Abstract
Background
The Gail model is widely used for the assessment of risk of invasive breast cancer based on recognized clinical risk factors. In recent years, a substantial number of single-nucleotide polymorphisms (SNPs) associated with breast cancer risk have been identified. However, it remains unclear how to effectively integrate clinical and genetic risk factors for risk assessment.
Methods
Seven SNPs associated with breast cancer risk were selected from the literature and genotyped in white non-Hispanic women in a nested case–control cohort of 1664 case patients and 1636 control subjects within the Women’s Health Initiative Clinical Trial. SNP risk scores were computed based on previously published odds ratios assuming a multiplicative model. Combined risk scores were calculated by multiplying Gail risk estimates by the SNP risk scores. The independence of Gail risk and SNP risk was evaluated by logistic regression. Calibration of relative risks was evaluated using the Hosmer–Lemeshow test. The performance of the combined risk scores was evaluated using receiver operating characteristic curves. The net reclassification improvement (NRI) was used to assess improvement in classification of women into low (<1.5%), intermediate (1.5%–2%), and high (>2%) categories of 5-year risk. All tests of statistical significance were two-sided.
Results
The SNP risk score was nearly independent of Gail risk. There was good agreement between predicted and observed SNP relative risks. In the analysis for receiver operating characteristic curves, the combined risk score was more discriminating, with area under the curve of 0.594 compared with area under the curve of 0.557 for Gail risk alone (P < .001). Classification also improved for 5.6% of case patients and 2.9% of control subjects, showing an NRI value of 0.085 (P = 1.0 × 10−5). Focusing on women with intermediate Gail risk resulted in an improved NRI of 0.195 (P = 8.6 × 10−5).
Conclusions
Combining validated common genetic risk factors with clinical risk factors resulted in modest improvement in classification of breast cancer risks in white non-Hispanic postmenopausal women. Classification performance was further improved by focusing on women at intermediate risk.
CONTEXT AND CAVEATS
Prior knowledge
It is not known whether adding genotyping data from single-nucleotide polymorphisms (SNPs) associated with breast cancer risk to the Gail model of breast cancer risk results in clinical gains.
Study design
Composite SNP relative risk scores (SNP risk) using genotyping data from seven SNPs previously associated with breast cancer risk were estimated among participants in a case–control study of white non-Hispanic postmenopausal women within the Women's Health Initiative Clinical Trial. The SNP risk estimates were combined multiplicatively with risk estimates from the Gail model and compared with the estimates from the Gail model.
Contribution
Breast cancer risk was more accurately estimated by combining Gail risk estimates and SNP risk.
Implications
The combined risk assessment model has clinical validity in white non-Hispanic postmenopausal women.
Limitations
The scope of this study at present is limited to white non-Hispanic postmenopausal women who share clinical characteristics of the women used in this cohort.
From the Editors
Breast cancer risk in women is strongly associated with both genetic and environmental factors. The discovery of the breast cancer susceptibility genes, breast cancer 1, early onset (BRCA1) and breast cancer 2, early onset (BRCA2), and a decade of subsequent clinical research led to substantial positive impact on the health of women affected with the Mendelian cancer predisposition syndromes, conferred by germline mutations in these genes (1–3). However, much of the familial aggregation of breast cancer remains unexplained. Building on previous work suggesting the existence of substantial polygenic influences on breast cancer risk (4), recent genome-wide association studies (5–10), and a candidate gene association study (11), have demonstrated that an expanding set of single-nucleotide polymorphisms (SNPs) are reproducibly associated with breast cancer risk in women of European ancestry, and in some cases, in women from other racial or ethnic backgrounds.
As with other disorders with complex inheritance, like prostate cancer, Crohn's disease, and hypercholesterolemia (12–15), the discovery and validation of SNPs associated with breast cancer risk have created an opportunity to explore whether a panel of SNPs can be used to predict disease risk and to assess the clinical relevance of such a panel. In the context of breast cancer, the assessment of invasive cancer risk has important clinical implications that can affect decisions about appropriate counseling, screening regimens, and risk reduction strategies (3,16–20). Thus, improvements in risk prediction have the potential to affect clinical care if they are demonstrated to have clinical validity and utility.
For sporadic (nonfamilial) breast cancer, the Gail model (21) has been commonly used to produce individual risk estimates in women. The model incorporates individual risk factors including age, family history (breast cancer among first-degree relatives), personal reproductive history (age at menarche and at first live birth), and personal medical history (number of previous breast biopsies and presence of biopsy-confirmed atypical hyperplasia) to estimate the 5-year risk and lifetime risk of invasive breast cancer in an individual. The model uses relative risk calculation based on logistic regression of these risk factors, followed by conversion to absolute risk based on epidemiological data for breast cancer incidence and competing risks. A projected Gail 5-year risk score (5-year absolute risk of breast cancer diagnosis) of greater than or equal to 1.66% is an important tool to identify women who have an increased risk of developing breast cancer and may benefit from risk reduction with selective estrogen receptor (ER) modulators (19,20). Therefore, this model has implications for primary prevention of invasive breast cancer. However, both the discriminatory accuracy of the Gail model and its calibration in certain populations have been challenged (22−23), and uptake of primary prevention strategies among physicians caring for women at increased risk for sporadic breast cancer has been modest (24).
