Abstract
Background
Cancer stem cells are presumed to have virtually unlimited proliferative and self-renewal abilities and to be highly resistant to chemotherapy, a feature that is associated with overexpression of ATP-binding cassette transporters. We investigated whether prolonged continuous selection of cells for drug resistance enriches cultures for cancer stem–like cells.
Methods
Cancer stem cells were defined as CD44+/CD24− cells that could self-renew (ie, generate cells with the tumorigenic CD44+/CD24− phenotype), differentiate, invade, and form tumors in vivo. We used doxorubicin-selected MCF-7/ADR cells, weakly tumorigenic parental MCF-7 cells, and MCF-7/MDR, an MCF-7 subline with forced expression of ABCB1 protein. Cells were examined for cell surface markers and side-population fractions by microarray and flow cytometry, with in vitro invasion assays, and for ability to form mammospheres. Xenograft tumors were generated in mice to examine tumorigenicity (n = 52). The mRNA expression of multidrug resistance genes was examined in putative cancer stem cells and pathway analysis of statistically significantly differentially expressed genes was performed. All statistical tests were two-sided.
Results
Pathway analysis showed that MCF-7/ADR cells express mRNAs from ABCB1 and other genes also found in breast cancer stem cells (eg, CD44, TGFB1, and SNAI1). MCF-7/ADR cells were highly invasive, formed mammospheres, and were tumorigenic in mice. In contrast to parental MCF-7 cells, more than 30% of MCF-7/ADR cells had a CD44+/CD24− phenotype, could self-renew, and differentiate (ie, produce CD44+/CD24− and CD44+/CD24+ cells) and overexpressed various multidrug resistance–linked genes (including ABCB1, CCNE1, and MMP9). MCF-7/ADR cells were statistically significantly more invasive in Matrigel than parental MCF-7 cells (MCF-7 cells = 0.82 cell per field and MCF-7/ADR = 7.51 cells per field, difference = 6.69 cells per field, 95% confidence interval = 4.82 to 8.55 cells per field, P < .001). No enrichment in the CD44+/CD24− or CD133+ population was detected in MCF-7/MDR.
Conclusion
The cell population with cancer stem cell characteristics increased after prolonged continuous selection for doxorubicin resistance.
CONTEXT AND CAVEATS
Prior knowledge
Cancer stem cells (ie, CD44+/CD24− cells that can self-renew, differentiate, invade, and form tumors in vivo) are characterized as having virtually unlimited proliferative and self-renewal abilities and being highly resistant to chemotherapy. It is unclear whether the population of cancer stem cells is actually increased by drug therapy.
Study design
Long-term doxorubicin-selected MCF-7/ADR cells, weakly tumorigenic parental MCF-7 cells, and MCF-7/MDR, an MCF-7 subline with forced expression of ABCB1 protein, were studied in culture and in xenograft tumors in mice.
Contribution
MCF-7/ADR cells expressed mRNAs from ABCB1 and other genes also found in breast cancer stem cells. In contrast to parental MCF-7 cell cultures (with a cancer stem cell phenotype in <0.05% of cells), more than 30% of cells in MCF-7/ADR cell cultures had a cancer stem cell phenotype (ie, could self-renew, differentiate, and overexpress various multidrug resistance–linked genes) and were more aggressive as shown in Matrigel assays, which measure invasiveness, and tumorigenesis assays in mice.
Implications
Prolonged continuous selection for doxorubicin resistance appeared to increase the population of cells with cancer stem cell characteristics.
Limitations
Only one highly drug-resistant breast cancer cell line, MCF-7/ADR, and its parental line MCF-7 were studied. Immunodeficient mice were used in tumorigenesis studies. The exact mechanism of cancer stem cell enrichment in MCF-7/ADR cultures is unclear.
From the Editors
A major obstacle to cancer treatment is the development of multidrug resistance. Cancer that relapses after an initial response or that presents as chemotherapy-resistant disease is usually resistant to several chemically unrelated drugs in addition to the initial compound used in therapy (1). The various mechanisms responsible for multidrug resistance include overexpression of certain ATP-binding cassette transporters (2). Cancer stem cells also appear to be highly resistant to various chemotherapies (3–8). This phenomenon of drug resistance may also result, in part, from the high levels of ATP-binding cassette transporters expressed by these cells (9). However, the relationship between exposure to chemotherapeutic agents and the expansion of drug-resistant cancer stem cell–like populations has not been fully explored.
Since the 1980s, investigators have generated many drug-resistant sublines to study the mechanisms of multidrug resistance. One line in particular, MCF-7/ADR-RES, has recently been the subject of much controversy and was finally identified as a doxorubicin-resistant derivative of OVCAR-8, an ovarian cancer cell line [see http://www.sanger.ac.uk/genetics/CGP/Genotyping/synlinestable.shtml and (10–12)]. Thus, much of the previous information obtained from the MCF-7/ADR-RES cell line cannot be used for comparison with the parental MCF-7 cells. In this study, we used doxorubicin-selected MCF-7/ADR cells, which have been fully characterized and validated as breast cancer cells that are derived from the original parental MCF-7 cells (13,14). Our chromosomal analysis of MCF-7/ADR cells verified their origin and the parental MCF-7 cells as breast cancer cells (15).
One unanswered question concerning cancer stem cells is whether this population is actually expanded by drug therapy. Levina et al. (16) evaluated the effects of a 3-day drug therapy regimen on lung cancer cells and concluded that the stem cell-like cell population was enriched. However, to our knowledge, there have been no reports characterizing putative cancer stem cells in cultures of highly drug-resistant cells that were established by a prolonged, continuous, multistep selection regimen. In this study, we investigated whether a population of cells with cancer stem cell characteristics (ie, cells that have CD44+/CD24− surface markers, are highly invasive, are able to self-renew [able to produce cells with the tumorigenic CD44+/CD24− phenotype], are able to differentiate, and are tumorigenic) was enriched in a breast cancer cell line that was selected with doxorubicin. The purpose of this study was to assess whether continuous drug selection could result in the expansion of a cancer stem cell–like population.
Materials and Methods
Drugs, Culture Medium, and Antibodies
Doxorubicin was purchased from Calbiochem (San Diego, CA). Etoposide was purchased from Sigma Chemicals Co (St Louis, MO). Real-time reverse transcription–polymerase chain reaction (RT-PCR) reagents were purchased from Roche Applied Sciences (Indianapolis, IN). Dulbecco’s modified Eagle medium (DMEM), Iscove modified Dulbecco's medium, L-glutamine, and a solution of penicillin and streptomycin were obtained from Invitrogen (Carlsbad, CA). Antibodies against ABCB1, CD24, CD44, or CD133 protein were purchased from BD Biosciences (San Jose, CA). Anti-ABCB1 antibody was conjugated with fluorescein isothiocyanate. Anti-CD44 antibodies were conjugated to fluorescein isothiocyanate for double-staining studies and to allophycocyanin for triple-staining studies. Anti-CD24 and anti-CD133 antibodies were conjugated with phycoerythrin.
Cells and Cell Culture
The MCF-7 breast cancer cell line and the multistep doxorubicin-selected subline, MCF-7/ADR, were gifts from Kapil Mehta (M.D. Anderson Cancer Center, Houston, TX) (14). MCF-7/ADR cells were cultured in high-dose doxorubicin (860 nM) every other passage, as described previously (14). No experiments were performed with cells that had been cultured without doxorubicin for more than 2 weeks, and all experiments were performed with cells grown in the absence of doxorubicin for at least 3 days to avoid drug-associated secondary effects. OVCAR-8, a human ovarian cancer cell line, and NCI-ADR-RES cells, a human drug-resistant ovarian cancer cell line, were obtained from the Developmental Therapeutics Program, National Cancer Institute. Multidrug-resistant MCF-7/MDR cells, which were derived from the parental MCF-7 cells that were transfected with ABCB1 cDNA, were established previously (17).
MCF-7 cells and their sublines (MCF7-FLV1000, which expresses wild-type ABCG2, and MCF7-AdVp3000, which expresses a mutant ABCG2) were maintained in DMEM supplemented with 10% fetal calf serum, penicillin (100 units/mL), and streptomycin (100 μg/mL). MCF-7 AdVP3000 (T482-ABCG2) cells were maintained in the presence of doxorubicin at 3 μg/mL and verapamil at 5 μg/mL, and MCF-7 FLV1000 (R482-ABCG2) cells were cultured in the presence of flavopiridol at 1 μg/mL (18,19). MCF-7/VP-16 cells, which overexpress ABCC1, were cultured as described previously (20). Briefly, the cells were generated by use of a stepwise selection with etoposide (from 200 nM to 10 μM) and maintained in 4 μM etoposide in DMEM supplemented with 10% fetal calf serum, penicillin (100 units/mL), and streptomycin (100 μg/mL). The estrogen-independent breast cancer cell line, MB-MDA-231, and its ABCB1-overexpressing subline, MDA-MB-231 VB100, were cultured as previously described (21). Briefly, MDA-MB-231 cells and MDA-MB-231 VB100 were grown in Iscove modified Eagle medium supplemented with 10% fetal bovine serum, 2 mm glutamine, penicillin (100 units/mL), and streptomycin (100 μg/mL). Medium for MDA-MB-231 VB100 cells was also supplemented with vinblastine (100 ng/mL).
The human cervical epidermal carcinoma cell line, KB-3-1, and its doxorubicin-resistant subline, KB-A1, which were established in the Gottesman laboratory, were cultured in DMEM with 10% fetal calf serum supplemented with 2 mM glutamine, penicillin (100 units/mL), and streptomycin (100 μg/mL) (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2 and 95% air. The drug-resistant subline KB-A1 was kept under constant selection in doxorubicin (1 μg/mL) (22).
The human large-cell lung tumor line, COR-L23P, and its doxorubicin-selected MRP1-overexpressing multidrug-resistant variant line, COR-L23R, as previously described by Barrand et al. (23), were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin (100 units/mL), and streptomycin (100 μg/mL) (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2 and 95% air. The COR-L23R cells were also maintained in doxorubicin (0.2 μg/mL) (23).
RNA Isolation
RNA was isolated from approximately 1 million MCF-7 and MCF-7/ADR cells grown in two six-well plates to characterize ATP-binding cassette transporter expression, as described previously (24). Briefly, the medium and any detached cells were first removed from the wells, and total RNA was isolated from the cells that remained attached to the culture dish by use of the Qiagen RNeasy kit (Qiagen, Valencia, CA) by the manufacturer's protocol. RNA was quantitated by use of a NanoDrop micro-volume spectrophotometer (Thermo Scientific, Wilmington, DE), and then the integrity of the mRNA was verified with an Agilent BioAnalyzer (Agilent Technologies, Santa Clara, CA).
