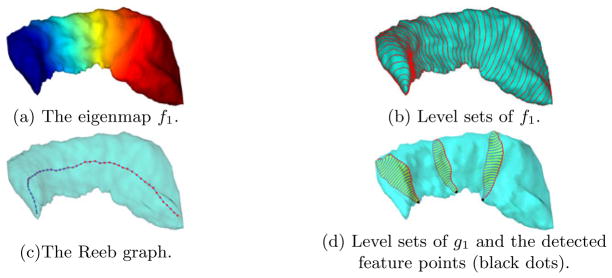
With the hippocampus in Fig. 1(a) as an example, we visualize in Fig. 2(a), (b), (c) the eigenmap f1, its level contours, and the Reeb graph, respectively. If we assume the hippocampus is aligned in Talairach orientation and fix the sign of f1 to be negative at the most posterior vertex of  , we have the Reeb graph as an explicit representation of the simple, tail-to-head structure of the hippocampus. As we traverse the graph from the node representing the level set f1 = α1 to the other end, we move from the tail of the hippocampus to its head. Interestingly, if we compute the Laplace-Beltrami eigenmap of the sphere, its Reeb graph also has a chain structure as in Fig. 2(c). Using the Reeb graph of the Laplace-Beltrami eigenmap, we thus have a richer and intrinsic characterization of structural similarities between shapes beyond topology. In this case, it provides a deeper justification of using spherical parameterizations for the mapping of hippocampal surfaces besides the fact they both have genus-zero topology. On the other hand, it might improve our understanding of the difficulties in building spherical parameterizations for more irregular shapes and eventually lead to the selection of more suitable parameterization domains.
, we have the Reeb graph as an explicit representation of the simple, tail-to-head structure of the hippocampus. As we traverse the graph from the node representing the level set f1 = α1 to the other end, we move from the tail of the hippocampus to its head. Interestingly, if we compute the Laplace-Beltrami eigenmap of the sphere, its Reeb graph also has a chain structure as in Fig. 2(c). Using the Reeb graph of the Laplace-Beltrami eigenmap, we thus have a richer and intrinsic characterization of structural similarities between shapes beyond topology. In this case, it provides a deeper justification of using spherical parameterizations for the mapping of hippocampal surfaces besides the fact they both have genus-zero topology. On the other hand, it might improve our understanding of the difficulties in building spherical parameterizations for more irregular shapes and eventually lead to the selection of more suitable parameterization domains.
Fig. 2.
The Laplace-Beltrami eigenmaps of a hippocampal surface.
With the Reeb graph providing information about the global structure of a surface, we compute a second Laplace-Beltrami eigenmap to detect the point on level contours that marks the salient local feature highlighted in Fig. 1. For this purpose, we represent a contour as a set of L points xl(1 ≤ l ≤ L) and build a smooth mesh in ℝ3 with the contour as the boundary. As a first step, we construct a Delaunay triangulation from these points using the software triangle [16]. The final smooth mesh 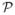 is then obtained by applying Laplacian smoothing to vertices belonging to the interior of this mesh while fixing the boundary points xl. Using the Neumann boundary condition, we map this surface patch to ℝ by computing the first eigenfunction of the Laplace-Beltrami operator on
is then obtained by applying Laplacian smoothing to vertices belonging to the interior of this mesh while fixing the boundary points xl. Using the Neumann boundary condition, we map this surface patch to ℝ by computing the first eigenfunction of the Laplace-Beltrami operator on 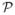 , which we denote as g1. As in the first eigenmap, we remove the ambiguity in the sign of g1 by fixing it to be negative at the most lateral vertex in
, which we denote as g1. As in the first eigenmap, we remove the ambiguity in the sign of g1 by fixing it to be negative at the most lateral vertex in 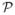 .
.