Two previous studies have analyzed the potential impact of adding genetic information from a panel of seven SNPs associated with breast cancer risk to the Gail model (25,26). One analysis used receiver operating characteristic (ROC) curves to predict that the area under the curve (AUC) would improve from 0.607 for the Gail model alone, as implemented in the National Cancer Institute's Breast Cancer Risk Assessment Tool (BCRAT), to 0.632 when risk information from the seven SNPs was combined with the Gail model (BCRATplus7) (25). In an editorial accompanying that article (25), Pepe and Janes (27) suggested that ROC curve analysis might not be the most useful method for assessing performance of these models and that reclassification tables would allow the assessment of the fraction of individuals with meaningful improvements in risk prediction. A second analysis showed that real gains, albeit modest, could be achieved in reclassification of risk, under the assumption that the model combining information from the seven SNPs with the Gail model was well calibrated (26).
In addition, other studies have begun to set modest expectations for potential clinical gains from combining SNP information with clinical risk factors (25,26,28,29). However, these studies have either been theoretical in nature (25,26,28) or they combined model building with evaluation (29), which may complicate evaluating the results in clinical context. We set out to empirically assess the clinical validity of a prespecified model combining genetic information with risk estimates from the Gail model in a nested case–control study from the Women's Health Initiative (WHI) Clinical Trial (30). We also assessed whether the improvement in risk assessment from incorporating genetic information might be larger in subsets of women at intermediate risk based on clinical risk factors. We focused our analyses on white non-Hispanic women because of the combined availability of Gail model validity information and accurate odds ratios for SNPs in this group.
Subjects and Methods
Ascertainment of Case Patients and Control Subjects
Of 68 132 participants, we identified 2166 women who developed invasive breast cancer between random assignment (September 1, 1993, to December 31, 1998) and the originally planned end of the intervention phase (March 31, 2005) of the WHI Clinical Trial (30). We matched one control subject without a cancer diagnosis for each case patient for baseline age at enrollment in the WHI Clinical Trial, self-reported ethnicity, trial intervention (hormone replacement therapy, dietary modification, or calcium and vitamin D supplementation), years since trial randomization, and whether or not they had had a hysterectomy. Written informed consent was obtained from each participant, and the study was approved by the Fred Hutchinson Cancer Research Center's Institutional Review Board. Participants in the WHI Clinical Trial had an opportunity to opt in or out of any collaboration involving commercial entities because some women may prefer not to participate in research involving commercial (as opposed to nonprofit) entities. We restricted our analyses to the subset of these case patients and control subjects that had consented for collaborations involving commercial entities. Approximately, 84% of all eligible case patients and control subjects provided such consent. The interventions used in the WHI Clinical Trial are independent of baseline genetic and clinical risk factors by study design (30), so the analyses presented here were not stratified by trial intervention. In this nested case–control study, we focused our analyses on 3300 white non-Hispanic women, representing 87% of the matched case patients and control subjects.
Estimation of Gail Risk Score
We used the Gail model to estimate the 5-year absolute risk of breast cancer (“Gail risk”) based on age, ethnicity, age at menarche, age at birth of first child, number of first-degree relatives with breast cancer, and the number of previous breast biopsies. We did not have information on biopsy histopathology (ie, whether atypical hyperplasia was present), and this was coded as “unknown.” We computed Gail risk for the subjects using a reimplementation of the current BCRAT version 2, based on program source code downloaded from the National Cancer Institute Web site (http://www.cancer.gov/bcrisktool/) on August 7, 2008.
SNP Selection and SNP Risk Score
In our model, we included SNPs that showed statistically significant associations with breast cancer risk in a genome-wide association study with correction for multiple testing and were confirmed in an independent set of case patients and control subjects. Seven SNPs were found to meet these criteria at the time that the study was initiated (5–8). Our model for SNP risk was based on allele frequencies and effect sizes (odds ratios) reported in these original studies (Table 1).
Table 1.