GeneChip Eukaryotic Expression Arrays
Isolated total RNA from the MCF-7 parental cells and MCF-7/ADR cells was used with the GeneChip Sample Cleanup Module to prepare biotinylated eukaryotic complementary RNA to hybridize to the GeneChip expression probe arrays, according to the manufacturer's protocol (Qiagen). Briefly, double-stranded cDNA was prepared from the isolated RNA by use of the SuperScript system (Invitrogen Life Technologies) with a T7-(dT)24 primer. This double-stranded cDNA was used to make the biotin-labeled complementary RNA by in vitro transcription with a labeling kit from Enzo (Farmingdale, NY). The biotinylated complementary RNA was fragmented for 35 minutes at 94°C in fragmentation buffer in the kit and then hybridized to the Affymetrix GeneChip U133A 2.0 arrays (Affymetrix, Santa Clara, CA) for 16 hours at 45°C. Each probe array was then washed and stained before being scanned twice with the GeneArray Scanner, according to the manufacturer's procedures (Affymetrix). Experiments with the arrays were performed in duplicate for each sample type (25). The raw microarray data have been posted online at http://cebs.niehs.nih.gov.
Microarray Analysis
The CEL files for hybridizations of the MCF-7/ADR cells and the MCF-7 cells were log2 transformed by use of RMA Express (26,27). Microarray data for the expression of 14 500 genes from MCF-7 and MCF-7/ADR were filtered with BRB-ArrayTools (version 3.8.0) (developed by Dr Richard Simon and the BRB-ArrayTools Development Team from the Biometric Research Branch at the National Cancer Institute, National Institutes of Health, Bethesda, MD). The array data were filtered by use of the exclusion parameters, spot intensities that were less than 10 and genes with a P value for the log-ratio variation that was greater than .01, and a total of 4976 genes were identified for further analysis. These 4976 genes were clustered by use of the Euclidean distance and complete linkage (Supplementary Figure 1, available online). The resulting dendrogram indicated that, for all experimental conditions, the duplicate microarrays form clusters, suggesting that the results were reproducible. A two-sample t test (with random variance model) was used to perform a class comparison and determine the statistically significant changes in gene expression between the MCF-7 and MCF-7/ADR cells. The univariate test random variance model parameters were a equals 1.68165, b equals 14.3422, and Kolmogorov–Smirnov statistic equals 0.03998. The nominal statistical significance level of each univariate test was .001. The first 419 genes listed were statistically significant at the nominal .001 level in the univariate test with a spot intensity fold change of 10. Genes with a spot intensity fold increase of greater than 10 are shown in Supplementary Table 1 (available online), and genes with spot intensity fold decrease of greater than −10 are shown in Supplementary Table 2 (available online). Parametric P values, false discovery rate, and the fold change as determined by the BRB-ArrayTools software are shown in Supplementary Tables 1 and 2 (available online). These genes were then analyzed by Ingenuity Pathways Analysis software (Ingenuity, Redwood City, CA) to determine which biological relationships exist between the genes present in this list. The reference set for this analysis was the Ingenuity Knowledge Base (Genes only), and the network analysis was set to direct relationships. All data sources were used to obtain data from patients and all human cell lines and tissues. The stringent filter was also used. Seventy molecules per network and 25 networks were used for this analysis. A numerical value was determined by the software to rank networks according to relevance to the genes in the input dataset. This score took into account the number of focus genes (genes in the original input dataset) in the network and the size of the network to approximate relevance of the network to the original list of focus genes. These calculations were based on the hypergeometric distribution that was calculated with a Fisher exact test for 2 × 2 contingency tables. The top three networks are shown in Supplementary Figure 2 (available online). Annotation was retrieved from the Affymetrix Netaffx site (www.affymetrix.com/analysis/index.affx) by use of DAVID (http://david.abcc.ncifcrf.gov/). The biological functions of genes with altered expression in the MCF-7/ADR cells are presented in Supplementary Table 3 (available online).
Quantitative RT-PCR Analysis
The mRNA expression levels of the selected genes showing statistically significant changes in the microarray analysis were validated with real-time quantitative RT-PCR. The LightCycler RNA Master SYBR Green Kit and LightCycler 480 instrument (Roche Biochemicals, Indianapolis, IN) were used in these studies as described previously (15). Specific PCR primer sequences for all validated genes and the reference gene (Supplementary Table 4, available online) were designed by use of LightCycler Probe Design (version 2) software, unless otherwise indicated. Crossing points (ie, cycle at which the fluorescence from a sample crosses above the background also known as the threshold) for each transcript were determined through second-derivative maximum analysis with arithmetic baseline adjustment (Roche LightCycler 480 software). Crossing point values for each transporter were normalized to the respective crossing point values for mRNA expression of the reference gene, plasma membrane calcium ATPase 4 (PMCA4) (28). Fold change in gene expression for drug-resistant cell lines compared with parental cells was determined from the same amount of total RNA by use of the delta, delta threshold cycle (Ct) method. A Ct of 40 was used for calculations when the gene expression was not detected.
Drug Resistance Gene Expression Analysis with the TaqMan Low-Density Array
The cDNA was synthesized from 1 μg of total RNA that had been isolated from CD44+/CD24+ and CD44+/CD24− MCF-7/ADR cells in a 20-μL reaction volume with a high-capacity cDNA kit and RNase inhibitor (Applied Biosystems, Foster City, CA) by the manufacturer's instructions. The reverse transcription conditions were as follows: 10 minutes at 25°C, 120 minutes at 37°C, and 5 seconds at 85°C. After reverse transcription, cDNA was stored at 4°C. Cells from two separate cell sortings were evaluated.
Expression levels of genes linked to drug resistance were measured with custom-made TaqMan low-density arrays (Applied Biosystems) that test for a total of 381 genes (Supplementary Table 6, available online), including all known ATP-binding cassette transporters and many other genes that are involved in drug resistance (From the Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute, [ J.-P. Gillet, A. M. Calcagno, S. V. Ambudkar, M. M. Gottesman]; Bioinformatics and Computational Biosciences Branch, National Institute of Allergy and Infectious Diseases, [S. Varma], NIH Bethesda, Maryland; Division of Pathology, Norwegian Radium Hospital, Oslo University Hospital, and The Medical Faculty, University of Oslo, Oslo, Norway [B. Davidson]; Section for Gynecologic Oncology, Division of Obstetrics and Gynecology, Norwegian Radium Hospital, Oslo University Hospital, Oslo, Norway [ M. Bunkholt Elstrand]; Cleveland Clinic Taussig Cancer Institute, Cleveland, Ohio [R. Ganapathi]; Departments of Gynecologic Oncology and Cancer Biology, and Center for RNA Interference and Non-Coding RNA, University of Texas M. D. Anderson Cancer Centre, Houston, Texas [A. A. Kamat, A. K. Sood ]; Fox Chase Cancer Center, Philadelphia, Pennsylvania [M. V. Seiden ]; Vincent Center for Reproductive Biology, Vincent Obstetrics and Gynecology Service, Massachusetts General Hospital, Boston, Massachusetts [ B. R. Rueda], unpublished results). The cDNA (1 μg) was mixed with 2× TaqMan Universal PCR Master Mix (Applied Biosystems) and loaded on the TaqMan low-density array. Expression was analyzed with an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) by the manufacturer's instructions.
Expression data from the TaqMan low-density array were analyzed with RQ documents and the RQ Manager Software for automated data analysis (Applied Biosystems) to provide Ct values, which were median normalized (28). Two-dimensional hierarchical cluster analysis, by use of the Manhattan distance with the complete linkage algorithm, was performed with the CIMminer tool (http://discover.nci.nihl.gov) to group expression data for the 381 genes (including genes linked to multidrug resistance on the basis of their expression profiles) from two biological replicates of the CD44+/CD24+ and CD44+/CD24− MCF-7/ADR cells.
Surface Marker Analysis by Flow Cytometry
We assessed expression of CD44, CD24, CD133, and ABCB1 surface markers on MCF-7, MCF-7/ADR, MCF-7/MDR, OVCAR-8, OVCAR-8/ADR, KB-A1, MCF7-FLV1000, MCF7-AdVp3000, MCF-7/VP-16, COR-L23P, COR-L23R, MDA-MB-231, and MDA-MB-231 VB100 cells. Cells were washed with phosphate-buffered saline (PBS) before they were removed from the culture dish with trypsin and EDTA, counted, resuspended in 5 mL of fresh medium, incubated at 37°C for 30 minutes, and then transferred to Eppendorf tubes (1 × 106 cells per tube). After a wash with a solution of PBS and 0.5% bovine serum albumin, cells were centrifuged at 500g, and the supernatant was removed. The appropriate fluorochrome-conjugated antibody against a surface marker was added at 1:10 dilution (5–50 μL) in PBS and 0.5% bovine serum albumin and incubated on ice in the dark for 30 minutes. All antibodies were used at a dilution of 1:10. Cells were washed with PBS and 0.5% bovine serum albumin. 7-Amino-actinomycin D (7-AAD) was added to all samples, except for the unstained and single-stained cells, to exclude dead cells from the analysis. Cells were analyzed by flow cytometry with an LSRII flow cytometer (BD Biosciences). Data were collected with FACSDiVa software (BD Biosciences) from no fewer than 30 000 cells. The following laser filters were used: fluorescence detector FL1, for fluorescein isothiocyanate; fluorescence detector FL2, for phycoerythrin; fluorescence detector FL3, for 7-AAD; and fluorescence detector FL5, for allophycocyanin (red laser). If compensation to adjust for spectral overlap was required, compensation beads were used. Briefly, 50 μL of the positive control beads and 50 μL of the negative control beads (BD Biosciences) were added, followed by 5 μL of the specific fluorochrome-conjugated antibody, and cells were processed as described above.
Mammosphere Formation and Limiting Dilution Assay
We used MCF-7 and MCF-7/ADR cells as well as the CD44+/CD24− and CD44+/CD24+ MCF-7/ADR cells to assess mammosphere formation in the absence of culture-plate attachment. Mammosphere identification was performed as described (29) and formation frequency for each cell line was determined by a limiting dilution assay (30) in which various numbers of cells (500 cells to one cell) per well were plated in 96-well low-binding plates (six wells per dilution; Fisher Scientific, Hampton, NH). The number of mammospheres was scored 10–14 days later, depending on the spheroid growth rates. Cells were cultured in serum-free DMEM/F12 medium supplemented with fibroblast growth factor (10 ng/mL) and epidermal growth factor (10 ng/mL) (Preprotech, Rocky Hill, NJ); insulin (50 μg/mL) (Sigma Chemical Co); and B27 (100 units/mL), N2 supplements (100 units/mL), penicillin (100 units/mL), and streptomycin (100 μg/mL) (Invitrogen).
