Abstract
A specific and sensitive serodiagnostic test for Mycobacterium ulcerans infection would greatly assist the diagnosis of Buruli ulcer and would also facilitate seroepidemiological surveys. By comparative genomics, we identified 45 potential M. ulcerans specific proteins, of which we were able to express and purify 33 in E. coli. Sera from 30 confirmed Buruli ulcer patients, 24 healthy controls from the same endemic region and 30 healthy controls from a non-endemic region in Benin were screened for antibody responses to these specific proteins by ELISA. Serum IgG responses of Buruli ulcer patients were highly variable, however, seven proteins (MUP045, MUP057, MUL_0513, Hsp65, and the polyketide synthase domains ER, AT propionate, and KR A) showed a significant difference between patient and non-endemic control antibody responses. However, when sera from the healthy control subjects living in the same Buruli ulcer endemic area as the patients were examined, none of the proteins were able to discriminate between these two groups. Nevertheless, six of the seven proteins showed an ability to distinguish people living in an endemic area from those in a non-endemic area with an average sensitivity of 69% and specificity of 88%, suggesting exposure to M. ulcerans. Further validation of these six proteins is now underway to assess their suitability for use in Buruli ulcer seroepidemiological studies. Such studies are urgently needed to assist efforts to uncover environmental reservoirs and understand transmission pathways of the M. ulcerans.
Author Summary
Buruli ulcer is a slowly progressive but potentially devastating disease of skin and subcutaneous tissue caused by the bacterium Mycobacterium ulcerans. The disease is widespread throughout West and Central Africa, and some countries in the region have established Buruli ulcer control programs. Buruli ulcer is difficult to distinguish from other chronic skin conditions that require different treatments, and there is an urgent need for an accurate point-of-care diagnostic test. In this study, we have used genomic techniques to identify 45 potential M. ulcerans–specific antigens, 33 of which we have been able to produce and purify. We tested these proteins against sera from patients, healthy people living in the same region as the patients and from individuals living in a region with no cases of Buruli ulcer. We found that seven proteins were able to elicit antibody responses that were significantly different between patients and the control subjects from the non-endemic region but not from the healthy individuals in the same Buruli ulcer endemic region. Further analysis showed that six of these M. ulcerans proteins might be useful as markers of exposure to M. ulcerans and could be developed into tools to uncover environmental reservoirs and understand transmission pathways of the bacterium.
Introduction
Mycobacterium ulcerans is the causative agent of the severe necrotizing skin disease known as Buruli ulcer (BU). The clinical presentation of the disease begins with the appearance of a small, painless, nodule or papule. As the infection progresses, necrosis of subcutaneous fat and eventual breakdown of skin occurs leading to the appearance of a characteristic ulcer with an undermined edge [1], [2]. BU has been reported in more than 30 countries world-wide, although the primary burden of disease is carried by those in western and sub-Saharan Africa [3]. Since the 1980s there has been a rapid re-emergence of the disease, where in some endemic regions it is now more common than the two other major mycobacterial diseases, tuberculosis and leprosy [4]. The victims of this disease are most commonly children, although any individual may be affected [5], [6]. Whilst control efforts are underway in many affected countries, a major shortcoming is the lack of a simple, rapid method to confirm infection with M. ulcerans.
The necrosis of subcutaneous fat seen in BU is due to the actions of a family of polyketide toxins known as mycolactones. These toxins possess cytotoxic and immunosuppressive activities and injection of purified mycolactone into animal models replicates the pathology seen in human BUs [7], [8]. Mycolactones are produced by large polyketide synthases encoded on the pMUM series of megaplasmids in members of the M. ulcerans/Mycobacterium marinum complex known as mycolactone producing mycobacteria (MPM) [9], [10]. Three polyketide synthase genes (mlsA1, mlsA2 and mlsB) encode the mycolactone synthesis machinery, whilst a number of accessory genes that appear essential for mycolactone production are also present on the pMUM plasmids [9]. Furthermore, these plasmids contain a number of predicted protein-coding DNA sequences (CDS), with products of unknown function, the majority of which are conserved amongst different strains of MPM [11]. Several studies indicate that all the MPM (including M. ulcerans) diverged from a common M. marinum progenitor, through acquisition of the pMUM plasmid [10], [12], [13]. M. marinum strains share greater than 98% DNA identity with M. ulcerans [10], [12], [13] but they do not contain pMUM and do not make mycolactone, although some MPM that were given species names prior to the discovery of pMUM plasmids are confusingly referred to as M. marinum, M. “liflandii” and M. pseudoshottsii [14], [15], [16]. For simplicity, here all mycolactone producing mycobacteria are referred to as M. ulcerans and the closely related, non-mycolactone producing strains are referred to as M. marinum, in accordance with recent recommendations [17].
There are several laboratory-based methods for the diagnosis of BU in humans. These strategies currently include the direct examination of Ziehl-Neelsen stained smears, culture of M. ulcerans and PCR from swabs or tissue samples. Microscopy based on the Ziehl-Neelsen stain from BU swabs or biopsies is quick, however, several studies have shown that the sensitivity of this method is highly variable (40 – 80%, depending on the laboratory) [18], [19]. Culture of M. ulcerans from a suspect lesion remains the gold standard for diagnosis, however, due to the long incubation times required (up to 12 weeks) and low sensitivity it is not appropriate for pre-treatment diagnosis [20]. PCR for the M. ulcerans specific insertion sequence IS2404 was first validated as a diagnostic test in 1997 [21] and several studies have shown it to be the most sensitive of the currently employed diagnostic techniques [20], [22], [23]. However, high reagent costs and the need for specialized equipment and trained staff to perform and interpret PCR restricts its use to larger, central laboratories. Thus none of these approaches are suited for use in rural African regions where BU is endemic and the World Health Organization has designated the development of new approaches to the diagnosis of M. ulcerans infection a research priority.
Early attempts to develop diagnostics for BU relied upon injection of Burulin (a crude M. ulcerans whole cell lysate) into individuals and waiting for the development of a delayed-type hypersensitivity response, akin to the Mantoux test [24]. Whilst those in the active stage of disease had strong reactions to Burulin, the majority also reacted to PPD, suggesting that there was significant cross-reactivity amongst mycobacterial antigens. Subsequently it was shown that BU patients develop serum antibodies to whole cell lysate of M. ulcerans [25], as well as culture filtrate proteins [26], [27]. However, these studies also found that up to 37% of control subjects had positive antibody responses to M. ulcerans proteins, again indicating that a significant degree of cross reactivity with other mycobacteria exists [25], [26], [27].