Seven genomic loci associated with invasive breast cancer*
| First author, year (reference) | SNP | Gene† | Chromosomal location | Frequency of high-risk allele | Reported, OR (95% CI)‡ |
| Easton, 2007 (5) | rs2981582 | FGFR2 | 10q26 | 0.38 | 1.26 (1.23 to 1.30) |
| Easton, 2007 (5) | rs3803662 | TOX3 | 16q12 | 0.25 | 1.20 (1.16 to 1.24) |
| Easton, 2007 (5) | rs889312 | MAP3K1 | 5q11 | 0.28 | 1.13 (1.10 to 1.16) |
| Stacey, 2007 (7) | rs13387042 | ― | 2q35 | 0.50 | 1.20 (1.14 to 1.26) |
| Easton, 2007 (5) | rs13281615 | ― | 8q24 | 0.40 | 1.08 (1.05 to 1.11) |
| Stacey, 2008 (8) | rs4415084 | FGF10 | 5p12 | 0.44 | 1.16 (1.10 to 1.21) |
| Easton, 2007 (5) | rs3817198 | LSP1 | 11p15 | 0.30 | 1.07 (1.04 to 1.11) |
Characteristics of the loci from the cited genome-wide association studies. CI = confidence interval; OR = odds ratio; SNP = single-nucleotide polymorphism. ― = a relevant gene has not been identified.
The seven SNPs are located in or near the following genes: FGFR2 (fibroblast growth factor receptor 2); TOX3 (TOX high-mobility group box family member 3, previously known as TNRC9); MAP3K1 (mitogen-activated protein kinase kinase kinase 1); FGF10 (fibroblast growth factor 10); LSP1 (lymphocyte-specific protein 1).
Odds ratio per copy of the high-risk allele, as reported in the cited study.
We defined an individual's composite SNP relative risk score (“SNP risk”) as the product of genotype relative risk values for the seven SNPs. Based on a log-additive risk model, the three genotypes AA, AB, and BB for a single SNP have relative risk values of 1, OR, and OR2, under a rare disease model, where OR is the previously reported disease odds ratio for the high-risk allele, B, vs the low-risk allele, A (Table 1). If the B allele has frequency p, then these genotypes have population frequencies of (1 − p)2, 2p(1 − p), and p2, assuming Hardy–Weinberg equilibrium. We scaled the genotype relative risk values for each SNP so that based on these frequencies the average relative risk in the population is 1. Specifically, given the unscaled population average relative risk (μ) = (1 − p)2 + 2p(1 − p)OR + p2OR2, we used the adjusted risk values 1/μ, OR/μ, and OR2/μ for AA, AB, and BB genotypes. Missing genotypes were assigned a relative risk of 1.
Combined Gail × SNP Risk Score
The formula for our combined 5-year Gail × SNP absolute risk score is:
where Gail is the Gail absolute risk score, and SNP1 to 7 are relative risk scores for the individual SNPs, each scaled to have a population average of 1 (Supplementary Methods and Supplementary Figure 1, available online). Because our SNP risk scores have been “centered” to have a population average risk of 1, if we assume independence among the seven SNPs, then the population average risk across all genotypes for the combined score is consistent with the underlying Gail risk estimate.
Statistical Analysis
We assessed the associations between different categories of clinical risk factors and breast cancer risk using Cochran–Armitage trend tests. We used logistic regression to assess association between individual SNPs and breast cancer risk. We assumed a log-additive allelic model for genetic associations, as described above. We also used logistic regression to test for interactions between pairs of SNPs by testing for statistical significance of model terms formed as the product of the individual SNP risk terms. We used likelihood ratio tests for comparing nested logistic regression models; these tests are one-sided but do not make assumptions about the sign of the relationship between the dependent and independent variables. All other P values were two-sided, and unless otherwise noted, P values less than or equal to .05 were considered statistically significant.
We used logistic regression to estimate odds ratios and 95% confidence intervals (CIs) associated with the logarithms of Gail risk and SNP risk separately, and as combined into the Gail × SNP risk score. The intercept term in the logistic regression was unrestricted and allowed the analysis to adapt to case–control sampling.
The Hosmer–Lemeshow test (31) is typically used to assess calibration of absolute risk estimates in cohort data. Instead, we used the test to assess agreement between expected and observed relative risks in our case–control study. We calculated the expected numbers of case patients and control subjects using a model in which the log-odds for breast cancer depended directly on the estimated log-odds ratio for a particular risk score. Here, we fit a logistic regression model with a coefficient (β) equal to 1.0 for the risk score, in conjunction with a freely estimated location parameter to match the overall number of case patients in the case–control sampling. This approach is justified by early work (32) showing that logistic regression parameter estimates under case–control sampling have targets that differ from those under prospective sampling only though a shift in the logistic regression location parameter.
We assessed classification performance of Gail risk, SNP risk, and Gail × SNP risk using ROC curves. Bootstrap resampling (1000 replicates) was used to estimate confidence intervals for AUC as well as empirical P values for differences in AUC. Each bootstrap replicate was formed by selecting at random from the original case patients and control subjects, allowing the same individual to be selected more than once, to obtain the same total numbers of case patients and control subjects. We used the pcvsuite software package (33) to compute age-adjusted ROC curves and AUC values by measuring performance within deciles of the cohort defined by age and averaging these results to obtain values for the entire cohort.