Invasion and Motility Assays
We used MCF-7, MCF-7/ADR, and MCF-7/MDR cells for invasion and motility assays. For invasion assays, cells were removed from the culture dish with trypsin and EDTA, resuspended in DMEM with 10% fetal calf serum, centrifuged at 500g, washed in Dulbecco's PBS, and resuspended in serum-free DMEM at 50 000 cells per milliliter. The Biocoat Matrigel invasion chamber (BD Biosciences) was used after a 2-hour rehydration step at 37°C with serum-free DMEM. The invasion chamber has six cell culture inserts, each of which contains a polyethylene terepthalate membrane with 8-μm pores that has been coated with Matrigel (one invasion chamber per well in a six-well plate). A total of 100 000 cells in 2 mL of serum-free DMEM was added to the apical side of an insert, and then 2.5 mL of DMEM plus 10% fetal calf serum was added to the basal side of the insert as the chemoattractant. Cells were kept at 37°C overnight. The next morning medium was poured off, and cells that had not invaded through the pores of the insert were scraped off the apical side of the inserts with a sterile cotton swab and discarded. Cell culture inserts were then placed into a Diff Quik staining set (Dade Behring, Newark, DE) and treated with fixative (triarylmethane and methanol, 1:1 vol/vol) for 2 minutes, with stain 1 (xanthenes dye) for 2 minutes, with stain 2 (thiazine dye) for 2 minutes, and then with two 2-minute water washes. For each insert, cells in five different fields were counted at a magnification of ×10, and values were averaged. Invasion of MCF-7 cells was compared with that of MCF-7/ADR and MCF-7/MDR cells. Wells used for invasion assays were photographed at ×40.
For motility studies, we used invasion chambers with control inserts that contained the same type of membrane but without the Matrigel coating (one chamber per well of a six-well plate). A total of 100 000 cells in 2 mL of serum-free DMEM was added to the apical side of each insert, and 2.5 mL of serum-free DMEM (ie, with no chemoattractant) was added to the basal side of each insert. The inserts were processed as described above for the invasion assay. Motility of the MCF-7 cells was compared with that of MCF-7/ADR and MCF-7/MDR cells.
Analysis of the Side-Population Cell Fraction
MCF-7 and MCF-7/ADR cells were used in the side-population assay to identify stem cells (ie, the side-population fraction) that overexpress an ABC transporter, such as ABCB1, and so can transport Hoechst 33342 dye. Briefly, cells were suspended in prewarmed RPMI 1640 medium containing 2% fetal bovine serum and 2 mM HEPES buffer at 1 × 106 cells per milliliter and then incubated at 37°C for 20 minutes; Hoechst 33342 dye (Invitrogen) was added to a final concentration of 5 μg/mL (8.1 μM) from a stock solution at 1 mg/mL and incubated with cells for 90 minutes in a 37°C shaking bath. Control cells were incubated with 50 μM verapamil or 10 μM cyclosporine A (Sigma Chemicals Co) for 15 minutes at 37°C before Hoechst 33342 dye addition to inhibit the ABCB1 transporter. All cells were immediately placed on ice, washed, and resuspended in ice-cold Hanks buffered salt solution containing 2% fetal bovine serum and 2 mM HEPES buffer (pH 7.4). 7-AAD (BD Pharmingen, Lexington, KY) was added to the cells at 20 μg/mL and red-stained dead cells were detected with FL3 detector and excluded from the analysis. Hoechst 33342-labeled cells were analyzed on a LSRII flow cytometer (BD Biosciences), and cell aggregates were discarded from the analysis by doublet discrimination. Side-population cells were visualized or sorted by use of red (FL8) vs blue (FL7) ultraviolet channels both in linear mode, as described previously (31).
Analysis of Aldehyde Dehydrogenase Activity
The expression of aldehyde dehydrogenase (ALDH) is enriched in cancer stem cells (32,33). For this reason, ALDH activity in MCF-7 and MCF-7/ADR cells was determined by use of an Aldeflour kit (StemCell Technologies, Inc, Vancouver, BC, Canada) as per the manufacturer's instructions. Briefly, 1 × 106 cells were suspended in the Aldeflour buffer containing the ALDH substrate dipyrromethene boron difluoride aminoacetaldehyde at 1 μM and incubated for 40 minutes at 37°C. A specific ALDH inhibitor, diethylaminobenzaldehyde, was used at 50 μM as a negative control and to establish the gate for ALDH-positive cells. Aldeflour fluorescence was detected by use of the FL1 detector in a LSRII flow cytometer.
Xenograft Tumor Studies in Mouse Models
We used 6- to 8-week-old female nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice aged 6–8 weeks (NCI Frederick animal facility) for these experiments. All mouse studies were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International–accredited facility, in compliance with the US Public Health Service guidelines for the care and use of animals in research. Mice received estrogen supplementation by subcutaneous implantation of a 90-day pellet because MCF-7 cells require estrogen supplementation for growth in vivo. Sham-sorted or MCF-7 and MCF-7/ADR cells sorted for indicated cell surface markers were resuspended in 100 μL of Dulbecco's PBS and injected into the inguinal mouse mammary pad (pad 4) to generate one tumor per mouse. A total of three or four mice per experimental group were used in three independent experiments. Thus, for all experiments in this study, we used a total of 52 mice selected at random. Mice were housed at five mice per cage on arrival. These mice were selected at random and placed into new cages for each experimental group. The other eight mice died because of fight wounds or had complications after estrogen pellet implantation. They were not used in any experiment. The mice were labeled specifically with an ear tag to identify each tumor type. The growth of tumors was monitored with a caliper twice a week, and the weight (in milligrams) of each mouse was also monitored twice a week. Tumor growth was measured biweekly, and the weights (in milligrams) were calculated by use of the formula for a prolate ellipsoid and by assuming a specific gravity of 1.0 g/cm3, as described by Plowman et al. (34): L × W2 × 0.5.
Statistical Methods
Statistical significance was determined with unpaired Student t tests, except as noted for analyses of microarray data below. P values of less than .05 were considered statistically significant. All statistical tests were two-sided. Means and 95% confidence intervals (CIs) are reported. A power calculation was not performed before the start of the experiments.
The microarray data were filtered by use of BRB-ArrayTools by excluding spot intensities of less than 10 and genes with a P value for log-ratio variation of greater than .01. Genes that remained in the analysis were then used to determine class comparisons. This class comparison analysis yielded 419 genes with the fold change of greater than plus or minus 10 and a P value of less than .001, as determined in a univariate test that compared gene expression of MCF-7/ADR cells with that of the MCF-7 cells. To identify which genes were statistically significantly overexpressed or underexpressed in the MCF-7/ADR CD44+/CD24− cells (in comparison with the MCF-7/ADR CD44+/CD24+) via data from the TaqMan low-density array, we used class comparisons with two-sample t tests and 10 000 random permutations (BRB-ArrayTools, version 3.7.0). Results were considered statistically significant at a P value of less than .05.
For the invasion and motility assays, a t test was performed for each pair to determine whether the number of cells that invaded the Matrigel or moved through the pores, respectively, was statistically significantly different between cell types. The mean, difference, and 95% confidence interval for the difference of means were calculated for each pair assayed.
Results
Microarray Analysis of Global Gene Expression in MCF-7/ADR Cells and Parental MCF-7 Cells
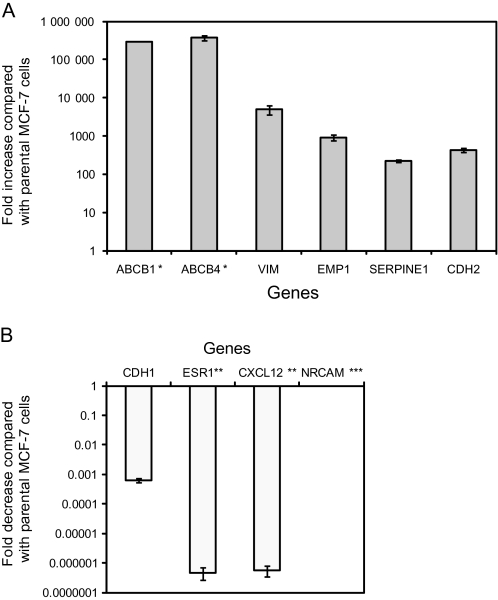
The MCF-7/ADR cells that have been selected in increasing concentrations of doxorubicin have a drug-resistance phenotype because they overexpress ABCB1 transporters (14). We investigated whether MCF-7/ADR cultures that have been subjected to long-term continuous drug selection contain an expanded cancer stem cell–like population. We first used microarray analysis of mRNA isolated from parental MCF-7 and MCF-7/ADR cells to evaluate the global differences in gene expression. We found that expression data from 4976 genes were clustered (Supplementary Figure 1, available online) into two distinct groups (ie, one from MCF-7 cells and the other formed from MCF-7/ADR cells). Using a class comparison analysis, we identified 419 differentially expressed genes between these two cell types by a univariate test (Supplementary Tables 1 and 2, available online). As expected, ABCB1 mRNA expression was higher in long-term doxorubicin-selected MCF-7/ADR cells than in parental MCF-7 cells. We also found statistically significant lower mRNA expression of estrogen receptor (ESR1) (−126.03-fold, 95% CI = −121.73 to −130.32-fold; P < .001), E-cadherin (−289.15-fold, 95% CI = −252.68 to −325.63-fold; P < .001), and CD24 (−35.6-fold, 95% CI = −9.9 to −61.3-fold; P < .001) but higher mRNA expression of ALD1A3 (4.84-fold, 95% CI = 4.12 to 5.56-fold; P = .005) and N-cadherin (18.99-fold, 95% CI = 13.01 to 24.97-fold; P = .028) in MCF-7/ADR cells than in parental MCF-7 cells. The gene expression profile of MCF-7/ADR cells thus mimics that of breast cancer stem cells (4,6,33). We also found a non-statistically significantly higher (1.99-fold, 95% CI = 1.26 to 2.72-fold; P = .095) expression of SNAI1 mRNA in MCF-7/ADR cells than in parental MCF-7 cells. It should be noted that the loss of E-cadherin expression followed by an increase in N-cadherin expression is a hallmark of the epithelial-to-mesenchymal transition and a more invasive phenotype that has been linked to cancer stem cells (35– 37).