We reasoned that the identification of specific M. ulcerans antigens may help to overcome some of the difficulties associated with cross-reactivity to conserved mycobacterial antigens and accelerate efforts to develop a rapid diagnostic test. A recent increase in the number of mycobacterial genome sequences available has provided an opportunity to comprehensively identify proteins that are unique to M. ulcerans. We identified 45 M. ulcerans candidate antigens and then used PCR to assess their distribution in 26 M. ulcerans and 30 M. marinum strains. We then determined the reactivity of these proteins to sera from BU patients by ELISA. Here we show that a subset of these proteins is conserved amongst all M. ulcerans strains and that some of these proteins may be useful in seroepidemiological studies of Buruli ulcer.
Methods
Ethics statement
Ethics approval for serum collection was obtained from the National Ethics Committee of the Ministry of Health in Benin and the Human Ethics Committee of the Faculté des Sciences de la Santé, University of Abomey-Calavi, Benin. Informed consent was obtained for every participant. For children involved in this study, written, informed consent was obtained from their parents or legal guardians.
Bacterial strains
Cloning of M. ulcerans unique sequences was performed in E. coli TOP10 cells (Invitrogen). The mycobacterial strains used in this study and their characteristics are listed in Table S1.
Identification of M. ulcerans unique genes
Sequences unique to M. ulcerans were identified first by bioinformatic comparisons between the M. ulcerans genome and other recently completed mycobacterial genomes, using BLASTCLUST as described [28]. The completed and unfinished genomes utilized in this study were (Name, Genbank accession number): M. abcessus CIP104536 (NC_010397), M. avium subsp. paratuberculosis K10 (NC_002944), M. bovis BCG str. Pasteur 1173P2 (NC_008769), M. gilvum PYR-GCK (NC_009338), M. intracellulare ATCC 13950 (NZ_ABIN00000000), M. kansasii ATCC 12748 (NC_ACBV00000000), M. leprae TN (NC_002677), M. marinum M (NC_010612), M. smegmatis mc2 155 (NC_008596), Mycobacterium sp. KMS (NC_008705), Mycobacterium sp. JLS (NC_009077), Mycobacterium sp. MCS (NC_008146), M. tuberculosis C (NZ_AAKR00000000), M. tuberculosis CDC 1551 (NC_002755), M. tuberculosis F11 (NC_009565), M. tuberculosis H37Rv (NC_000962), M. tuberculosis H37Ra (NC_009525), M. tuberculosis Haarlem (NZ_AASN00000000), M. avium 104 (GenBank accession NC_008595), M. ulcerans Agy99 (NC_008611), and M. vanbaalenii PYR-1 (NC_008726). Forty-seven chromosomally encoded sequences and 54 plasmid-encoded sequences were initially selected as being M. ulcerans specific with no ortholog present in any other mycobacterial genome. Orthologues were defined as predicted amino acid sequences with greater than 85% amino acid identity to M. ulcerans proteins. Further criteria were applied to restrict the number of coding sequences (CDS) to investigate (see results section). Due to the high level of genetic relatedness between M. ulcerans and M. marinum strains, M. marinum and M. ulcerans strains spanning the known genetic diversity were then tested for these CDS by PCR using standard conditions and the addition of 5% dimethyl sulfoxide.
DNA cloning and sequencing
Primers were designed to selected CDS to include a 5′ CACC sequence for cloning into the Gateway entry vector pENTR/SD/D-TOPO (Invitrogen) (see Table S2 for a list of oligonucleotides used in this study). CDS were PCR amplified using either Pfu Turbo (Stratagene) or KOD (Novagen) DNA polymerases under standard reaction conditions. Gateway cloning reactions were performed according to the manufacturer's instructions into the entry vector pENTR/SD/D-TOPO and positive clones were confirmed to be in-frame by DNA sequencing. Subcloning of the sequences of interest into one of three different E. coli expression vectors, pDEST17, pET-DEST42 or pBAD-DEST49 (Invitrogen), was accomplished using LR clonase enzyme (Invitrogen) according to manufacturer's instructions.
Protein expression
For pDEST17 or pET-DEST42 constructs, overnight cultures of E. coli Rosetta 2 (Novagen), Rosetta-gami (Novagen), or C43 [29] containing the cloned expression constructs were grown and used to inoculate 200 ml volumes of Luria-Bertani broth containing 100 µg/ml ampicillin and 30 µg/ml chloramphenicol. Cells were grown at 37°C with shaking until they reached an optical density at 600 nm of approximately 0.8, at which point they were induced with IPTG (Isopropyl β-D-1-thiogalactopyranoside) at a final concentration of 1 mM. Cultures were incubated for a further 4 hrs at 37°C with shaking. Cells were then harvested by centrifugation.
For expression of proteins from pBAD-DEST49, E. coli DH10B cultures containing the appropriate constructs were grown as described above (without the addition of chloramphenicol) and were induced with a final concentration of 0.2% L-arabinose.
Polyhistidine-tagged protein purification
Recombinant M. ulcerans fusion proteins that displayed high levels of expression from either of the three expression vectors were purified under denaturing conditions using Talon metal affinity resin (Clontech). Briefly, cell pellets from 200 ml cultures post-induction were resuspended in 10 ml wash buffer (300 mM NaCl, 50 mM NaH2PO4, 8M urea, pH 7.0). Samples were sonicated on ice for 3×30s and the sonicate was then pelleted for 20 min at 12,500× g. The clarified lysate was added to 1 ml prepared resin and incubated at room temperature for 30 min. The solution was drained from the column and the column was washed with 5 ml of wash buffer for 10 min at room temperature with gentle agitation. This was repeated twice, after which the protein was eluted from the column in wash buffer containing 150 mM imidazole, followed by a final elution using wash buffer containing 200 mM imidazole. Eluates were analyzed by SDS-PAGE and dialyzed against wash buffer to remove imidazole.
Sera collection
Serum samples were collected from 93 individuals, comprising three groups. Group 1 (Patient group) consisted of 30 BU patients living in villages near the Ouémé river in the Ouémé province of Benin, where Buruli ulcer is highly prevalent (8–20 cases per 1000 individuals [30]). The patients (11 female and 19 male, aged 3–74 years) were recruited from the Centre de Diagnostic et de Traitement de l'Ulcère de Buruli in Pobè, Benin. Diagnosis of Buruli ulcer was made by Ziehl-Neelsen staining of material taken from swabs of the lesions or directly from biopsy material and subsequently confirmed by IS2404 PCR. Group 2 (Endemic Controls) consisted 24 participants (14 female and 10 male aged 5–72 years) with no history of BU but from the same villages in the Ouémé province as the patients. Group 3 (Non-endemic Controls) consisted of 30 participants (15 female and 15 male), living in villages around Ouidah (Atlantic province), a non-endemic coastal area for BU in Benin, approximately 150 km southwest of Pobè [30]. BCG vaccination exposure (based on the presence of a scar) for all three groups was approximately 60%. Serum was prepared from 8 ml of blood for each participant and aliquots of each sample were stored at −80°C.