We also evaluated classification accuracy using reclassification tables (34,35) and quantified differences in classification by net reclassification improvement (NRI) (36). Although reclassification tables are most intuitive in population-based studies, the NRI metric remains useful in case–control studies because it is unaffected by case–control sampling. NRI is the sum of the proportion of case patients moving to a higher-risk category minus the proportion of case patients moving to a lower risk category, and the proportion of control subjects moving to a lower risk category minus the proportion of control subjects moving to a higher-risk category (36). For simple tests of the hypothesis (NRI = 0), we used an asymptotic Z test (36). We also used bootstrap resampling to evaluate 95% confidence intervals for NRI and to determine empirical P values for differences in NRI. We used the same bootstrap samples for each classifier and used paired tests to compare classification performance to preserve the correlation structure of the classifiers.
We recently learned (N. Cook, personal communication) that NRI may be biased when calculated within a limited range of risk scores. Where this bias would be relevant, we determined an empirical distribution of NRI under the null hypothesis using permuted risk scores and evaluated statistical significance of the actual NRI by comparison with this distribution.
Results
Characteristics of Case Patients and Control Subjects in the Nested Case–Control Cohort
Clinical characteristics of the 1664 case patients and 1636 control subjects selected from the WHI Clinical Trial that were successfully genotyped are summarized in Table 2. Most breast cancer risk factors that contribute to the Gail model were differentially distributed in the case patient and control subject groups, including age at menarche (Ptrend = .02), age at birth of first child (Ptrend = .004), first-degree relatives with breast cancer (Ptrend = .0001), and number of previous breast biopsies (Ptrend = 1 × 10−5). Age was not associated with case patient vs control subject status (Ptrend = .84) because ages were matched in case patients and control subjects.
Table 2.
Demographic characteristics and Gail model clinical risk factors for breast cancer in the nested case–control cohort*
| Characteristics | Case patients, No. (%) | Control subjects, No. (%) | Ptrend† |
| Total | 1664 (100) | 1636 (100) | |
| ER status | |||
| ER-positive tumor | 1218 (73.2) | ||
| ER-negative tumor | 208 (12.5) | ||
| Unknown‡ or missing§ | 238 (14.3) | ||
| Age at screening, y | .84 | ||
| 50–59 | 510 (30.6) | 510 (31.2) | |
| 60–69 | 789 (47.4) | 767 (46.9) | |
| 70–79 | 365 (21.9) | 359 (21.9) | |
| Missing§ | 0 (0.0) | 0 (0.0) | |
| Age at menarche, y | .02 | ||
| <12 | 379 (22.8) | 349 (21.3) | |
| 12 | 441 (26.5) | 407 (24.9) | |
| 13 | 497 (29.9) | 481 (29.4) | |
| ≥14 | 338 (20.3) | 395 (24.1) | |
| Missing§ | 9 (0.5) | 4 (0.2) | |
| Age at birth of first child, y | .004 | ||
| <20 | 182 (10.9) | 214 (13.1) | |
| 20–24 | 625 (37.6) | 665 (40.6) | |
| 25–29 | 370 (22.2) | 349 (21.3) | |
| ≥30 | 151 (9.1) | 117 (7.2) | |
| No term pregnancy | 200 (12.0) | 166 (10.1) | |
| Missing§ | 136 (8.2) | 125 (7.6) | |
| Age at menopause, y | .52 | ||
| <45 | 300 (18.0) | 294 (18.0) | |
| 45–49 | 355 (21.3) | 390 (23.8) | |
| 50–54 | 646 (38.8) | 596 (36.4) | |
| >54 | 238 (14.3) | 237 (14.5) | |
| Missing§ | 125 (7.5) | 119 (7.3) | |
| First-degree relatives with breast cancer | .0001 | ||
| 0 | 1253 (75.3) | 1319 (80.6) | |
| >1 | 309 (18.6) | 226 (13.8) | |
| Missing§ | 102 (6.1) | 91 (5.6) | |
| Number of previous breast biopsies | 1 × 10−5 | ||
| 0 | 1056 (63.5) | 1148 (70.2) | |
| 1 | 298 (17.9) | 188 (11.5) | |
| >2 | 102 (6.1) | 83 (5.1) | |
| Missing§ | 208 (12.5) | 217 (13.3) | |
The nested case–control cohort was selected from the Women's Health Initiative (WHI) Clinical Trial. ER = estrogen receptor.
Ptrend values were calculated using the two-sided Cochran–Armitage test for trend.
Pathology data on tumor ER status was unavailable or indeterminate.
Data for the specified characteristic was not available from the WHI Clinical Trial.