The 419 genes that were identified as being differentially expressed between MCF-7/ADR cells and parental MCF-7 cells by the microarray analysis were further examined with Ingenuity Pathway Analysis software to identify biological networks that were active in these cells. In MCF-7/ADR cells, the highest scoring network was the transforming growth factor β (TGFB1) network, which is involved in regulating cell proliferation, apoptosis, and induction of the epithelial-to-mesenchymal transition (Supplementary Table 3, available online) and includes ESR1 and p53 proteins (Supplementary Figure 2, available online). This result was consistent with the microarray analysis because SNAI1 protein has recently been reported to mediate the TGFB1 signaling pathway and to decrease expression of E-cadherin and the estrogen receptor in breast cancer cells (35). Other high scoring networks (Supplementary Table 3, available online) were related to survival (ANXA1, TGFB1, and SERPINE1), loss of adhesion (CDH2, EGFR, and LOX), and the epithelial-to-mesenchymal transition (eg, TWIST1) (36,38). Cancer stem cells have been associated with an invasive phenotype (ie, MMP1, CDH2, and CDH1) and increased survival (ie, TGFB1 and ALDH1), similar to the characteristics that were found in the pathway analysis (39).
We used real-time RT-PCR to validate the expression profiles for 10 genes (ABCB1, ABCB4, CDH1, CDH2, CXCL12, EMP1, ESR1, NRCAM, SERPINE2, and VIM) that were associated with drug resistance and stem cell characteristics in both the MCF-7 and MCF-7/ADR cells (Supplementary Table 4, available online). Patterns of gene expression were similar between the microarray analysis and real-time RT-PCR analysis (Figure 1), although the fold change varied between the real-time RT-PCR analysis and the microarray analysis. Thus, after the long-term drug selection process, gene expression patterns observed in MCF-7/ADR cells are similar to those in breast cancer stem cells (4,6,33).
Figure 1.
Differential gene expression in long-term doxorubicin-selected MCF-7/ADR cells and parental MCF-7 cells. A) Six genes with higher expression in MCF-7/ADR cells than in parental MCF-7 cells. B) Four genes with lower expression in MCF-7/ADR cells than in parental MCF-7 cells. Real-time quantitative reverse transcription–polymerase chain reaction was used to measure the mRNA expression levels of genes with statistically significantly different expression, as detected in the microarray analysis, between MCF-7/ADR and MCF-7 cells. * = Not detected in parental MCF-7 cells; ** = not detected in MCF-7/ADR cells; *** = not detected in either cell line. Data are the average fold change values in MCF-7/ADR cells compared with MCF-7 cells and are from at least two independent experiments, with at least two replicates per gene. Error bars = 95% confidence intervals.
The CD44+/CD24− Cell Population in MCF-7/ADR and MCF-7 Cell Cultures
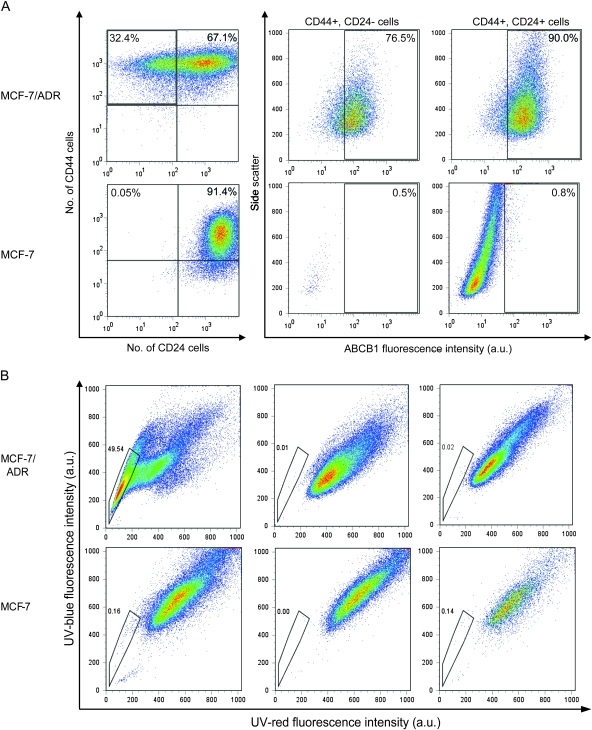
We further investigated whether MCF-7/ADR and parental MCF-7 cells possessed the putative breast cancer stem cell markers, CD24 and CD44, which have been reported in BRCA1-mutated murine cancer stem cells (29,30). Virtually all cells in parental MCF-7 cultures expressed both CD44 and CD24 (ie, CD44+/CD24+ cells) (Figure 2, A), with a negligible fraction (<0.05%) expressing only CD44 but not CD24 (ie, CD44+/CD24− cells). In contrast, cultures of MCF-7/ADR cells had a substantial fraction (32.4%) of cells that expressed CD44 but not CD24 (ie, CD44+/CD24− cells), consistent with the decreased expression of CD24 observed in the gene expression analysis. In addition, cultures of MCF-7/ADR cells, but not of parental MCF-7 cells, had a substantial side-population cell fraction. Treatment of MCF-7/ADR cultures with verapamil or cyclosporine A, which inhibit ABCB1 transporters, eliminated the side population in these cultures (Figure 2, B), which indicates that the function of ABCB1 transporter can be used to identify the side population. These results indicate that cultures of drug-selected MCF-7/ADR cells contain a substantial population of cells with the CD44+/CD24− phenotype and a side-population fraction with functional ABCB1.
Figure 2.
Analysis of mammary stem cell markers in cultures of MCF-7/ADR cells and parental MCF-7 cells. We used CD44, CD24, and ABCB1 expression as stem cell markers and transport of Hoechst 33342 dye to indicate ABCB1 function. Cells were sorted for these markers by flow cytometry. A) Analysis of the cell surface expression of CD44 and CD24 protein on MCF-7 parental cells and multistep doxorubicin-selected MCF-7/ADR cells. CD44 and CD24 protein were detected with fluorochrome-conjugated antibodies. Flow cytometry was used to sort cells into CD44+/CD24− and CD44+/CD24+ populations, and the expression of ABCB1 protein was assessed in each population, also by flow cytometry. Histograms from a representative experiment of a total of four experiments are shown. Percentages indicate the amount of cells in that quadrant. Side scatter is reflected and refracted light intensity related to cell granularity and complexity. B) Analysis of the side-population cell fraction in MCF-7 and MCF-7/ADR cell cultures. Left) Hoechst 33342 dye treatment only. Middle) Verapamil treatment. Right) Cyclosporine A treatment. All cells were incubated with Hoechst 33342 dye, which was excited at 357 nm and its fluorescence was analyzed at two wavelengths (blue = 402–446 nm; and red = 650–670 nm), both in linear mode. Cells were incubated with 50 μM verapamil or 10 μM cyclosporine A before addition of Hoechst dye; dead cells were excluded by incubation with 7-AAD (20 μg/mL) immediately before analysis. Cell aggregates were discarded from the analysis by doublet discrimination. Side-population cells are shown by the outlined area. a.u. = arbitrary units.
We also compared cell surface expression of ABCB1 protein between populations of CD44+/CD24− and CD44+/CD24+ MCF-7/ADR cells by flow cytometry. ABCB1 protein has been shown previously to be overexpressed on the cell surface of MCF-7/ADR cells (14), and cell surface expression of ABCB1 protein has been shown to be associated with hematopoietic stem cells (40) and solid tumor cancer stem cells (39). As expected, nearly all MCF-7/ADR cells expressed ABCB1 protein, with essentially no difference between the CD44+/CD24− and CD44+/CD24+ cell populations, but ABCB1 was not detected on MCF-7 parental cells. In addition, MCF-7/ADR cell cultures had a higher fraction of ALDH-positive cells (5.6%, average of two independent experiments), a characteristic that has been previously associated with cancer stem cells (29,32,33), than in the parental MCF-7 cells (0.9%) (difference = 4.7%, 95% CI = −1.4% to 10.8%). Aldeflour-positive cells were found to be lower in MCF-7 cell cultures than in MCF-7/ADR cell cultures (Supplementary Figure 3, A, available online). Previously, we reported that breast tumors with BRCA1 mutations contain CD133+ cells (30). In this work, we also evaluated cell surface expression of CD133 in MCF-7 and MCF-7/ADR cell cultures. Neither culture expressed CD133 (Supplementary Figure 3, B, available online). Thus, only CD44+/CD24− cells were associated with the drug-resistant phenotype; however, the role played by CD44+/CD24− cells in the development of drug resistance is not clear.
We then examined cultures of other drug-selected cells for cell surface expression of CD24, CD44, and CD133 proteins, including doxorubicin-resistant and verapamil-resistant MCF-7 AdVP3000 cells, doxorubicin-resistant KB-A1 cells, doxorubicin-resistant NCI-ADR-RES cells (also known as OVCAR-8/ADR cells), doxorubicin-resistant COR-L23R cells, the estrogen receptor-negative breast cancer cell line MDA-MB-231, and vinblastine-resistant MDA-MB-231 VB100 breast cancer cells and their respective parental cell lines, but found none that were enriched in CD44+/CD24− cells. We also evaluated another drug-selected breast cancer cell line, MDA-MB-231 VB100 (21) and found that approximately 50% of the parental MDA-MB-231 and MDA-MB-231 VB100 cells had the CD44+/CD24− phenotype, and so drug-selected MDA-MB-231 VB100 cells were not enriched in CD44+/CD24− cells. In addition, cultures of all other cell lines examined (ie, MCF-7 AdVP3000, KB-A1, NCI-ADR-RES, COR-L23R, and MDA-MB-231 VB100) were not enriched in CD133+ cells (data not shown). In particular, no enrichment of CD133+ cells was found in drug-resistant ovarian cell cultures, in contrast to a recent report that identified CD133 as a cancer stem cell marker in ovarian tumors (41).
To further examine whether the presence of ABCB1 protein alone could enrich for cancer stem cell markers, we examined MCF-7/MDR cells, which had previously been derived from parental MCF-7 cells by transfection with cDNA for the ABCB1 transporter (17). We found that MCF-7/MDR cell cultures were not enriched in CD44+/CD24− cells and that the invasion ability of MCF-7/MDR cells was not enhanced (data not shown). Thus, ABCB1 protein expression alone does not appear to stimulate the expression of cancer stem cell markers.