SDS-PAGE and Western blotting
For analysis of purified protein reactivity by western blotting, purified proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 12% polyacrylamide gels under reducing conditions, as described previously [31]. The gel was then transferred to nitrocellulose membranes in Tris-glycine buffer containing 20% methanol. Membranes were blocked in 5% skim milk, PBS with 0.1% Tween-20. Membranes were then incubated with human sera at 1/500 dilution. Prior to incubation with anti-human IgG horseradish-peroxidase (HRP) conjugated secondary antibody, the membranes were washed repeatedly in PBS-0.1% Tween-20. Bands were detected by chemiluminescence using the Western lightning kit (Perkin Elmer).
Detection of human antibodies against potential M. ulcerans antigens by ELISA
Purified recombinant M. ulcerans proteins were adsorbed to polystyrene 96-well plates (Costar, Corning) at 0.5 µg per well in carbonate coating buffer (pH 9.6) and incubated at 4°C overnight. Plates were washed five times with PBS (pH 7.2) containing 0.05% Tween 20 (Sigma-Aldrich) (PBS-T) and then five times with PBS before being blocked with 100 µl 5% fetal bovine serum in PBS. Plates were washed as described above and 50 µl human sera (diluted 1/100 in PBS with 0.5% BSA) was added to each well. Plates were incubated for 2 hrs at room temperature before being washed as described above. 50 µl of goat anti-human IgG-HRP (Millipore) diluted at 1/2500 in PBS with 0.5% BSA was added to each well and incubated at room temperature for 1 hr, followed by washing as described above. Plates were developed by the addition of 50ul of ABTS solution (Invitrogen) to each well, incubated for 5 minutes and the reaction was stopped by the addition of 50 µl of 0.01% sodium azide, 0.1M citric acid. The colour change reaction was measured at 405 nm in Multiskan Ascent microplate reader (Thermo Fisher Scientific). Positive control wells were incubated with anti-6xHis-HRP antibody (Roche Applied Science).
Data analysis
ELISA test results were analysed using GraphPad Prism version 5.0c (GraphPad Software, San Diego California USA). One-way ANOVA with Bonnferroni's post-test was used to compare OD values for patient, endemic controls and non-endemic control groups whose distributions approximated normality. ELISA data with non-gaussian distributions were analysed by the Kruskal-Wallis test with Dunn's post test. Cut-off scores for determination of inclusion of proteins for analysis were determined as the mean OD of the control population. Receiver-operator characteristic (ROC) analysis was used to determine the best cut-off value and associated sensitivity and specificity that discriminated individuals living in BU endemic areas from those living in non-endemic areas.
Results
Identification of potential M. ulcerans antigens
Forty-seven chromosomally-encoded CDS, potentially unique to M. ulcerans were identified by bioinformatic comparison of the M. ulcerans genome to 21 other mycobacterial genomes (see Materials and Methods for genomes used). We then excluded CDS <50 codons in length and the insertion sequence elements IS2404 and IS2606 and also applied the following criteria to include only those CDS likely to generate an immune response: i) predicted membrane association; ii) predicted secretion signal; or iii) previously confirmed as expressed in a proteomic study [32], resulting in 34 chromosomal CDS of interest. We also identified 33 unique CDS on the pMUM001 plasmid and in addition we included in our analysis the sequences encoding the 12 unique functional domains of the mycolactone polyketide synthases (MlsA1, MlsA2 & MlsB) as representative of the entire genetic variability present in this locus [9].
Due to the close genetic relationship between M. ulcerans and M. marnium and the knowledge that M. marinum M, does not reflect the genetic diversity in the M. marinum complex [10], 30 genetically diverse M. marinum isolates were tested for the presence of the selected chromosomal CDS by PCR. The M. marinum strains do not contain pMUM plasmids, so these isolates were not tested for the presence of any of the plasmid CDS. Eleven of the CDS identified as M. ulcerans specific by bioinformatic comparisons were found to be present in at least one of the other M. marinum strains tested and were thus excluded from further analyses (Table 1).
Table 1. Percentage of M. marinum and M. ulcerans strains containing selected M. ulcerans Agy99 sequences.
| CDS | % MMb strains | % MUc strains | % MUc African strains | CDS | % MUc strains | % MUc African strains |
| MUL_0027 | 3.7 | - | - | MUP002 | 100 | 100 |
| MUL_0076 | 51.8 | - | - | MUP003 | 100 | 100 |
| MUL_0503 | 92.6 | - | - | MUP004 | 100 | 100 |
| MUL_0508 | 0 | 88.5 | 100 | MUP006 | 80.8 | 100 |
| MUL_0510 | 0 | 88.5 | 100 | MUP007 | 92.3 | 100 |
| MUL_0511 | 0 | 92.3 | 100 | MUP013 | 92.3 | 100 |
| MUL_0512 | 0 | 84.6 | 100 | MUP014 | 88.5 | 100 |
| MUL_0513 | 0 | 84.6 | 100 | MUP015 | 96.2 | 100 |
| MUL_0515 | 0 | 76.9 | 100 | MUP016 | 80.8 | 100 |
| MUL_0516 | 0 | 88.5 | 100 | MUP017 | 61.5 | 100 |
| MUL_0517 | 0 | 96.2 | 100 | MUP018 | 80.8 | 100 |
| MUL_0526 | 29.6 | - | - | MUP019 | 73.1 | 100 |
| MUL_0527 | 48.1 | - | - | MUP020 | 80.8 | 100 |
| MUL_0551 | 0 | 73.1 | 100 | MUP021 | 88.5 | 100 |
| MUL_0552 | 0 | 88.5 | 100 | MUP023 | 100 | 100 |
| MUL_0998 | 0 | 92.3 | 100 | MUP024 | 100 | 100 |
| MUL_0999 | 0 | 92.3 | 100 | MUP038 | 92.3 | 90 |
| MUL_1001 | 0 | 92.3 | 100 | MUP045 | 100 | 100 |
| MUL_1135 | 7.5 | - | - | MUP046 | 96.2 | 100 |
| MUL_2590 | 44.4 | - | - | MUP057 | 46.2 | 100 |
| MUL_2831 | 0 | 100 | 100 | MUP064 | 46.2 | 100 |
| MUL_2832 | 0 | 96.2 | 100 | MUP065 | 53.8 | 100 |
| MUL_3210 | 74.1 | - | - | MUP066 | 88.5 | 100 |
| MUL_3212 | 0 | 100 | 100 | MUP067 | 73.1 | 100 |
| MUL_3214 | 0 | 96.2 | 100 | MUP068 | 65.4 | 100 |
| MUL_3215 | 0 | 92.3 | 100 | MUP070 | 65.4 | 100 |
| MUL_3216 | 0 | 96.2 | 100 | MUP071 | 88.5 | 100 |
| MUL_3217 | 0 | 100 | 100 | MUP074 | 88.5 | 100 |
| MUL_3218 | 0 | 69.2 | 100 | MUP075 | 88.5 | 100 |
| MUL_3230 | 0 | 100 | 100 | MUP076 | 84.6 | 100 |
| MUL_3440 | 66.6 | - | - | MUP078 | 92.3 | 100 |
| MUL_3828 | 0 | 69.2 | 100 | MUP079 | 92.3 | 100 |
| MUL_4213 | 44.4 | - | - | MUP080 | 96.2 | 100 |
| MUL_4217 | 44.4 | - | - | PKS domainsa | 100 | 100 |
= All sequences encoding the 12 PKS domains are found in 100% of mycolactone producing mycobacteria.