Assessment of Association Between Individual SNPs and Invasive Breast Cancer Risk
The seven selected SNPs were genotyped across genomic DNA samples from trial participants using a custom array (Affymetrix, Inc, Santa Clara, CA) and/or the Sequenom MassARRAY iPLEX platform (Supplementary Methods, available online). There were few missing genotypes because of unsuccessful genotype measurements (Table 3, and Supplementary Methods, available online).
Table 3.
Associations between single-nucleotide polymorphisms (SNPs) and invasive breast cancer in case patients and control subjects from the Women's Health Initiative Clinical Trial*
| SNP | Call rate† | PHWE‡ | Frequency of high-risk allele | Observed, OR (95% CI)§ | P║ |
| rs2981582 | 1.000 | .29 | 0.41 | 1.35 (1.22 to 1.50) | 2.9 × 10−9 |
| rs3803662 | 1.000 | .85 | 0.29 | 1.20 (1.08 to 1.34) | 8.1 × 10−4 |
| rs889312 | 0.996 | .64 | 0.28 | 1.24 (1.12 to 1.38) | 6.7 × 10−5 |
| rs13387042 | 0.910 | .58 | 0.51 | 1.16 (1.05 to 1.29) | 3.6 × 10−3 |
| rs13281615 | 1.000 | .43 | 0.42 | 1.08 (0.98 to 1.20) | .11 |
| rs4415084 | 0.997 | .07 | 0.40 | 1.18 (1.06 to 1.30) | 1.5 × 10−3 |
| rs3817198 | 1.000 | .60 | 0.32 | 1.11 (1.00 to 1.23) | .06 |
Associations were analyzed by univariate logistic regression for predicting breast cancer from the specified SNP genotype under a log-additive allelic model. CI = confidence interval; HWE = Hardy–Weinberg equilibrium; OR = odds ratio.
The proportion of individuals for whom a genotype was successfully determined, for a particular SNP.
P values for Hardy–Weinberg equilibrium were calculated using a one-sided likelihood ratio test.
Odds ratio per copy of the high-risk allele in the case–control sample.
P values for tests of association were calculated using a one-sided likelihood ratio test.
We verified that the seven SNPs in this panel showed associations with breast cancer risk that were consistent with the prespecified parameters in our SNP risk model (Table 1). Five of the seven SNPs showed statistically significant association with breast cancer risk in our case–control cohort (Table 3). All seven SNPs had estimated allelic odds ratios that were well within the range of previously reported confidence intervals (Table 3).
We used logistic regression to test for pairwise interactions among the seven SNPs. We did not detect any statistically significant interactions; 21 distinct pairwise tests showed one test with P < .05 and none with P < .01 (data not shown). However, this study was only powered to detect relatively strong pairwise interactions.
Independence of Gail Risk and SNP Risk
The validity of our combined Gail × SNP risk score relied on the independence of Gail risk and SNP risk. Gail risk and SNP risk were tested separately for association with breast cancer incidence by logistic regression with log-transformed predictors. Both were strongly and statistically significantly associated with breast cancer (Table 3). Estimation of breast cancer risk with the SNP risk score gives relative risk estimates roughly proportional to observed disease rates (Table 4). The Gail risk and SNP risk were weakly but statistically significantly correlated (Pearson correlation [r] = 0.042, P = .02). The combined predictor formed by multiplying the Gail risk by the SNP risk (Gail × SNP risk) was more strongly associated with breast cancer risk than either component alone (Table 4). Including log-transformed Gail risk and SNP risk as separate terms in the logistic regression model further improved the fit (P = 2.3 × 10−5) by accommodating the difference in calibration of the two terms. However, in a model with these separate terms, an interaction term did not further improve prediction of breast cancer status (P = .5) (data not shown in Table 4).
Table 4.
Association of Gail risk, single-nucleotide polymorphism (SNP) risk, and Gail × SNP risk with invasive breast cancer*
| Predictor† | β‡ (95% CI) | OR (95% CI) per twofold increase in risk score§ | P║ |
| log (Gail risk) | 0.46 (0.30 to 0.63) | 1.38 (1.23 to 1.54) | 1.8 × 10−8 |
| log (SNP risk) | 1.11 (0.86 to 1.36) | 2.16 (1.82 to 2.57) | 6.4 × 10−19 |
| log (Gail risk × SNP risk) | 0.65 (0.51 to 0.78) | 1.57 (1.42 to 1.72) | 3.3 × 10−21 |
Univariate logistic regression analysis for predicting invasive breast cancer from the specified risk scores. CI = confidence interval; OR = odds ratio.
The risk score used to predict invasive breast cancer.
The logistic regression coefficient for the specified predictor.
The fitted odds ratio corresponding to a twofold increase in the risk score, equal to 2β.
The P values were calculated using a one-sided likelihood ratio test for including the predictor term in the model vs not including the predictor term.