As an illustration, we visualize the second eigenmap in Fig. 2(d) by plotting the level sets of the eigenmap g1 on three interpolated surface patches. From the results we can see the second eigenmap g1 projects each patch along the medial-to-lateral direction for brains in Talairach orientation, and we can locate the feature point on the ridge by picking the point on the level contour attaining the maximal values in g1. To detect the whole ridge line, we compute the above eigenmap g1 for the level sets of f1 at the value αk for K1 ≤ k ≤ K2 and connect their feature points sequentially to form the landmark curve 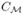 shown as the red contour in Fig. 1(a). For the subcortical surfaces under study here, the ridge feature becomes less salient as we approach the tail or head part, thus we choose K1 > 1 and K2 < K to pick out the most distinguished part of the ridge line and use it to guide the mapping process. For all surfaces tested so far in our work, we sample K = 100 level contours from the first eigenmap and find that choosing K1 = 20, K2 = 85 gives robust results in the second embedding. More generally, these parameters may as well be determined with a learning-based approach.
shown as the red contour in Fig. 1(a). For the subcortical surfaces under study here, the ridge feature becomes less salient as we approach the tail or head part, thus we choose K1 > 1 and K2 < K to pick out the most distinguished part of the ridge line and use it to guide the mapping process. For all surfaces tested so far in our work, we sample K = 100 level contours from the first eigenmap and find that choosing K1 = 20, K2 = 85 gives robust results in the second embedding. More generally, these parameters may as well be determined with a learning-based approach.
3 Spherical Mapping With Landmark Constraints
Following the landmark detection process with Laplace-Beltrami eigenmaps, we compute a harmonic map from each surface to the sphere. By combining these harmonic maps, we can obtain diffeomorphic maps between different surfaces[17].
To ensure the one-to-one correspondences on the landmark curve 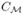 of different surfaces are maintained in their spherical parameterizations, we sample a curve
of different surfaces are maintained in their spherical parameterizations, we sample a curve 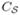 = {(θ, φ)|0.25π ≤ θ ≤ 0.75π, φ = 0} on the unit sphere
= {(θ, φ)|0.25π ≤ θ ≤ 0.75π, φ = 0} on the unit sphere 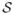 with K2 − K1 + 1 evenly spaced points and define a boundary condition by fixing the map from each landmark point on
with K2 − K1 + 1 evenly spaced points and define a boundary condition by fixing the map from each landmark point on 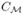 to the corresponding point on
to the corresponding point on 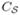 with the same index. Besides the boundary condition, the Laplace-Beltrami eigenmaps also provide a convenient way of estimating a good initial map from
with the same index. Besides the boundary condition, the Laplace-Beltrami eigenmaps also provide a convenient way of estimating a good initial map from  to
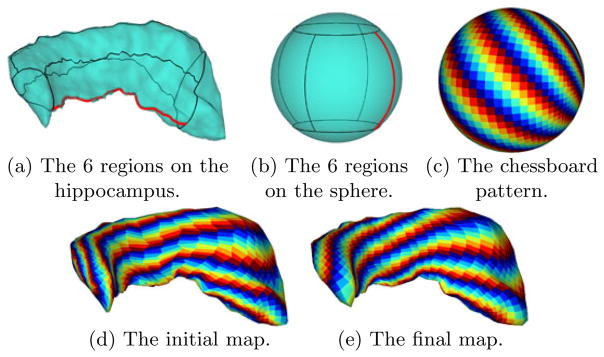
to 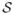 . Using the detected feature point as the origin, we divide each level contour of f1 with its index K1 ≤ k ≤ K2 into 4 segments of equal length and connect the corresponding end points of these segments to divide the hippocampus into 6 regions as shown in Fig. 3(a). Similarly we divide the sphere into 6 regions as shown in Fig. 3(b). For a point in a specific region on
. Using the detected feature point as the origin, we divide each level contour of f1 with its index K1 ≤ k ≤ K2 into 4 segments of equal length and connect the corresponding end points of these segments to divide the hippocampus into 6 regions as shown in Fig. 3(a). Similarly we divide the sphere into 6 regions as shown in Fig. 3(b). For a point in a specific region on  , we find its initial map as the point in the corresponding region on
, we find its initial map as the point in the corresponding region on 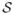 that correlates best with it in terms of their distances to the 4 boundary segments of the regions. The quality of the map is visualized in Fig. 3(d), which is obtained by using the initial map to project a colored chessboard on the sphere shown in Fig. 3(c) to the hippocampus.
that correlates best with it in terms of their distances to the 4 boundary segments of the regions. The quality of the map is visualized in Fig. 3(d), which is obtained by using the initial map to project a colored chessboard on the sphere shown in Fig. 3(c) to the hippocampus.
Fig. 3.
The computation of the harmonic map from a hippocampus to the unit sphere.