= M. marinum;
= M. ulcerans.
The pMUM plasmids are known to vary in both size and gene content between M. ulcerans strains [11], [33], and so we tested a selection of 26 geographically distinct M. ulcerans isolates, by PCR, for the presence of each of the plasmid sequences (Table 1). Additionally, we examined these strains for the presence of each of the selected chromosomal sequences by PCR, and found that six of the plasmid CDS and four of the chromosomally encoded CDS were present in all of the M. ulcerans strains tested (Table 1). Because the focus of our study is to develop diagnostic reagents for use in African patients where the need for BU control is most urgent, we particularly focused on the distribution of sequences of interest in African isolates of M. ulcerans. The 23 M. ulcerans specific chromosomal CDS were found in all 10 of the African M. ulcerans strains tested. As expected, the plasmid-borne mycolactone polyketide synthase (mls) sequences were found in all M. ulcerans strains tested, and also 32 of 33 non-mls plasmid sequences were present in all African strains. MUP038 was absent by PCR from a single African strain (M. ulcerans strain Kob), which confirms previous findings for this strain showing it has a plasmid DNA deletion [33] (Table 1).
The completed list of M. ulcerans unique CDS selected for further study included 13 chromosomal and 30 plasmid CDS (12 mls and 18 non-mls) and is shown in Table S3. In addition we included as positive controls two CDS encoding proteins Hsp65 and Hsp18 (MUL_2232) which have been reported to be antigenic but which are present in other mycobacteria species [34], [35]. This resulted in a total of 45 proteins under investigation.
Cloning, expression and purification of M. ulcerans sequences in E. coli
All 45 of these selected M. ulcerans sequences were amplified from strain Agy99 genomic DNA and 44 were cloned using the Gateway system into at least one of three different expression vectors, pDEST17 (N-terminal 6xHis tag), pET-DEST42 (C-terminal 6xHis tag) or pBAD-DEST49 (N-terminal thioredoxin tag, C-terminal 6xHis tag). We then successfully expressed 37 of 44 (84.1%) of the target sequences in E. coli as either N-terminal or C-terminal 6xHis tagged proteins or both at or near their predicted molecular weight (Table S3). Whilst 37 proteins could be expressed in quantities sufficient for western blotting, only 33 of these 37 were able to be produced in quantities sufficient for ELISA analysis.
Seven proteins were unable to be expressed in E. coli as either N- or C-terminal 6xHis tagged fusions. Expression of two of these proteins (MUP016 and MUP017) from either pDEST17 or pET-DEST42 proved to be toxic to E. coli. Sequencing of the remaining five constructs showed in-frame fusions had been made and western blots on WCLs of E. coli bearing these constructs showed an absence of recombinant protein expression after induction (data not shown). Further attempts were made to salvage these seven proteins by cloning into pBAD-DEST49, a vector with arabinose inducible expression, an N-terminal thioredoxin tag and a C-terminal 6xHis tag, designed to improve protein expression and solubility, and by using E. coli strain C43, which has been optimized for the expression of toxic or membrane proteins [29], [36]. However, these efforts were also unsuccessful and so these proteins were unable to be produced to levels sufficient for purification.
Diagnostic potential of M. ulcerans specific antigens
To evaluate the potential for these proteins to be used to diagnose M. ulcerans infection or assess exposure to the bacterium, sera was collected from 39 IS2404 PCR confirmed BU patients (designated: patient) and 24 controls with no past or current diagnosis of BU (designated: endemic control). Both these groups came from the high BU prevalence Ouémé region in Benin. Sera were also obtained from 30 additional control subjects who lived in the BU non-endemic Ouidah region of Benin (designated: control). A pre-study decision was taken to analyze in these three groups rather than only patients versus non-endemic controls as previous studies had indicated the possibility of asymptomatic exposure to M. ulcerans in BU endemic areas [34].
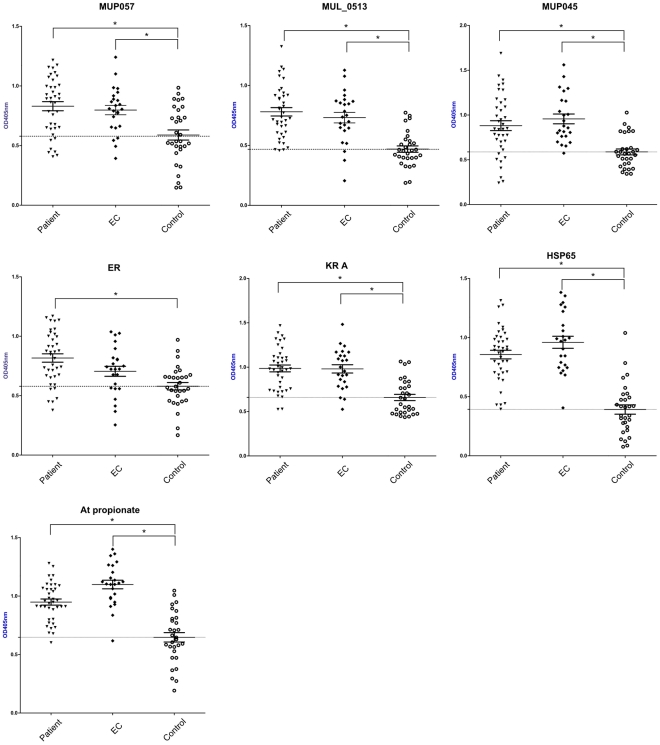
Screening of 33 proteins by ELISA uncovered significantly higher IgG antibody responses in patients compared to non-endemic controls for the following seven proteins; MUP045, MUP057, MUL_0513, Hsp65, and the mycolactone Mls domains (AT-propionate, ER, and KR-A domains) (p<0.05, Figure 1). However, IgG responses of the endemic controls were not significantly different to those of patients for any of the seven proteins (Figure 1). Endemic controls had IgG responses that were significantly higher than those of non-endemic controls for the six of these seven proteins, suggesting that residents of endemic regions with no history of BU might have been exposed to M. ulcerans. The remaining 26 proteins, including the positive control antigen MUL_2232 (Hsp18), had no ability to elicit discriminatory antibody responses between any groups (Figure S1).
Figure 1. ELISA results for seven M. ulcerans proteins capable of discriminating between patients and controls.