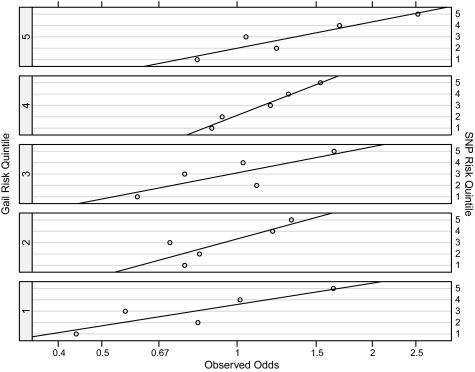
To visualize the properties of SNP risk for different Gail risk categories, we binned the Gail risk and SNP risk scores into quintiles and evaluated the relationship between SNP risk and the odds of developing breast cancer within each Gail risk stratum (Figure 1). The SNP risk was consistently associated with breast cancer within each stratum. This showed further evidence that Gail risk and SNP risk provided independent information about risk.
Figure 1.
Observed odds of breast cancer for quintiles of single-nucleotide polymorphism (SNP) risk, stratified by Gail risk quintile (open circles). The overall odds of breast cancer in our study for case patients and control subjects were 1:1 because equal numbers of case patients and control subjects were selected. Quintiles of Gail and SNP risk are labeled 1–5 from lowest to highest risk. The straight lines represent linear least squares fits to the data within each Gail risk stratum. SNP risk was consistently associated with breast cancer odds within the Gail risk strata.
A linear regression model was used to test whether the log-transformed SNP risk score was predictive of any of the clinical risk factors contributing to the Gail model, while adjusting for case–control status. The SNP risk score was not statistically significantly associated with age at menarche (P = .96), age at menopause (P = .78), number of first-degree relatives with breast cancer (P = .20), or number of previous breast biopsies (P = .41). The SNP risk score was possibly associated with age at first birth (P = .10), and this association appeared to be specifically mediated by rs2981582 in the fibroblast growth factor receptor 2 (FGFR) gene. This SNP alone showed stronger evidence for association (P = .008) and was the only SNP associated with any clinical risk factor of the Gail model with P < .05 (data not shown). When only the control subjects were analyzed, age at first birth was still statistically significantly associated with SNP risk (P = .03) and with SNP rs2981582 (P = .002), and there were no other associations of clinical risk factors and SNP risk with a nominal P value of less than .05. These findings would not remain statistically significant after correction for multiple testing; hence, they require replication and validation in other datasets.
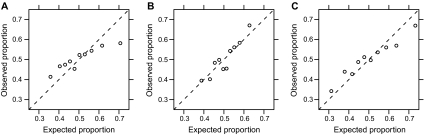
Calibration of the SNP Risk Score
To assess calibration of the SNP risk score, we used the Hosmer–Lemeshow test (31), which compares expected and observed counts of case patients and control subjects in deciles of risk (Supplementary Table 1, available online). As shown in Figure 2, the SNP risk score appeared to be well calibrated (P = .18). The Gail risk score, however, was not well calibrated (P = 1.6 × 10−6), and this lack of calibration carried over to the Gail × SNP risk score to an intermediate extent (P = .003). These results were consistent with our analysis using a logistic regression model, where a twofold increase in Gail risk showed only a 1.38-fold increase in cancer incidence (Table 4). The relatively poor calibration of the Gail risk score is consistent with a previous report from the WHI observational study (23). We did not observe an improvement in calibration in the subset of women with no missing observations for Gail risk factors.
Figure 2.
Observed vs expected proportions of case patients, for deciles of risk scores. A) Gail risk. B) The single-nucleotide polymorphism (SNP) risk score. C) The combined Gail × SNP risk score (risk score = open circles). If the risk scores are calibrated, the open circles should fall along the dashed line with a slope of 1.
Classification Performance of the Gail × SNP Risk Score
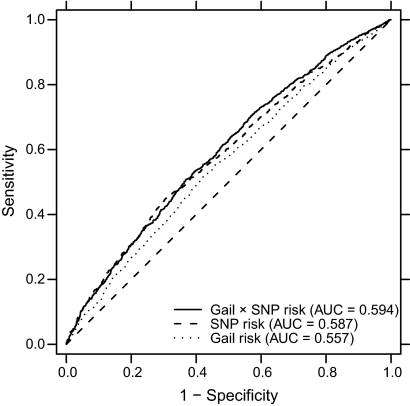
To assess the ability of the Gail × SNP risk score to better classify women's breast cancer risk, we used the ROC curve analysis. We compared classification performance using the Gail risk score, the SNP risk score, and the Gail × SNP risk score (Figure 3). We observed an AUC of 0.594 (95% CI = 0.575 to 0.612) for the combination of Gail × SNP risk compared with AUC of 0.557 (95% CI = 0.537 to 0.575) for Gail risk alone and AUC of 0.587 (95% CI = 0.567 to 0.607) for SNP risk alone. The difference in AUC for the combined risk score vs Gail risk score alone was statistically significant (difference in AUC = 0.037, 95% CI of the difference = 0.025 to 0.051, empirical P < .001). We also computed age-adjusted ROC curves, and all age-adjusted AUC values were within 0.001 of the unadjusted values (data not shown).