Starting from the initial map, the numerical algorithm we developed previously [18] is then used to compute the harmonic map from  to
to 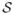 while respecting the boundary condition. By representing the surface
while respecting the boundary condition. By representing the surface  and the sphere
and the sphere 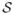 implicitly as a signed distance function ϕ and ψ, respectively, we compute the harmonic map u:
implicitly as a signed distance function ϕ and ψ, respectively, we compute the harmonic map u:  →
→ 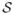 by solving the following PDE iteratively:
by solving the following PDE iteratively:
| (4) |
where is the intrinsic Jacobian of the map u with Ju denoting the regular Jacobian in ℝ3. When discretizing the gradient operators in (4), adaptive numerical schemes were developed in [18], where more numerical details can be found, to take into account the boundary condition on the landmark curve. For the hippocampus in Fig. 3, the result of the harmonic map is visualized in Fig. 3(e). The quality of the map is illustrated in its ability of preserving the regularity of the chessboard pattern.
4 Experimental Results
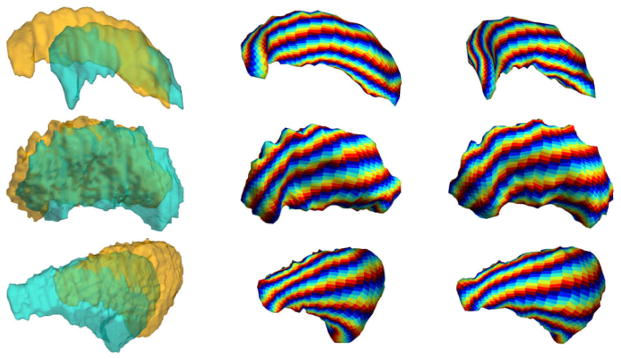
In this section we present experimental results to demonstrate the application of our algorithm in brain mapping. In our method, the only assumption on the input data is that they are from brains in Talairach orientation. Both the landmark detection step using Laplace-Beltrami eigenmaps and the harmonic mapping process are intrinsic to the surface geometry, so natural pose variations among shapes can be handled automatically. In the first experiment, we demonstrate this property of our algorithm and its ability in aligning common geometric features on three subcortical structures: the hippocampus, putamen, and caudate nucleus. Two examples from each structure are used as input data and shown in the first column of Fig. 4. The whole mapping process is completely automated, and it takes around 10 minutes on a PC for each surface. For each structure, the harmonic maps of the two surfaces are visualized in the second and third column of Fig. 4 by using the maps to project the chessboard pattern in Fig. 3(c) onto the surfaces. For all examples, we can see geometrically salient features are correctly aligned with our mapping algorithm even though they have different poses.
Fig. 4.
Mapping results of subcortical structures with different poses. Row one: hippocampi; row two: putamens; row three: caudate nuclei.
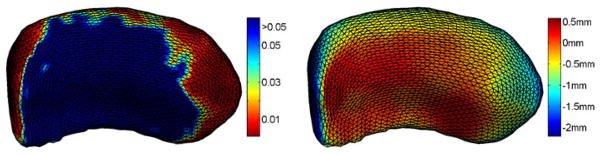
In the second experiment, we apply our method to study changes in hippocampal morphometry for patients with secondary progressive multiple sclerosis (SPMS). The input data are the left hippocampi from a group of 16 normal controls and 11 patients with SPMS shown in their natural poses in Fig. 5. Once the mapping to the sphere is completed, we project a regular triangular mesh of the sphere to each surface, establishing one-to-one correspondences that are critical for group analyses. After factoring out rotation and translation, we compute an atlas 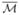 , shown in Fig. 6, by averaging shapes from the control group. By aligning each surface rigidly with the atlas, we compute the displacement from each vertex on the surface to the corresponding vertex on
, shown in Fig. 6, by averaging shapes from the control group. By aligning each surface rigidly with the atlas, we compute the displacement from each vertex on the surface to the corresponding vertex on 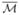 . To test group differences, a Wilcoxon rank-sum test is applied to the displacement values of the control and patient group at each vertex. The map of P values on all vertices are visualized in Fig. 6(a), together with the average displacement of the patient group in Fig. 6(b). From the results we can see large areas of volume losses in the hippocampi of patients with SPMS are successfully localized. To correct for multiple comparisons, we apply a permutation test [1] for 1 million times and an overall P value of 0.000076 is obtained, which clearly shows the significance of our mapping results.