Comparison of patient, endemic control (EC) and non-endemic control sera reactivity to seven M. ulcerans antigens that showed an ability to discriminate between patient and control sera. Mean OD405nm readings for each group and standard error of the mean are shown. The horizontal dotted line in each figure represents the mean OD405nm reading of the control group. * = p<0.05.
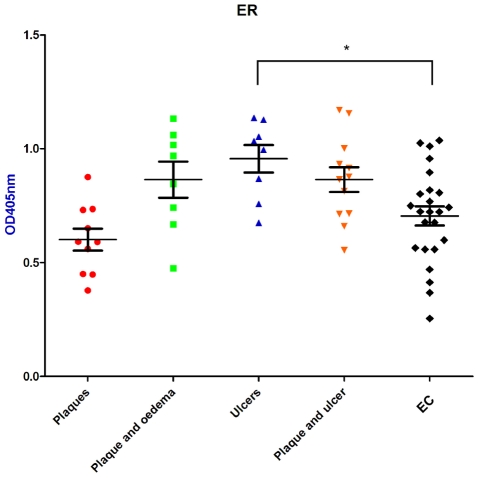
When serum antibody responses for the proteins shown in Figure 1 were analysed according to disease state (i.e, plaque, plaque and oedema, ulcer, or plaque and ulcer), there was no significant difference in reactivity between disease state and endemic controls for each protein except for ER (Figure 2). Mean ELISA absorbance values for sera from patients with ulcers were significantly higher in their reactivity to the ER domain of the mycolactone PKS than endemic controls and other lesion types (p<0.05, Figure 2).
Figure 2. Analysis of BU patient serum reactivity by disease state.
Patient antibody responses were analysed by disease state for the mycolactone polyketide synthase ER domain. Means and SEM of each group are shown. * = p<0.05.
Seroepidemiological potential of M. ulcerans specific antigens
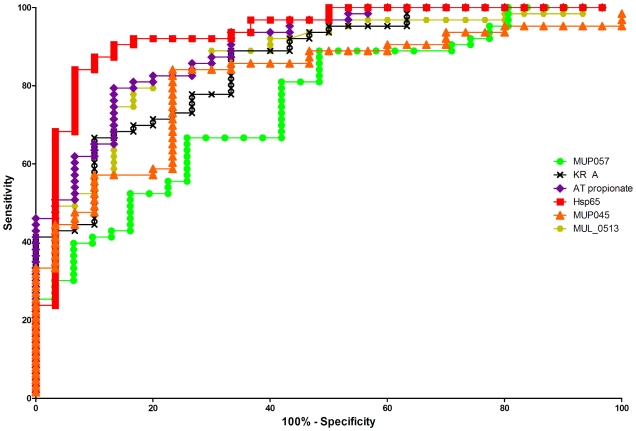
We examined six of the seven proteins shown in Figure 1 to test if they could effectively distinguish subjects from BU endemic and BU non-endemic regions of Benin. We performed receiver-operator curve (ROC) analyses for each of these six antigens by combining the ELISA absorbance values for patient and endemic control groups (collectively referred to as BU-endemic) (Table 2) to calculate sensitivity and the non-endemic control group for specificity. ROC curves for each of the six proteins are shown in Figure 3. The ELISA OD cutoffs that maximized the accuracy of each of the six antigens are shown in Table 2 and the results indicated that a number of these proteins might be useful in determining new M. ulcerans endemic areas. In particular MUL_0513 and two of the Mls domains (AT-propionate, KR-B) all produced high area under curve (AUC, >0.8), and good sensitivity (>70%) and specificity (>80%). However the best antigen was Hsp65, with AUC of 0.932, 84.1% sensitivity and 93.3% specificity at an OD cut-off of 0.693. Hsp65 also had the highest likelihood ratio (12.6), indicating that an individual living in a BU endemic area is 12.6 times more likely to have a positive Hsp65 test than an individual residing in a non-endemic area.
Table 2. Receiver-operator characteristics for proteins with predictive value for living in a BU endemic area.
| Antigen | AUCa | 95% CI | Cut-off | Sensitivity (%) | Specificity (%) | Likelihood ratiob | NPVc |
| MUP045 | 0.810 | 0.720–0.901 | 0.824 | 57.1 | 90.0 | 5.7 | 0.61 |
| MUP057 | 0.753 | 0.652–0.854 | 0.893 | 39.7 | 93.5 | 6.2 | 0.57 |
| MUL_0513 | 0.873 | 0.799–0.946 | 0.622 | 74.6 | 86.7 | 5.6 | 0.70 |
| Hsp65 | 0.932 | 0.873–0.991 | 0.693 | 84.1 | 93.3 | 12.6 | 0.77 |
| AT propionate | 0.897 | 0.832–0.962 | 0.896 | 79.4 | 86.7 | 6.0 | 0.70 |
| KR A | 0.856 | 0.777–0.934 | 0.850 | 69.8 | 83.3 | 4.2 | 0.68 |
AUC, area under curve;
Likelihood ratio represents the likelihood that someone with a positive test will live in a BU endemic area, i.e. a likelihood ratio of 5.0 means someone with a positive test is 5 times more likely live in BU endemic area than an individual with a negative test;
NPV, negative predictive value.
Figure 3. Receiver operator characteristic (ROC) analysis of six M. ulcerans specific proteins.
ROC curves are shown for the six M. ulcerans specific proteins detailed in Figure 1 (MUP045, MUP057, MUL_0513, Hsp65, AT propionate and KR A).
Discussion
The current diagnostic delay experienced by Buruli ulcer patients contributes to morbidity and increased economic hardship [37]. A diagnostic test that is rapid, inexpensive, sensitive, specific and suitable for in-field use would facilitate the timely diagnosis of BU and is a World Health Organization (WHO) research priority. Current laboratory diagnostics including culture, PCR, AFB microscopy and histopathology all have various limitations. We have used comparative genomics to identify M. ulcerans specific sequences to explore the usefulness of the encoded proteins for use in the serodiagnosis of Buruli Ulcer in endemic areas of West Africa.
Whilst bioinformatic methods were initially used to identify M. ulcerans specific CDS, eleven of these “M. ulcerans specific” CDS were found in at least one of the M. marinum strains tested (Table 1). These CDS are distributed across the M. ulcerans genome, and were all situated within regions of DNA that are not present in the M. marinum M genome. It is interesting to note that some of these sequences appear restricted to certain M. marinum sequence types that are genetically closer to M. ulcerans than the sequenced M. marinum M strain. For example MUL_4213 was restricted to the ST3 and ST5 strains tested and MUL_0027 was restricted to the ST3 strains tested. Some of these sequences may be useful as genotyping tools to more clearly delineate the relationships between M. ulcerans and M. marinum strains. These findings are consistent with the known high genetic relatedness of M. ulcerans and M. marinum [10], [13], and underlies the need for experimental validation of bioinformatic results.