Figure 3.
Receiver operating characteristic curves and area under the curve (AUC) for Gail risk, single-nucleotide polymorphism (SNP) risk, and combined Gail × SNP risk.
We used a reclassification table to assess the assignment of women to low, intermediate, and elevated risk categories. We chose 5-year absolute risk thresholds of 1.5% and 2% (<1.5% for below average risk, 1.5%–2.0% for intermediate risk, and >2.0% for elevated risk) and evaluated reclassification for the Gail × SNP risk score vs Gail risk score alone (Table 5). The Gail × SNP risk score assigned more individuals to the tails of the risk distribution: it placed 22% fewer case patients and 29% fewer control subjects in the intermediate 1.5%–2.0% bin compared with the Gail score alone. The NRI for this table was 0.085 (Z = 4.3, P = 1.0 × 10−5), indicating that these changes are often in the right direction. Classification improved for 5.6% of case patients (P = 4.8 × 10−5) and 2.9% of control subjects (P = .018). Next, we investigated the extent to which reclassification performance might be distorted by poor calibration of the Gail model in the WHI cohort. We used a logistic regression model to fit breast cancer status against the logarithm of the 5-year Gail risk estimate to obtain recalibrated Gail risk estimates. This has no effect on model discrimination, but the Hosmer–Lemeshow test suggested that the scores were now well calibrated (P = .87, compared with the previous P = 1.6 × 10−6). We adjusted the classification cut points to represent the same quantiles on this new risk scale. The NRI for this table was 0.099 (Z = 4.2, P = 1.3 × 10−5), with improvement for 7.9% of case patients (P = 1.4 × 10−6) and 2.0% of control subjects (P = .11).
Table 5.
Reclassification for Gail risk vs combined Gail × single-nucleotide polymorphism (SNP) risk*
| Gail risk | Gail × SNP risk |
|||
| <1.5% | 1.5%–2.0% | >2.0% | Overall† | |
| <1.5% | ||||
| Women | 1060 | 241 | 64 | 1365 |
| Case patients | 455 | 133 | 41 | 629 |
| Control subjects | 605 | 108 | 23 | 736 |
| Proportion of cases | 0.429 | 0.552 | 0.641 | 0.461 |
| 1.5%–2.0% | ||||
| Women | 351 | 342 | 263 | 956 |
| Case patients | 155 | 172 | 157 | 484 |
| Control subjects | 196 | 170 | 106 | 472 |
| Proportion of cases | 0.442 | 0.503 | 0.597 | 0.506 |
| >2.0% | ||||
| Women | 43 | 129 | 807 | 979 |
| Case patients | 19 | 64 | 468 | 551 |
| Control subjects | 24 | 65 | 339 | 428 |
| Proportion of cases | 0.442 | 0.496 | 0.580 | 0.563 |
| Overall‡ | ||||
| Women | 1454 | 712 | 1134 | 3300 |
| Case patients | 629 | 369 | 666 | 1664 |
| Control subjects | 825 | 343 | 468 | 1636 |
| Proportion of cases | 0.433 | 0.518 | 0.587 | 0.504 |
Reclassification of case patients and control subjects selected from the Women's Health Initiative clinical trial as a result of incorporating genetic information from seven SNPs into the Gail model. Reclassification was calculated for strata of absolute 5-year risk of <1.5%, 1.5%–2.0%, and >2.0%.
Counts of women, case patients, and control subjects, and the proportion of case patients aggregated across the three strata of Gail × SNP risk.
Counts of women, case patients, and control subjects, and the proportion of case patients, aggregated across the three strata of Gail risk.
Reclassification performance is sensitive to the number of clinically meaningful risk categories because binning of risk scores conceals improvements in risk estimates that do not cross the prespecified thresholds. If risk thresholds at 1.2%, 1.5%, 1.8%, and 2.4% are chosen to distribute women into quintiles of Gail risk, then NRI increases to 0.141 (Z = 5.63, P = 9.0 × 10−9). For deciles of Gail risk, NRI is 0.182 (Z = 6.2, P = 2.1 × 10−10). The NRI is only weakly dependent on the precise placement of the cut points (Supplementary Figure 2, available online).