Formation of Mammospheres by MCF-7/ADR Cells
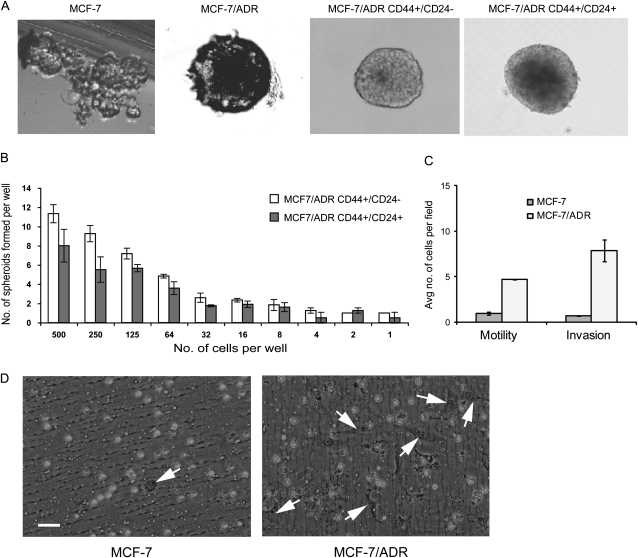
Formation of nonadherent spherical clusters of cells or mammospheres in three-dimensional culture is a characteristic of cancer stem cells (42). We found that MCF-7/ADR cells had a high capacity to form mammospheres, with approximately one in five cells capable of forming mammospheres. The MCF-7/ADR mammospheres were large and grew more rapidly than MCF-7 mammospheres, which were small cell clusters that adhered to the culture dish and were difficult to score in the limiting dilution assay (Figure 3, A). Although the CD44+/CD24− cell population isolated by flow cytometry from MCF-7/ADR cells had a tendency to form mammospheres more efficiently than the CD44+/CD24+ cell population, the two types of cells gave similar results in the limiting dilution assay (Figure 3, B).
Figure 3.
Characterization of MCF-7 and MCF-7/ADR cells in three-dimensional culture. Mammosphere formation by unsorted and sorted (CD44+/CD24− and CD44+/CD24+) cell populations of MCF-7/ADR cells and parental MCF-7 cells was assessed by the limiting dilution assay. A) Morphologic appearance of mammospheres. A representative mammosphere from a total of 50 countable mammospheres from each experimental condition is shown. B) Spheroid formation by sorted cells in anchorage-independent culture. Long-term spheroid assays were performed by limiting dilution with 500 cells to one cell per well of a 96-well low-binding plate. The number of colonies was scored at the end of 14 days. This experiment was performed twice, with six wells per cell dilution. Error bars = 95% confidence intervals. C) Motility and invasiveness of MCF-7/ADR cells and parental MCF-7 cells. Cell culture inserts were used for both assays. Membranes coated with Matrigel were used for invasion assays, and membranes without a Matrigel coating were used for motility assays. For each insert, cells in five different ×10 fields were counted. Cell numbers from two experiments were averaged. Number of replicates (wells) for each bar is four. Differences in motility or invasiveness were compared with a two-sided t test (both P < .001). D) Representative inserts from invasion assays for MCF-7/ADR cells and parental MCF-7 cells. White circles are pores. MCF-7 cell in the field captured on the left contains only one cell (arrow). MCF-7/ADR cells in the right panel are more numerous and elongated, as indicated by arrows, with a mesenchymal appearance. Cultures were photographed at a magnification of ×40. Scale bar = 50 μm.
Invasiveness and Morphology of MCF-7/ADR Cells in Culture
It has been shown (39) for other mammary cancer cells (ie, MCF7-S and B3R cell lines and primary cells isolated from the human breast cancer tissue) that the ability of cancer stem cells to migrate in cell culture and to metastasize in NOD/SCID mice is enhanced, compared with cell populations that do not contain cancer stem cells. In vitro Matrigel assays in culture dishes that contain Matrigel-coated membrane inserts have been used to investigate the invasive properties of various cell types including breast cancer cell lines (43). We used this assay system to assess motility (without Matrigel-coated membranes) and invasiveness (with Matrigel-coated membranes) between MCF-7/ADR cells and their parental MCF-7 cells. MCF-7/ADR cells were statistically significantly more motile than parental MCF-7 cells (MCF-7 cells = 0.96 motile cells per field and MCF-7/ADR cells = 4.72 motile cells per field; difference = 3.76 motile cells per field, 95% CI = 2.41 to 5.11 motile cells per field; P < .001) (Figure 3, C) and statistically significantly more invasive (MCF-7 cells = 0.82 invasive cells per field; MCF-7/ADR = 7.51 invasive cells per field; difference = 6.69 invasive cells per field; 95% CI = 4.82 to 8.55 invasive cells per field; P < .001) (Figure 3, C and D). The morphology of MCF-7/ADR cells was elongated (which has been associated with increased invasiveness and increased motility) and cultured cells did not form the cobblestone pattern, that is characteristic of MCF-7 cells (Supplementary Figure 4, available online).
Tumorigenicity of MCF-7/ADR Cells in Mice
MCF-7 cells are weakly tumorigenic in NOD/SCID mice and require estrogen supplementation to generate a tumor (44). In preliminary studies, we found that 2.8 × 106 MCF-7 cells could form tumors in two of four mammary fat pad injection sites after 65 days (data not shown) but that 0.5 × 106 or 1.0 × 106 MCF-7 cells injected in a single mouse could form tumors in only one of the four sites after 85 days (Table 1). However, these MCF-7 tumors did not grow larger during the 120-day observation period, probably because the estrogen source (ie, pellets) had been exhausted. In contrast, MCF-7/ADR cells were more tumorigenic, and 1 × 106 cells injected in fat pad 4 could form a tumor after a mean of 45 days and 0.5 × 106 cells could form tumors after a mean of 66 days. At a constant number of cells per injection, MCF-7/ADR cells formed larger tumors in a single mouse (Table 1) in less time than MCF-7 parental cells. In addition, by 94 days after injection, tumors were formed by 0.05 × 106 MCF-7/ADR cells in two of the three injection sites and after 108 days a tumor was formed by 0.005 × 106 MCF-7/ADR cells in one of the three injection sites, but no tumors were formed by the same number of parental MCF-7 cells. Although tumorigenicity of CD44+/CD24− and sham-sorted MCF-7/ADR cells was similar, no tumors were formed by CD44+/CD24+ or CD44−/CD24+ MCF-7/ADR cells, indicating that the increased tumorigenicity of MCF-7/ADR cells could be attributed to the CD44+/CD24− MCF-7/ADR cell population (Table 1).
Table 1.
Evaluation of tumorigenicity in vivo*
| Cell line and estrogen supplementation | No. of cells injected | No. of tumors/No. of mice | Mean latency, days (95% CI) | Maximum tumor volume, mm3 | Day of tumor harvest |
| With estrogen supplementation | |||||
| MCF-7/ADR | 1 × 106 | 4/7 | 45 (38.3 to 53.2) | 1160 | 74 |
| 0.5 × 106 | 2/3 | 66 (54.2 to 77.8) | 180 | 74 | |
| 0.05 × 106 | 2/3 | 94 (83.7 to 105.3) | 470 | 120 | |
| 0.005 × 106 | 1/3 | 108 | 190 | 120 | |
| MCF-7 | 1 × 106 | 4†/7 | 85 (64.7 to 105.3) | 80 | 120 |
| 0.5 × 106 | 2†/3 | 116 (108.2 to 123.8) | 20 | 120 | |
| 0.05 × 106 | 0/3 | ‡ | ‡ | ‡ | |
| 0.005 × 106 | 0/3 | ‡ | ‡ | ‡ | |
| MCF-7/ADR CD44+/CD24– | 1 × 106 | 1/2 | 65 | 670 | 88 |
| 0.005 × 106 | 1/2 | 120 | 200 | 120 | |
| MCF-7/ADR CD44+/CD24+ | 1 × 106 | 1/3 | ‡ | 20 | 120 |
| MCF-7/ADR CD44–/CD24+§ | 0.005 × 106 | 0/2 | ‡ | ‡ | ‡ |
| Without estrogen supplementation | |||||
| MCR-7/ADR SHAM | 0.25 × 106 | 2/4 | 90 (80 to 100) | 520 | 100 |
| 0.1 × 106 | 0/4 | ‡ | ‡ | ‡ | |
| MCF-7/ADR CD44+/CD24– | 1 × 106 | 1/2 | 75 | 1630 | 120 |
| 0.2 × 106 | 1/2 | 65 | 320 | 74 | |
| MCF-7/ADR SHAM p2 | 0.2 × 106 | 1/1 | 60 | 180 | 100 |
| 0.1 × 106 | 1/1 | 65 | 80 | 74 |
Nonobese diabetic/severe combined immunodeficient mice (n = three to four mice per group; some mice died from unrelated causes, such as bite wounds, etc, and were not scored for this reason; some groups as indicated had only one and others had two, three, or four mice) with or without estrogen supplementation in the form of estrogen pellets were used for these experiments. Tumor cells, as indicated, were injected in 100 μL of phosphate-buffered saline bilaterally into mouse mammary fat pads (one tumor per mouse). Data are from three independent experiments, except as noted. MCF-7/ADR sham-sorted cells (SHAM) p2 were cells from one tumor injected with 0.5 × 106 MCF-7/ADR cells. Sham-sorted cells (SHAM) and cells sorted by use of CD44 and CD24 expression were injected within 1–2 hours after sorting. The mean latency was calculated as the time from tumor cell injection to appearance of measurable tumors (at least 5 mm × 5 mm). The 95% confidence interval (CI) was calculated for each tumor group with more than one tumor. Day of tumor harvest was determined from the day of cell injection into the mouse fat pad.
Palpable tumors only; no further growth.
No palpable tumors were present after 120 days.
Because there were few CD44–/CD24+ MCF-7/ADR cells, tumorigenicity assays with these cells could be performed only once.
We found decreased ESR1 mRNA expression in MCF-7/ADR cells, compared with MCF-7 cells, in a global gene expression analysis and in an RT-PCR analysis, and so we examined whether MCF-7/ADR cells had become estrogen independent during the course of these experiments by studying their ability to form tumors in mice without estrogen supplementation. Indeed, MCF-7/ADR and CD44+/CD24− MCF-7/ADR cells formed tumors without estrogen supplementation (Table 1). In addition, as few as 0.1–0.2 × 106 MCF-7/ADR cells could form a tumor within 60 days of injection (at four sites per mouse) without estrogen supplementation. Thus, cultures of MCF-7/ADR cells not only were enriched in cells with the CD44+/CD24− phenotype but the MCF-7/ADR cells also became estrogen independent.
Self-renewal of CD44+/CD24− and CD44+/CD24+ MCF-7/ADR Cell Populations
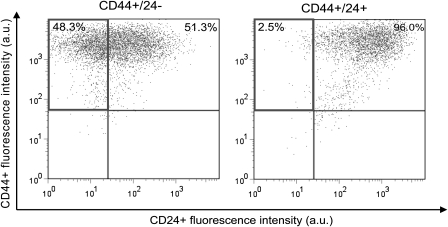
We assessed the ability of CD44+/CD24− and CD44+/CD24+ MCF-7/ADR cell populations to self-renew (ie, able to produce cells with the tumorigenic CD44+/CD24− phenotype) and to differentiate (ie, capacity to generate the phenotypically diverse nontumorigenic CD44+/CD24+ MCF-7/ADR cells) in vitro in the absence of doxorubicin by culturing each cell type for 14–16 days in low-attachment cell culture flasks so that mammospheres could be formed. We then determined cell surface expression of CD44 and CD24 proteins on these mammospheres to assess self-renewal. The CD44+/CD24− MCF-7/ADR cells were able to self-renew and differentiate into CD44+/CD24+ cells (Figure 4, Left), but the CD44+/CD24+ MCF-7/ADR cells essentially did not change, with nearly all cells remaining CD44+/CD24+ cells (Figure 4, Right). Similar results were obtained when cells were sorted for CD44 and for CD24 and grown as a monolayer for 14 days in the absence of doxorubicin (results not shown).