Only nine M. ulcerans specific sequences were conserved amongst all mycolactone producing mycobacterial strains tested. However, in agreement with previous findings that African M. ulcerans strains form a close genetic complex with relatively little variation at the whole genome level [38], [39], [40], all selected chromosomal genes were present in all African M. ulcerans isolates that were examined. As pMUM plasmids are known to vary in size and gene content between M. ulcerans strains [11], [33], it is not surprising that only six of 33 non-PKS CDS were completely conserved. However, plasmid CDS may be the most specific to M. ulcerans as they appear to be restricted to mycolactone producing mycobacteria.
During the course of this work the sequences of two other pMUM plasmids from MPM became available [11] and revealed a number of differences between these plasmids with regards to the potential antigens chosen for this study. These differences can explain the inability to detect some of the selected CDS in MPM strains. For example, MUP006 orthologs in pMUM002 and pMUM003 contain a different 5′ nucleotide sequence, explaining the absence of a PCR product in some MPM strains. Conversely, MUP068 and MUP070 orthologs in pMUM001 and pMUM002 contain different 3′ nucleotide sequences. Furthermore, MUP065 appears to be a pseudogene in pMUM001, as its ortholog in pMUM002 and pMUM003 are significantly longer, and MUP067 is deleted from pMUM002 and pMUM003. All of these examples serve to highlight the genetic variability in the non-PKS component of these pMUM plasmids.
In this study 44 M. ulcerans specific sequences were cloned with 37 (84%) of these successfully expressed in E. coli. Seven proteins were unable to be expressed in multiple E. coli strains, and the lack of overexpressed protein was confirmed by western blotting (data not shown). Similar studies that have attempted to identify novel CDS in M. avium subsp. paratuberculosis have also shown variable rates of production of 6 x His tagged proteins (between 25 – 69% of cloned CDS able to be expressed) [41], [42], indicating that this tag may reduce expression of certain proteins. Furthermore, all proteins in this study were found to be insoluble under the expression conditions used, and six of the seven proteins that were unable to be expressed were predicted to be integral membrane proteins, with overexpression of these proteins possibly leading to toxicity. Both of these problems have been previously described in other mycobacterial protein expression studies [43], [44], [45]. Clearly, further optimization of the expression of these proteins, including the use of alternate proteins tags or different host-based expression systems (including mycobacteria) may be necessary to obtain useable amounts of protein for future testing. Recently, a Gateway-based vector has been developed for the overexpression of proteins in M. smegmatis and has shown that proteins which were previously insoluble in E. coli could be expressed in soluble form in M. smegmatis [46]. Furthermore, M. tuberculosis proteins have been successfully produced in the methylotrophic yeast Pichia pastoris with enhanced patient serum reactivity compared with those produced in E. coli [47], [48]. The future use of either of these systems for the proteins that proved difficult to produce in this study may be warranted to investigate their antigenicity.
We were able to confirm that patients with BU had significantly greater IgG antibody responses to seven of 33 purified M. ulcerans proteins as determined by ELISA compared with a control group (Figure 1). The inability of the remaining 26 proteins to discriminate between patients and controls is likely due to either the inability of a particular protein to elicit an antibody response or through shared epitopes with proteins from other sources leading to cross-reactive antibody responses. However, none of the proteins were able to discriminate between patients and control subjects from the same BU-endemic area and are therefore unlikely to be useful as a diagnostic test for M. ulcerans infection. By combining measurements of antibody responses to six antigens from people living in a BU endemic area, we were able to discriminate between subjects resident in endemic areas of Benin and controls living in a non-endemic area. Although it remains a possibility that these data reflect cross-reactive antibody responses to other mycobacteria, we suggest that the most likely interpretation of these findings is that many residents of BU-endemic areas have been exposed to M. ulcerans.
Somewhat surprisingly, the most discriminatory antigen was Hsp65, which has 94% homology to the same protein from M. tuberculosis, whilst antigens with no orthologues in other mycobacteria had lower sensitivity and specificity. Hsp65 is known to be an immunodominant antigen in the antibody response to mycobacteria [35] and we anticipated that all groups would have had significant levels of exposure to other mycobacteria from either BCG vaccination, tuberculosis infection, and from exposure to other environmental mycobacteria. We therefore anticipated that Hsp65 would be a sensitive but non-specific antigen. There may be several explanations for our findings. Firstly, patients and endemic controls were both from a region of high BU prevalence and therefore may have been recently and/or frequently exposed to M. ulcerans, leading to continual boosting of antibody responses that does not occur in the non-endemic control group. The fact that M. ulcerans elicits responses dominated by antibodies to Hsp65 at all stages of the infection in mice supports this hypothesis (35). Alternatively, it is possible that our patient and control groups differed in their exposure to mycobacteria other than M. ulcerans, and therefore the results do not reflect M. ulcerans specific antibody responses.
The small heat shock protein, Hsp18, had previously been identified as an immunodominant antigen that may be useful for predicting exposure to M. ulcerans [34]. Results from the current study, however, showed that Hsp18 was not able to discriminate between individuals living in an endemic area and those who reside in a non-endemic area. The reasons for these differences are unclear, however, a possible explanation may be that here, antibody responses were quantified by ELISA, as opposed to qualitative western blot analysis in the study by Diaz et al [34].
Although the other antigens we identified as M. ulcerans specific by our comparative genomic approach did not appear to be as sensitive or specific as Hsp65 in predicting individuals living in a Buruli endemic area, they nevertheless did show that significant antibody responses were detectable, and three antigens (MUL_0513, AT-propionate, KR-B) showed significant ability to discriminate individuals living in a BU endemic versus non-endemic area. Their use in combination may show additive effects and, in conjunction with case-control studies, may permit the development of an assay for detecting individuals exposed to M. ulcerans. More research in this direction would greatly assist efforts to uncover environmental reservoirs and understand transmission pathways of the bacterium.
Although a number of M. ulcerans specific sequences were investigated in this study, there were still a large number that were initially ruled out based on predictions of their cellular location or function. It is possible that some of these potential antigens may be good candidates for further analysis. Based on our findings of high response rates among patient and endemic control groups it appears unlikely that serological approaches will be useful for diagnosing Buruli ulcer. Indeed, the development of serodiagnostics for other mycobacterial diseases, such as tuberculosis, has proved difficult and many of the currently available serodiagnostics for mycobacteria have limited usefulness in the clinic [49], [50].
An alternative to BU serodiagnosis may be possible via direct antigen detection approaches. The extent and distribution of antigens within M. ulcerans lesions or within a patients circulation has not been defined, however, there is the possibility that some of the unique proteins described in this present study may be able to be developed as the basis for antigen capture assays. A diagnostic test based on this methodology would overcome the problems of cross-reactivity seen with tests based upon antibody responses, and would be more appropriate for resource limited areas than tests based on T-cell responses. This project has helped define the antigenic repertoire of M. ulcerans and research is continuing in this vein to investigate the feasibility of other diagnostic modalities.