. To test group differences, a Wilcoxon rank-sum test is applied to the displacement values of the control and patient group at each vertex. The map of P values on all vertices are visualized in Fig. 6(a), together with the average displacement of the patient group in Fig. 6(b). From the results we can see large areas of volume losses in the hippocampi of patients with SPMS are successfully localized. To correct for multiple comparisons, we apply a permutation test [1] for 1 million times and an overall P value of 0.000076 is obtained, which clearly shows the significance of our mapping results.
Fig. 5.
The hippocampal surfaces of (a) the control group; (b) the patient group.
Fig. 6.
The mapping results. (a) The map of P values. (b) The map of the average displacement of the patient group.
5 Conclusions
In this paper we have developed a novel surface mapping algorithm applicable to a class of subcortical structures. The maps from our method are diffeomorphic, and also correctly align geometric features, both locally and globally, that are automatically detected with Laplace-Beltrami eigenmaps. An application of our method in group analyses has also been presented to demonstrate its robustness and ability of detecting anatomical changes due to pathology.
References
- 1.Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22(4):1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 2.Gerig G, Styner M, Jones D, Weinberger D, Lieberman J. Shape analysis of brain ventricles using SPHARM. Proc Workshop on Mathematical Methods in Biomedical Image Analysis. 2001:171–178. [Google Scholar]
- 3.Davies RH, Twining CJ, Allen PD, Cootes TF, Taylor CJ. Shape discrimination in the hippocampus using an MDL model. Proc IPMI. 2003:38–50. doi: 10.1007/978-3-540-45087-0_4. [DOI] [PubMed] [Google Scholar]
- 4.Gu X, Wang Y, Chan TF, Thompson PM, Yau ST. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Trans Med Imag. 2004;23(8):949–958. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Thompson PM, de Zubicaray G, Rose SE, Tu Z, Dinov I, Toga AW. Direct mapping of hippocampal surfaces with intrinsic shape context. NeuroImage. 2007;37(3):792–807. doi: 10.1016/j.neuroimage.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans Imag Process. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 7.Joshi S, Miller MI. Landmark matching via large deformation diffeomorphisms. IEEE Trans Imag Process. 2000;9(8):1357–1370. doi: 10.1109/83.855431. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG. Abnormalities of hippocampal surface structure in very mild dementia of the alzheimer type. NeuroImage. 2006;30(1):52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pizer SM, Fritsch DS, Yushkevich PA, Johnson VE, Chaney EL. Segmentation, registration, and measurement of shape variation via image object shape. IEEE Trans Med Imag. 1999;18(10):851–865. doi: 10.1109/42.811263. [DOI] [PubMed] [Google Scholar]
- 10.Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Med Image Anal. 2003;7(3):207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- 11.Yushkevich PA, Zhang H, Gee JC. Continuous medial representation for anatomical structures. IEEE Trans Med Imag. 2006;25(12):1547–1564. doi: 10.1109/tmi.2006.884634. [DOI] [PubMed] [Google Scholar]
- 12.Reuter M, Wolter F, Peinecke N. Laplace-Beltrami spectra as Shape-DNA of surfaces and solids. Computer-Aided Design. 2006;38:342–366. [Google Scholar]
- 13.Qiu A, Bitouk D, Miller MI. Smooth functional and structural maps on the neocortex via orthonormal bases of the Laplace-Beltrami operator. IEEE Trans Med Imag. 2006;25(10):1296–1306. doi: 10.1109/tmi.2006.882143. [DOI] [PubMed] [Google Scholar]
- 14.Uhlenbeck K. Generic properties of eigenfunctions. Amer J of Math. 1976;98(4):1059–1078. [Google Scholar]
- 15.Reeb G. Sur les points singuliers d’une forme de Pfaff completement integrable ou d’une fonction nemérique. Comptes Rendus Acad Sciences. 1946;222:847–849. [Google Scholar]
- 16.Shewchuk JR. Delaunay refinement algorithms for triangular mesh generation. Comput Geom Theory & Applications. 2002;22(1–3):21–74. [Google Scholar]
- 17.Eells J, Sampson JH. Harmonic mappings of Riemannian manifolds. Ann J Math. 1964;86:109–160. [Google Scholar]
- 18.Shi Y, Thompson PM, Dinov I, Osher S, Toga AW. Direct cortical mapping via solving partial differential equations on implicit surfaces. Med Image Anal. 2007;11(3):207–223. doi: 10.1016/j.media.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]