Figure 4.
Self-renewal and differentiation abilities of sorted MCF-7/ADR cell population. Self-renewal was defined as maintenance of cells with cancer stem cell markers, CD44+/CD24–. Differentiation was defined as loss of these markers and appearance of CD44+/CD24+ double-positive populations. Cultures of MCF-7/ADR cells were sorted by flow cytometry into CD44+/CD24− or CD44+/CD24+ populations, each cell type was grown as mammospheres for 16 days, and then cells in the mammospheres were examined for cell surface expression of CD44 and CD24 by flow cytometry. Percentages in a quadrant indicate the relative numbers of cells in that quadrant at the end of the experiment. In both panels, upper left quadrant = CD44+/CD24– cells; upper right = CD44+/CD24+ cells; lower left = CD44–/CD24− cells, and lower right = CD44−/CD24+ cells. Data are from one experiment with duplicate samples; a total of two experiments were performed.
Gene Expression in CD44+/CD24− MCF-7/ADR Cells and Drug Resistance
We evaluated CD44+/CD24− and CD44+/CD24+ MCF-7/ADR cells for the expression of 380 genes that have been associated with drug resistance (J.-P. Gillet, A. M. Calcagno, S. Varma, B. Davidson, M. Bunkholt Elstrand, R. Ganapathi, A. A. Kamat, A. K. Sood, S. V. Ambudkar, M. V. Seiden, B. R. Rueda, and M. M. Gottesman, unpublished results) by use of a custom-made TaqMan low-density array. Values for the differential expression of each gene between these cell types were then averaged between replicate samples, and class comparisons were obtained with BRB-ArrayTools, version 3.6.0, software and two-sample t tests, with 10 000 random permutations. Results were considered statistically significant at a P value of less than .05. From this analysis, we developed a 16-gene signature that could distinguish between CD44+/CD24− and CD44+/CD24+ cells (Table 2); six of the 16 genes encoded ABC transporters (including the ABCA1 transporter) and five encoded members of the solute carrier superfamily of transporters. Another study (31) has reported increased ABCA1 mRNA expression in murine primitive hematopoietic cells. TRAF1 (45) and FAS (46) proteins are activators of nuclear factor κB, and RAD18 and MLH3 proteins participate in DNA repair. UGCG gene expression is of particular interest because it has been linked to breast cancer prognosis (47) and multidrug resistance (48).
Table 2.
Comparison of gene signatures for CD44+/CD24– and CD44+/CD24+ MCF-7/ADR cells*
| Gene symbol | Parametric P† | Geometric mean expression (95% CI) |
Fold change | |
| CD44+/CD24− cells | CD44+/CD24+ cells | |||
| GSTM4 | .001 | 0.044 (0.041 to 0.047) | 0.010 (0.0095 to 0.0105) | 4.41 |
| TRAF1 | .006 | 0.027 (0.025 to 0.029) | 0.011 (0.01 to 0.012) | 2.33 |
| SLC19A3 | .013 | 0.015 (0.012 to 0.018) | 0.006 (0.005 to 0.007) | 2.66 |
| ABCA1 | .013 | 0.673 (0.483 to 0.863 | 0.125 (0.094 to 0.156) | 5.39 |
| CCND1 | .016 | 26.278 (24.349 to 28.207 | 13.667 (11.697 to 15.637) | 1.92 |
| SLC1A5 | .017 | 12.092 (11.278 to 12.906) | 6.345 (5.374 to 7.316) | 1.91 |
| FAS | .023 | 0.933 (0.865 to 1.001) | 0.379 (0.278 to 0.48) | 2.46 |
| GGT1 | .029 | 0.068 (0.043 to 0.093) | 0.023 (0.0225 to 0.0235) | 2.91 |
| AHR | .031 | 0.354 (0.219 to 0.489 | 1.101 (0.957 to 1.245 | 0.32 |
| SLC29A1 | .031 | 0.410 (0.359 to 0.461 | 0.196 (0.151 to 0.241) | 2.09 |
| MLH3 | .033 | 0.318 (0.259 to 0.377 | 0.144 (0.112 to 0.176) | 2.22 |
| UGCG | .034 | 7.153 (4.874 to 9.432 | 3.017 (2.947 to 3.087) | 2.37 |
| RAD18 | .037 | 5.722 (4.292 to 7.152 | 2.991 (2.971 to 3.011) | 1.91 |
| SLC04A1 | .042 | 0.960 (0.77 to 1.15) | 0.379 (0.253 to 0.505) | 2.53 |
| SLC25A30 | .046 | 0.808 (0.677 to 0.939) | 0.418 (0.318 to 0.518) | 1.93 |
| CLDN5 | .047 | 0.013 (0.0129 to 0.0131 | 0.007 (0.005 to 0.009) | 1.98 |
CI = confidence interval.
Class comparisons were performed with BRB-ArrayTools, version 3.7.0, software and two-sample t tests with 10 000 random permutations. Results were considered statistically significant at P value of less than .05.
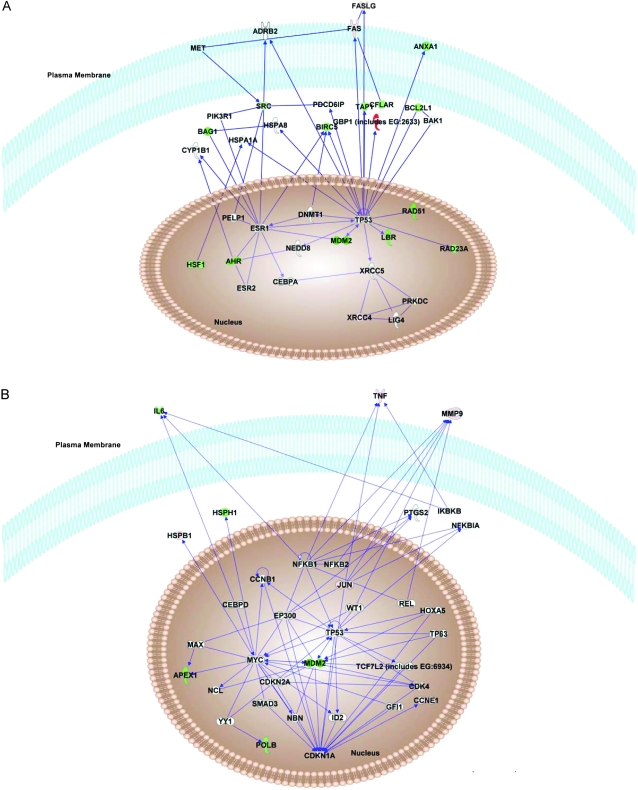
The 63 genes with a differential expression of greater than twofold between CD44+/CD24− MCF-7/ADR cells and CD44+/CD24+ MCF-7/ADR cells (Supplementary Table 7, available online) were also examined via Ingenuity Pathway Analysis (Figure 5). This analysis showed that CD44+/CD24− MCF-7/ADR cells have increased expression of CCNE1 and MMP9 as well as activated p53 and p21 (CDKN1A) pathways but decreased expression of many antiapoptotic genes (including BCL2L1, BAG1, BIRC5, and CFLAR). Because cultures of CD44+/CD24− MCF-7/ADR cells differentiate as shown by their capacity to produce both tumorigenic CD44+/CD24− cells and nontumorigenic CD44+/CD24+ MCF-7/ADR cells, respectively, we examined whether specific genes that have been associated with pluripotency (including POU5F1, NOTCH1, and NANOG) were differentially expressed between sorted CD44+/CD24− MCF-7/ADR cells and CD44+/CD24+ MCF-7/ADR cells by use of TaqMan real-time PCR. Essentially no changes were observed between these two types of cells (Supplementary Table 5, available online).
Figure 5.
Analysis of gene expression in CD44+/CD24− MCF-7/ADR cell population by TaqMan Low-Density Array and Ingenuity Pathway analysis. Genes in CD44+/CD24− MCF-7/ADR cells with expression levels that were greater than twofold higher or lower than their expression in CD44+/CD24+ MCF-7/ADR cells (Supplementary Table 7, available online) were analyzed by Ingenuity Pathways software to identify any biological relationships among these genes. A) Highest ranked pathway. B) Second highest ranked pathway. Green shapes = decreased gene expression; red shapes = increased gene expression. Pathways were drawn by use of the Path Designer feature in Ingenuity Pathways Analysis software, version 8.0 (Ingenuity).
Discussion
Long-term continuous exposure of MCF-7 cell cultures to doxorubicin not only selects for cells with a drug-resistant phenotype (ie, MCF-7/ADR cells), as previously described (14), but also induced a cancer stem cell phenotype in a population of the cells. Specifically, we found that such MCF-7/ADR cell cultures were enriched in cells with the cancer stem cell phenotype (ie, CD44+/CD24− cells), which has been associated with a breast cancer stem cell phenotype (29), and had a stem cell-like gene expression profile. MCF-7/ADR cells also were more invasive than parental MCF-7 cells, formed mammospheres, proliferated in three-dimensional cultures, and were more tumorigenic in mice than parental MCF-7 cells. Thus, cells with cancer stem cell characteristics appear to be generated by long-term drug selection in MCF-7/ADR cell cultures. The aim of this study was to determine the effects of continuous drug selection on the establishment of cancer stem cells in a population of cancer cells. The MCF-7/ADR cells were selected by resistance to increasing concentrations of doxorubicin and require selection pressure with exposure to high-dose doxorubicin (860 nM) every other passage to maintain their drug-resistant phenotype. MCF-7/ADR cells overexpress ABCB1 protein and have been widely used for studies of the multidrug-resistant phenotype (13,14,49).