Supporting Information
ELISA results for 26 M. ulcerans proteins that showed no significant difference in reactivity between patients, endemic controls or non-endemic controls. Reactivity of individual patient and endemic control (EC) samples are shown. Mean reactivity of non-endemic control sera group is represented by a dotted horizontal line across each graph. Mean OD405 nm readings for each group and standard error of the mean are also shown.
(0.15 MB PDF)
List of bacterial strains used in this study.
(0.12 MB DOC)
Oligonucleotides used in this study.
(0.08 MB DOC)
M. ulcerans genes tested in this study.
(0.06 MB DOC)
Acknowledgments
We thank the participating children and their families; school directors and teachers at the primary schools; the UR216 team of Cotonou, for performing field activities; and A. Massougbodji, for interceding with local authorities.
Footnotes
The authors have declared that no competing interests exist.
This research was supported by the UBS-Optimus Foundation Stop Buruli initiative (www.stopburuli.org) and the Association Francaise Raoul Follereau (www.raoul-follereau.org). TPS was supported by a fellowship from the National Health and Medical Research Council of Australia (www.nhmrc.gov.au). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Guarner J, Bartlett J, Whitney EA, Raghunathan PL, Stienstra Y, et al. Histopathologic features of Mycobacterium ulcerans infection. Emerg Infect Dis. 2003;9:651–656. doi: 10.3201/eid0906.020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayman J. Clinical features of Mycobacterium ulcerans infection. Australas J Dermatol. 1985;26:67–73. doi: 10.1111/j.1440-0960.1985.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Buruli ulcer disease. Wkly Epidemiol Rec. 2004;79:194–199. [PubMed] [Google Scholar]
- 4.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, et al. Mycobacterium ulcerans disease (Buruli ulcer) in rural hospital, Southern Benin, 1997-2001. Emerg Infect Dis. 2004;10:1391–1398. doi: 10.3201/eid1008.030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, et al. Mycobacterium ulcerans disease: role of age and gender in incidence and morbidity. Trop Med Int Health. 2004;9:1297–1304. doi: 10.1111/j.1365-3156.2004.01339.x. [DOI] [PubMed] [Google Scholar]
- 6.WHO. Buruli ulcer: progress report, 2004-2008. Wkly Epidemiol Rec. 2008;83:145–154. [PubMed] [Google Scholar]
- 7.George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, et al. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 8.Coutanceau E, Decalf J, Martino A, Babon A, Winter N, et al. Selective suppression of dendritic cell functions by Mycobacterium ulcerans toxin mycolactone. J Exp Med. 2007;204:1395–1403. doi: 10.1084/jem.20070234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci USA. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yip MJ, Porter JL, Fyfe JA, Lavender CJ, Portaels F, et al. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol. 2007;189:2021–2029. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidot SJ, Hong H, Seemann T, Porter JL, Yip MJ, et al. Deciphering the genetic basis for polyketide variation among mycobacteria producing mycolactones. BMC Genomics. 2008;9:462. doi: 10.1186/1471-2164-9-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stinear TP, Jenkin GA, Johnson PD, Davies JK. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J Bact. 2000;182:6322–6330. doi: 10.1128/jb.182.22.6322-6330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ucko M, Colorni A. Mycobacterium marinum infections in fish and humans in Israel. J Clin Microbiol. 2005;43:892–895. doi: 10.1128/JCM.43.2.892-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trott KA, Stacy BA, Lifland BD, Diggs HE, Harland RM, et al. Characterization of a Mycobacterium ulcerans-like infection in a colony of African tropical clawed frogs (Xenopus tropicalis). Comp Med. 2004;54:309–317. [PubMed] [Google Scholar]
- 16.Rhodes MW, Kator H, McNabb A, Deshayes C, Reyrat JM, et al. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int J Syst Evol Microbiol. 2005;55:1139–1147. doi: 10.1099/ijs.0.63343-0. [DOI] [PubMed] [Google Scholar]
- 17.Pidot SJ, Asiedu K, Kaser M, Fyfe JA, Stinear TP. Mycobacterium ulcerans and other mycolactone-producing mycobacteria should be considered a single species. PLoS Negl Trop Dis. 2010;4:e663. doi: 10.1371/journal.pntd.0000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bretzel G, Siegmund V, Nitschke J, Herbinger KH, Thompson R, et al. External quality assurance for the laboratory diagnosis of Buruli ulcer disease in Ghana. Trop Med Int Health. 2006;11:1688–1693. doi: 10.1111/j.1365-3156.2006.01722.x. [DOI] [PubMed] [Google Scholar]
- 19.Mensah-Quainoo E, Yeboah-Manu D, Asebi C, Patafuor F, Ofori-Adjei D, et al. Diagnosis of Mycobacterium ulcerans infection (Buruli ulcer) at a treatment centre in Ghana: a retrospective analysis of laboratory results of clinically diagnosed cases. Trop Med Int Health. 2008;13:191–198. doi: 10.1111/j.1365-3156.2007.01990.x. [DOI] [PubMed] [Google Scholar]
- 20.Bretzel G, Siegmund V, Nitschke J, Herbinger KH, Thompson W, et al. A stepwise approach to the laboratory diagnosis of Buruli ulcer disease. Trop Med Int Health. 2007;12:89–96. doi: 10.1111/j.1365-3156.2006.01761.x. [DOI] [PubMed] [Google Scholar]
- 21.Ross BC, Johnson PD, Oppedisano F, Marino L, Sievers A, et al. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl Environ Microbiol. 1997;63:4135–4138. doi: 10.1128/aem.63.10.4135-4138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbinger KH, Adjei O, Awua-Boateng NY, Nienhuis WA, Kunaa L, et al. Comparative study of the sensitivity of different diagnostic methods for the laboratory diagnosis of Buruli ulcer disease. Clin Infect Dis. 2009;48:1055–1064. doi: 10.1086/597398. [DOI] [PubMed] [Google Scholar]
- 23.Phillips RO, Sarfo FS, Osei-Sarpong F, Boateng A, Tetteh I, et al. Sensitivity of PCR for M. ulcerans on Fine Needle Aspirates for Diagnosis of Buruli ulcer. J Clin Microbiol. 2009. doi: 10.1128/JCM.01842-08. [DOI] [PMC free article] [PubMed]
- 24.Stanford JL, Revill WD, Gunthorpe WJ, Grange JM. The production and preliminary investigation of Burulin, a new skin test reagent for Mycobacterium ulcerans infection. J Hyg (Lond) 1975;74:7–16. doi: 10.1017/s0022172400046659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gooding TM, Johnson PD, Campbell DE, Hayman JA, Hartland EL, et al. Immune response to infection with Mycobacterium ulcerans. Infect Immun. 2001;69:1704–1707. doi: 10.1128/IAI.69.3.1704-1707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobos KM, Spotts EA, Marston BJ, Horsburgh CR, Jr, King CH. Serologic response to culture filtrate antigens of Mycobacterium ulcerans during Buruli ulcer disease. Emerg Infect Dis. 2000;6:158–164. doi: 10.3201/eid0602.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okenu DM, Ofielu LO, Easley KA, Guarner J, Spotts Whitney EA, et al. Immunoglobulin M antibody responses to Mycobacterium ulcerans allow discrimination between cases of active Buruli ulcer disease and matched family controls in areas where the disease is endemic. Clin Diagn Lab Immunol. 2004;11:387–391. doi: 10.1128/CDLI.11.2.387-391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 30.Wagner T, Benbow ME, Brenden TO, Qi J, Johnson RC. Buruli ulcer disease prevalence in Benin, West Africa: associations with land use/cover and the identification of disease clusters. Int J Health Geogr. 2008;7:25. doi: 10.1186/1476-072X-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Tafelmeyer P, Laurent C, Lenormand P, Rousselle JC, Marsollier L, et al. Comprehensive proteome analysis of Mycobacterium ulcerans and quantitative comparison of mycolactone biosynthesis. Proteomics. 2008;8:3124–3138. doi: 10.1002/pmic.200701018. [DOI] [PubMed] [Google Scholar]
- 33.Stinear TP, Hong H, Frigui W, Pryor MJ, Brosch R, et al. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J Bact. 2005;187:1668–1676. doi: 10.1128/JB.187.5.1668-1676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz D, Dobeli H, Yeboah-Manu D, Mensah-Quainoo E, Friedlein A, et al. Use of the immunodominant 18-kiloDalton small heat shock protein as a serological marker for exposure to Mycobacterium ulcerans. Clin Vaccine Immunol. 2006;13:1314–1321. doi: 10.1128/CVI.00254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coutanceau E, Legras P, Marsollier L, Reysset G, Cole ST, et al. Immunogenicity of Mycobacterium ulcerans Hsp65 and protective efficacy of a Mycobacterium leprae Hsp65-based DNA vaccine against Buruli ulcer. Microbes Infect. 2006;8:2075–2081. doi: 10.1016/j.micinf.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Dumon-Seignovert L, Cariot G, Vuillard L. The toxicity of recombinant proteins in Escherichia coli: a comparison of overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr Purif. 2004;37:203–206. doi: 10.1016/j.pep.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 37.Grietens KP, Boock AU, Peeters H, Hausmann-Muela S, Toomer E, et al. “It is me who endures but my family that suffers”: social isolation as a consequence of the household cost burden of buruli ulcer free of charge hospital treatment. PLoS Negl Trop Dis. 2008;2:e321. doi: 10.1371/journal.pntd.0000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rondini S, Kaser M, Stinear T, Tessier M, Mangold C, et al. Ongoing genome reduction in Mycobacterium ulcerans. Emerg Infect Dis. 2007;13:1008–1015. doi: 10.3201/eid1307.060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaser M, Rondini S, Naegeli M, Stinear T, Portaels F, et al. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol Biol. 2007;7:177. doi: 10.1186/1471-2148-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaser M, Gutmann O, Hauser J, Stinear T, Cole S, et al. Lack of insertional-deletional polymorphism in a collection of Mycobacterium ulcerans isolates from Ghanaian Buruli ulcer patients. J Clin Microbiol. 2009;47:3640–3646. doi: 10.1128/JCM.00760-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bannantine JP, Hansen JK, Paustian ML, Amonsin A, Li LL, et al. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol. 2004;42:106–114. doi: 10.1128/JCM.42.1.106-114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paustian ML, Amonsin A, Kapur V, Bannantine JP. Characterization of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis: implications for diagnosis of Johne's Disease. J Clin Microbiol. 2004;42:2675–2681. doi: 10.1128/JCM.42.6.2675-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bannantine JP, Stabel JR, Bayles DO, Geisbrecht BV. Characteristics of an extensive Mycobacterium avium subspecies paratuberculosis recombinant protein set. Protein Expr Purif. 2010;72:223–233. doi: 10.1016/j.pep.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 44.Bannantine JP, Waters WR, Stabel JR, Palmer MV, Li L, et al. Development and use of a partial Mycobacterium avium subspecies paratuberculosis protein array. Proteomics. 2008;8:463–474. doi: 10.1002/pmic.200700644. [DOI] [PubMed] [Google Scholar]
- 45.Araoz R, Honore N, Cho S, Kim JP, Cho SN, et al. Antigen discovery: a postgenomic approach to leprosy diagnosis. Infect Immun. 2006;74:175–182. doi: 10.1128/IAI.74.1.175-182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldstone RM, Moreland NJ, Bashiri G, Baker EN, Shaun Lott J. A new Gateway vector and expression protocol for fast and efficient recombinant protein expression in Mycobacterium smegmatis. Protein Expr Purif. 2008;57:81–87. doi: 10.1016/j.pep.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Benabdesselem C, Barbouche MR, Jarboui MA, Dellagi K, Ho JL, et al. High level expression of recombinant Mycobacterium tuberculosis culture filtrate protein CFP32 in Pichia pastoris. Mol Biotechnol. 2007;35:41–49. doi: 10.1385/mb:35:1:41. [DOI] [PubMed] [Google Scholar]
- 48.Benabdesselem C, Fathallah DM, Huard RC, Zhu H, Jarboui MA, et al. Enhanced patient serum immunoreactivity to recombinant Mycobacterium tuberculosis CFP32 produced in the yeast Pichia pastoris compared to Escherichia coli and its potential for serodiagnosis of tuberculosis. J Clin Microbiol. 2006;44:3086–3093. doi: 10.1128/JCM.02672-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steingart KR, Henry M, Laal S, Hopewell PC, Ramsay A, et al. Commercial serological antibody detection tests for the diagnosis of pulmonary tuberculosis: a systematic review. PLoS Med. 2007;4:e202. doi: 10.1371/journal.pmed.0040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson BL, Welch RJ, Litwin CM. Assessment of three commercially available serologic assays for detection of antibodies to Mycobacterium tuberculosis and identification of active tuberculosis. Clin Vaccine Immunol. 2008;15:1644–1649. doi: 10.1128/CVI.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ELISA results for 26 M. ulcerans proteins that showed no significant difference in reactivity between patients, endemic controls or non-endemic controls. Reactivity of individual patient and endemic control (EC) samples are shown. Mean reactivity of non-endemic control sera group is represented by a dotted horizontal line across each graph. Mean OD405 nm readings for each group and standard error of the mean are also shown.
(0.15 MB PDF)
List of bacterial strains used in this study.
(0.12 MB DOC)
Oligonucleotides used in this study.
(0.08 MB DOC)
M. ulcerans genes tested in this study.
(0.06 MB DOC)