In results from the microarray study of global gene expression, we first noted that MCF-7/ADR cells had a lower level of CD24 gene expression than parental MCF-7 cells. In an analysis of cell surface markers, we found that statistically significantly fewer MCF-7/ADR cells expressed CD24 and more expressed CD44+/CD24− cells compared with MCF-7 cells. Although both CD44+/CD24− and CD44+/CD24+ populations in long-term drug-selected MCF-7/ADR cultures expressed ABCB1 protein, only CD44+/CD24− cells could self-renew (ie, were able to produce cells with the tumorigenic CD44+/CD24− phenotype) and differentiate (ie, produce nontumorigenic CD44+/CD24+ cells), another feature that is consistent with the cancer stem cell phenotype (33). In addition, we found that the gene expression profile of CD44+/CD24– cells was associated with enhanced cell survival (Figure 5, Table 1, and Supplementary Table 5, available online) and multidrug resistance (as shown by results from the TaqMan low-density array). Thus, we have shown that the CD44+/CD24− cell population has the same phenotype as cancer stem cells. However, we were not able to determine whether the CD44+/CD24– cells with the drug-resistant phenotype were true cancer stem cells or were only mimicking the cancer stem cell phenotype. We further demonstrated that forced expression of ABCB1 protein in the same parental MCF-7 cells did not expand the CD44+/CD24− cell population (data not shown), indicating that long-term drug selection, not ABCB1 gene expression, was likely responsible for the emergence of the cancer stem cell phenotype.
Microarray analysis of MCF-7/ADR cells and parental MCF-7 cells indicated that continuous drug selection, in addition to increasing expression of cancer stem cell–related genes, produced cell populations with a more invasive phenotype that have undergone the epithelial-to-mesenchymal transition (ie, loss of expression of cell-to-cell adhesion genes, including that of E-cadherin genes, and increased expression of N-cadherin genes), which occurs during tumor invasion (50). During progression of most carcinoma types, loss of E-cadherin function and/or expression has also been described (51,52). We found that MCF-7/ADR cells were more invasive in Matrigel assays than parental MCF-7 cells, results that are consistent with these observations (50– 52) and also with those in a recent report (36) that the number of breast cancer stem cells increases after an epithelial-to-mesenchymal transition is induced.
Stem cells are presumed to be drug resistant because they have an enhanced capacity for DNA repair, decreased apoptosis, and higher expression of ATP-binding cassette transporters (9). We compared expression of a panel of multidrug resistance genes in CD44+/CD24− MCF/ADR cells and CD44+/CD24+ MCF/ADR cells by use of a TaqMan low-density array. With this array, we investigated the expression of a total of 381 genes, including all known ATP-binding cassette transporters and many other genes that are involved in drug resistance (J.-P. Gillet, A. M. Calcagno, S. Varma, B. Davidson, M. Bunkholt Elstrand, R. Ganapathi, A. A. Kamat, A. K. Sood, S. V. Ambudkar, M. V. Seiden, B. R. Rueda and M. M. Gottesman, unpublished results). The gene expression profile of CD44+/CD24− cells had characteristics that were consistent with those of cancer stem cells, including activated p53 and p21 pathways and expression of genes that are associated with slower cell cycle progression and xenotoxic damage (eg, RAD18, AHR, and MLH3) (53). Consistent with results previously reported by Clarke et al. (54), we found that CD44+/CD24− cells expressed approximately twice as much p21 mRNA as CD44+/CD24+ cells. In addition, the mRNA expression of two other DNA repair genes, MLH3 and RAD18, was also higher in CD44+/CD24− cells, which supports a previous report (55) that cancer stem cells have activated DNA proofreading mechanisms that include these two genes. Moreover, we observed that CD44+/CD24− cells had increased mRNA expression of UGCG, which has been associated with poor prognosis among patients with breast cancer (47) and drug resistance in leukemia patients (48). Pathway analysis of these genes showed that increased JUN protein expression was associated with the subsequent increased expression of MMP9 and CDKN1A proteins (Figure 5). When we compared gene expression profiles of CD44+/CD24− cells and CD44+/CD24+ cells by use of TaqMan low-density arrays, we found that the expression of both MMP9 and CDKN1A mRNAs was more than twofold higher in CD44+/CD24− cells. Loss of RAD51 expression and increased CCNE1 expression could also be linked to the tumorigenic properties of the CD44+/CD24− cell population (56,57). Furthermore, the nuclear factor κB signaling pathway, which has been associated with resistance to apoptosis in tumors undergoing epithelial-to-mesenchymal transition (58,59), was activated in CD44+/CD24− cells. Thus, differences in gene expression that we observed in CD44+/CD24− cells, compared with CD44+/CD24+ cells, are similar to those described previously in mammary cancer stem cells (4,9,31) and appear to be responsible for the substantial differences in tumorigenicity in mice between CD44+/CD24− and CD44+/CD24+ cells. When we further characterized the cancer stem cell phenotype of drug-selected MCF-7/ADR cells by comparing gene expression profiles of the CD44+/CD24− and CD44+/CD24+ cell populations, we found differences in the expression of various genes that could account for the ability of these cells to self renew (eg, CCND1 and HOXA5) and differentiate (eg, FAS).
The results with MCF-7 and MCF-7/ADR cells are also consistent with those reported by Li et al. (60). In tumor tissue isolated from patients with HER2-negative breast cancer before chemotherapy and after 12 weeks of treatment with docetaxel or with doxorubicin and cyclophosphamide, they found that the population of tumorigenic cells was higher after than before chemotherapy, the ability of the cells to form mammospheres was enhanced after chemotherapy, and xenograft tumors could be produced by 50% of tumor samples obtained after chemotherapy but by only 29% obtained before chemotherapy. Unfortunately, the study by Li et al. did not allow quantification of tumorigenicity by cell number.
Estrogen receptor–negative breast cancers have poorer clinical outcomes, with an increased frequency of metastasis, recurrence, and multidrug resistance, than estrogen receptor–positive breast cancers (35). We found that estrogen receptor mRNA in MCF-7/ADR cells could not be detected by real-time PCR, which supports the estrogen-independent tumorigenicity that we observed with these cells. In contrast, MCF-7 parental cells express estrogen receptor mRNA as detected by real-time PCR, have low tumorigenicity, and produce xenograft tumors only with estrogen supplementation, a well-established characteristic of these cells (61). It has been previously reported (35) that forced expression of SNAI1 protein in MCF-7 cells, a cell line that does not normally express SNAI1, resulted in decreased estrogen receptor expression, decreased cell–cell adhesion, and a more invasive phenotype (ie, as shown by formation of fewer cell aggregates and more cells invading through Matrigel). These characteristics are consistent with cells in which the epithelial-to-mesenchymal transition has been induced. We also found that MCF-7/ADR cells had a highly invasive phenotype, including loss of E-cadherin expression, increased N-cadherin expression, and greater Matrigel invasiveness. Our microarray data showed that SNAI1 expression was higher (1.99-fold, 95% CI = 1.26 to 2.72) in MCF-7/ADR cells than in parental MCF-7 cells. The relationship of SNAI1 expression to estrogen receptor expression and to the epithelial-to-mesenchymal transition in the MCF-7/ADR cells warrants additional study.
The ALDHs are a group of NAD(P)+-dependent enzymes that are involved in oxidizing aldehydes into their corresponding carboxylic acids and in endobiotic and xenobiotic metabolism (62). Increased ALDH activity, as measured with the Aldefluor reagent, is found in cord blood and leukemia cells (63), lung and neck cancer (64), head and neck cancer (65), breast cancer (66), liver cancer (67), and other tumor types (68) and is enriched in their respective cancer stem cell populations (69). Furthermore, overexpression of HER2 in human breast cancer cells has been shown to increase the percentage of cells with ALDH activity and has been associated with the increased tumorigenicity of ALDH-positive cells compared with ALDH-negative cells (70). ALDH expression is a characteristic associated with cancer stem cells (32,33). We found that MCF-7/ADR cell cultures had more ALDH-positive cells than parental MCF-7 cells (Supplementary Figure 3, A, available online).
Finally, this study had several limitations. We studied only one highly drug-resistant breast cancer cell line, MCF-7/ADR, that was established by a prolonged, continuous, multistep selection with doxorubicin and its parental line MCF-7. Other drug-resistant cell lines isolated from other types of cancers need to be studied for the enrichment of cells with characteristics of cancer stem cells. Immunodeficient mice were used in tumorigenesis studies. Some groups in the tumorigenesis studies had only one mouse and some groups had only one tumor for analysis. The exact mechanism of cancer stem cell enrichment in MCF-7/ADR cultures is unclear. Drug treatment may cause a decrease in the number of differentiated tumor cells, leading to an increase in the total fraction of cells with cancer stem cell functional and genomic characteristics. Although our findings suggest that enrichment of cancer stem cells in response to prolonged treatment with anticancer drugs is associated with a phenotype similar to that of cells undergoing an epithelial-to-mesenchymal transition, these observations should be further validated in several in vivo model systems. Such studies would help to establish whether there is a relationship among multidrug resistance, epithelial-to-mesenchymal transition, metastasis, and cancer stem cell phenotypes. Our result that a highly drug-resistant population of cancer stem cells is generated after prolonged drug selection may be helpful for the design of clinical trials to increase the efficiency of chemotherapeutic regimens.
In conclusion, we demonstrated that breast cancer MCF-7/ADR cells overexpressed drug resistance genes and that the proportion of cells with cancer stem cell characteristics increased after prolonged selection with doxorubicin. From these findings, we speculate that prolonged drug treatment of patients with breast cancer may also result in increased numbers of cells with a highly chemotherapy-resistant cancer stem cell–like phenotype that may even be cancer stem cells.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. A.M.C. was supported by the NIGMS Pharmacology Research Associate (PRAT) Program. J.F. was supported by the Intramural Research Program of the NIH and National Institutes of Environmental Health Sciences, contract HHSN273200700046U.
Supplementary Material
Footnotes
All rights, title and interest are assigned to the United States Government, and there are no conflicts of interest. The sponsors had no role in the study design, data collection and analysis, interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication.
Concerning Supplementary Table 6 (available online), citation details: J.-P. Gillet, M. M. Gottesman, A. M. Calcagno, S. V. Ambudkar, S. Varma, B. R. Rueda, A. Sood, R. Ganapathi, M. Seiden, and B. Davidson. Methods for prediction of clinical outcome to treatment of ovarian cancer patients. U.S. Provisional Patent Application No. 61/308,946, filed February 27, 2010. We thank Dr Kapil Mehta (M.D. Anderson Cancer Center, Houston, TX) for the MCF-7 breast cancer cell line and the multistep doxorubicin-selected subline, MCF-7/ADR, Dr Ty Voss (Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, Bethesda, MD) for the help with statistical analyses and Mr George Leiman for editorial assistance.
References
- 1.Gottesman M, Fojo T, Bates S. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 2.Calcagno AM, Kim IW, Wu CP, Shukla S, Ambudkar SV. ABC drug transporters as molecular targets for the prevention of multidrug resistance and drug-drug interactions. Curr Drug Deliv. 2007;4(4):324–333. doi: 10.2174/156720107782151241. [DOI] [PubMed] [Google Scholar]
- 3.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mine T, Matsueda S, Li Y, et al. Breast cancer cells expressing stem cell markers CD44+ CD24 lo are eliminated by Numb-1 peptide-activated T cells. Cancer Immunol Immunother. 2009;58(8):1185–1194. doi: 10.1007/s00262-008-0623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien CS, Farnie G, Howell SJ, Clarke RB. Are stem-like cells responsible for resistance to therapy in breast cancer? Breast Dis. 2008;29(2007, 2008):83–89. doi: 10.3233/bd-2008-29109. [DOI] [PubMed] [Google Scholar]
- 8.Trumpp A, Wiestler OD. Mechanisms of disease: cancer stem cells—targeting the evil twin. Nat Clin Pract Oncol. 2008;5(6):337–347. doi: 10.1038/ncponc1110. [DOI] [PubMed] [Google Scholar]
- 9.Dean M. ABC Transporters, Drug Resistance, and Cancer Stem Cells. J Mammary Gland Biol Neoplasia. 2009;14(1):3–9. doi: 10.1007/s10911-009-9109-9. [DOI] [PubMed] [Google Scholar]
- 10.Liscovitch M, Ravid D. A case study in misidentification of cancer cell lines: MCF-7/AdrR cells (re-designated NCI/ADR-RES) are derived from OVCAR-8 human ovarian carcinoma cells. Cancer Lett. 2007;245(1–2):350–352. doi: 10.1016/j.canlet.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Mehta K, Devarajan E, Chen J, Multani A, Pathak S. Multidrug-resistant MCF-7 cells: an identity crisis? J Natl Cancer Inst. 2002;94(21):1652–1654. doi: 10.1093/jnci/94.21.1652-b. [DOI] [PubMed] [Google Scholar]
- 12.Pirnia F, Breuleux M, Schneider E, et al. Uncertain identity of doxorubicin-resistant MCF-7 cell lines expressing mutated p53. J Natl Cancer Inst. 2000;92(18):1535–1536. doi: 10.1093/jnci/92.18.1535. [DOI] [PubMed] [Google Scholar]
- 13.Devarajan E, Chen J, Multani AS, Pathak S, Sahin AA, Mehta K. Human breast cancer MCF-7 cell line contains inherently drug-resistant subclones with distinct genotypic and phenotypic features. Int J Oncol. 2002;20(5):913–920. [PubMed] [Google Scholar]
- 14.Mehta K. High levels of transglutaminase expression in doxorubicin-resistant human breast carcinoma cells. Int J Cancer. 1994;58(3):400–406. doi: 10.1002/ijc.2910580316. [DOI] [PubMed] [Google Scholar]
- 15.Calcagno AM, Fostel JM, To KK, et al. Single-step doxorubicin-selected cancer cells overexpress the ABCG2 drug transporter through epigenetic changes. Br J Cancer. 2008;98(9):1515–1524. doi: 10.1038/sj.bjc.6604334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3(8):e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke R, Currier S, Kaplan O, et al. Effect of P-glycoprotein expression on sensitivity to hormones in MCF-7 human breast cancer cells. J Natl Cancer Inst. 1992;84(19):1506–1512. doi: 10.1093/jnci/84.19.1506. [DOI] [PubMed] [Google Scholar]
- 18.Honjo Y, Hrycyna CA, Yan Q-W, et al. Acquired mutations in the MXR/BCRP/ABCP gene alter substrate specificity in MXR/BCRP/ABCP-overexpressing cells. Cancer Res. 2001;61(18):6635–6639. [PubMed] [Google Scholar]
- 19.Robey RW, Medina-Perez WY, Nishiyama K, et al. Overexpression of the ATP-binding cassette half-transporter, ABCG2 (MXR/BCRP/ABCP1), in flavopiridol-resistant human breast cancer Cells. Clin Cancer Res. 2001;7(1):145–152. [PubMed] [Google Scholar]
- 20.Schneider E, Horton JK, Yang CH, Nakagawa M, Cowan KH. Multidrug resistance-associated protein gene overexpression and reduced drug sensitivity of topoisomerase II in a human breast carcinoma MCF7 cell line selected for etoposide resistance. Cancer Res. 1994;54(1):152–158. [PubMed] [Google Scholar]
- 21.Huff LM, Lee J-S, Robey RW, Fojo T. Characterization of gene rearrangements leading to activation of MDR-1. J Biol Chem. 2006;281(48):36501–36509. doi: 10.1074/jbc.M602998200. [DOI] [PubMed] [Google Scholar]
- 22.Shen DW, Cardarelli C, Hwang J, et al. Multiple drug-resistant human KB carcinoma cells independently selected for high-level resistance to colchicine, adriamycin, or vinblastine show changes in expression of specific proteins. J Biol Chem. 1986;261(17):7762–7770. [PubMed] [Google Scholar]
- 23.Barrand MA, Heppell-Parton AC, Wright KA, Rabbitts PH, Twentyman PR. A 190-kilodalton protein overexpressed in non-P-glycoprotein-containing multidrug-resistant cells and its relationship to the MRP gene. J Natl Cancer Inst. 1994;86(2):110–117. doi: 10.1093/jnci/86.2.110. [DOI] [PubMed] [Google Scholar]
- 24.Wu CP, Calcagno AM, Hladky SB, Ambudkar SV, Barrand MA. Modulatory effects of plant phenols on human multidrug-resistance proteins 1, 4 and 5 (ABCC1, 4 and 5) FEBS J. 2005;272(18):4725–4740. doi: 10.1111/j.1742-4658.2005.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein JN, Myers TG, O’Connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 26.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Calcagno AM, Chewning KJ, Wu CP, Ambudkar SV. Plasma membrane calcium ATPase (PMCA4): a housekeeper for RT-PCR relative quantification of polytopic membrane proteins. BMC Mol Biol. 2006;7:29. doi: 10.1186/1471-2199-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10(1):R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peeters SD, van der Kolk DM, de Haan G, et al. Selective expression of cholesterol metabolism genes in normal CD34+CD38− cells with a heterogeneous expression pattern in AML cells. Exp Hematol. 2006;34(5):622–630. doi: 10.1016/j.exphem.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakarala M, Wicha MS. Cancer stem cells: implications for cancer treatment and prevention. Cancer J. 2007;13(5):271–275. doi: 10.1097/PPO.0b013e318156da4e. [DOI] [PubMed] [Google Scholar]
- 34.Plowman J, Dykes D, Hollingshead M, Simpson-Herren L, Alley M. Human tumor xenograft models in NCI drug development. In: Teicher BA, editor. Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials, and Approval. Totowa, NJ: Humana Press: 1997. pp. 101–125. [Google Scholar]
- 35.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21(12):2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer—observations in vitro and in vivo. Cells Tissues Organs. 2007;185(1–3):191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1896. [DOI] [PubMed] [Google Scholar]
- 40.Chaudhary PM, Roninson IB. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell. 1991;66(1):85–94. doi: 10.1016/0092-8674(91)90141-k. [DOI] [PubMed] [Google Scholar]
- 41.Ferrandina G, Bonanno G, Pierelli L, et al. Expression of CD133-1 and CD133-2 in ovarian cancer. Int J Gynecol Cancer. 2008;18(3):506–514. doi: 10.1111/j.1525-1438.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- 42.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 43.Sheridan C, Kishimoto H, Fuchs R, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue W, Brodie A. MCF-7 human breast carcinomas in nude mice as a model for evaluating aromatase inhibitors. J Steroid Biochem Mol Biol. 1993;44(4–6):671–673. doi: 10.1016/0960-0760(93)90278-5. [DOI] [PubMed] [Google Scholar]
- 45.Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001;13(6):389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 46.Wajant H, Pfizenmaier K, Scheurich P. Non-apoptotic Fas signaling. Cytokine Growth Factor Rev. 2003;14(1):53–66. doi: 10.1016/s1359-6101(02)00072-2. [DOI] [PubMed] [Google Scholar]
- 47.Ruckhaberle E, Karn T, Hanker L, et al. Prognostic relevance of glucosylceramide synthase (GCS) expression in breast cancer. J Cancer Res Clin Oncol. 2009;135(1):81–90. doi: 10.1007/s00432-008-0436-9. [DOI] [PubMed] [Google Scholar]
- 48.Xie P, Shen Y-F, Shi Y-P, et al. Overexpression of glucosylceramide synthase in associated with multidrug resistance of leukemia cells. Leuk Res. 2008;32(3):475–480. doi: 10.1016/j.leukres.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Mann AP, Verma A, Sethi G, et al. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66(17):8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 50.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current Opin Cell Biol. 2005;17(5):548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198(1):11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 52.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Current Opin Cell Biol. 1993;5(5):806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 53.Zabierowski SE, Herlyn M. Melanoma stem cells: the dark seed of melanoma. J Clin Oncol. 2008;26(17):2890–2894. doi: 10.1200/JCO.2007.15.5465. [DOI] [PubMed] [Google Scholar]
- 54.Clarke RB, Spence K, Anderson E, Howell A, Okano H, Potten CS. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277(2):443–456. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 55.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 56.Akli S, Van Pelt CS, Bui T, et al. Overexpression of the low molecular weight cyclin E in transgenic mice induces metastatic mammary carcinomas through the disruption of the ARF-p53 pathway. Cancer Res. 2007;67(15):7212–7222. doi: 10.1158/0008-5472.CAN-07-0599. [DOI] [PubMed] [Google Scholar]
- 57.Richardson C. RAD51, genomic stability, and tumorigenesis. Cancer Lett. 2005;218(2):127–139. doi: 10.1016/j.canlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 58.Rennebeck G, Martelli M, Kyprianou N. Anoikis and survival connections in the tumor microenvironment: is there a role in prostate cancer metastasis? Cancer Res. 2005;65(24):11230–11235. doi: 10.1158/0008-5472.CAN-05-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizki A, Weaver VM, Lee S-Y, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68(5):1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 61.Osborne CK, Hobbs K, Clark GM. Effect of estrogens and antiestrogens on growth of human breast cancer cells in athymic nude mice. Cancer Res. 1985;45(2):584–590. [PubMed] [Google Scholar]
- 62.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36(2):279–299. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 63.Pearce DJ, Taussig D, Simpson C, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23(6):752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 64.Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact. 2009;178(1–3):48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clay MR, Tabor M, Owen JH, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32(9):1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16(1):45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park KS, Cho SY, Kim H, Paik YK. Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int J Cancer. 2002;97(2):261–265. doi: 10.1002/ijc.1585. [DOI] [PubMed] [Google Scholar]
- 68.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5(4):e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun S, Wang Z. ALDH high adenoid cystic carcinoma cells display cancer stem cell properties and are responsible for mediating metastasis. Biochem Biophys Res Commun. 2010;396(4):843–848. doi: 10.1016/j.bbrc.2010.04.170. [DOI] [PubMed] [Google Scholar]
- 70.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27(47):6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.