The usefulness and cost-effectiveness of a genetic test can be improved by avoiding testing individuals whose status is unlikely to change as a result of the test. Individuals who are far from the classification cut points are unlikely to be reclassified as a result of the test, and therefore, it is less efficient to test them. Therefore, excluding the tails of the risk distribution from the tested population should result in an improved NRI, with reclassification of a higher proportion of tested individuals (37). We evaluated NRI across a grid of possible lower and upper bounds of Gail risk. Excluding women in the tails of the risk distribution resulted in higher NRI (Supplementary Figure 3, available online). If all women with Gail risk less than 1.5% or greater than 2.0% were excluded, then NRI improved to 0.195 (Z = 3.8, P = 8.6 × 10−5). To address a possible bias in the NRI estimate, we determined the empirical distribution of NRI from 25 000 replicate datasets, where we recalculated Gail × SNP risk using permuted SNP risk scores. The mean NRI for women with Gail risk between 1.5% and 2.0% in these replicates was 0.024. Our observed NRI adjusted for this bias was 0.171 and remained statistically significant (empirical P = .0004).
Performance in Women With Previous Breast Biopsies
Because women with previous breast biopsies are at intermediately elevated risk of breast cancer and in particular need of risk stratification to guide future screening and preventative strategies, we also assessed the impact of the Gail × SNP risk score in this subset. Given the loss of an important risk stratifier, the Gail model had an AUC of only 0.514 (95% CI = 0.471 to 0.561) in this subset. The Gail × SNP risk score had an AUC of 0.571 (95% CI = 0.526 to 0.614). We also computed reclassification metrics in the biopsy subset (Supplementary Table 2, available online). In this subset, the NRI is 0.175, which is statistically significant despite the smaller number of events (Z = 3.9, P = 4.9 × 10−5). The classification improved for only 2.8% of case patients (P = .16) but for 14.8% of control subjects (P = 1.5 × 10−5). We used bootstrap resampling to evaluate whether the difference in NRI between the full cohort, and the biopsy subset was statistically significant. Based on 1000 bootstrap replicates, a 95% confidence interval for the improvement in NRI in the biopsy subset extended from 0.02 to 0.16 (empirical P = .03). This increase in NRI is not simply a consequence of conditioning on an important Gail risk factor; in the subset of individuals without a previous breast biopsy, NRI was reduced to 0.065.
Risk Assessment and Tumor ER Status
We evaluated risk assessment among subsets of breast cancer case patients defined by ER status (Supplementary Results, available online). Gail risk and Gail × SNP risk were substantially more predictive for ER-positive (Gail risk: P = 1.1 × 10−9; Gail × SNP risk: P = 2.4 × 10−22) than for ER-negative tumors (Gail risk: P = 0.89; Gail × SNP risk: P = 0.32) (Supplementary Tables 3 and 4, available online). We saw suggestive evidence that an SNP risk model using ER-specific odds ratios may improve performance for ER-negative tumors (improvement in AUC = 0.022, empirical P = 0.10) (Supplementary Table 5, available online).
Discussion
The major finding from this nested case–control cohort from the WHI Clinical Trial is that genetic risk information may be combined multiplicatively with Gail risk scores to improve breast cancer risk estimation in postmenopausal white non-Hispanic women. This finding is based both on the observation that breast cancer risk may be accurately estimated by combining published SNP risk estimates and also the observation that correlation between SNP risk scores and Gail risk for individuals is weak, allowing breast cancer risk to be more accurately estimated by combining SNP risk and Gail risk multiplicatively. Thus, this study supports the claim that the combined risk estimation model approach has clinical validity in the broad sense in postmenopausal white non-Hispanic women.
The calibration and discrimination of the Gail model in the WHI cohort are known to be somewhat worse than those seen in other large studies (23). This is likely because of a combination of factors, including higher mammography rates, differences in age distributions, and changes in breast biopsy procedures, with more common use of image-guided percutaneous core biopsy procedures that have a lower threshold for use than open biopsy (23). The lack of data on atypical hyperplasia may also contribute to lower calibration and discrimination in the WHI cohort. Additionally, as the WHI Clinical Trial tested the impact of hormone replacement therapy on breast cancer risk, a higher percentage of women in our case–control cohort may have received hormone replacement therapy than in the studies in which the Gail model was previously validated. Last, the age matching in our nested case–control study is likely to have contributed to the reduced performance of the Gail model because age plays a substantial role in that model. This limitation does not directly affect our assessment of independence and calibration of the SNP risk scores but may affect the quantitative metrics of improvement in risk assessment in the combined model. Reclassification performance is sensitive to model calibration as well as discrimination, and it will need to be further characterized in population-based cohorts.
When we assessed the performance of a combined risk predictor incorporating both the Gail risk and SNP risk, the combined risk predictor performed better in predicting breast cancer risk than either Gail risk or SNP risk alone. By ROC curve analysis, the AUC for Gail and SNP risk combined was 0.594 (95% CI = 0.575 to 0.612) as compared with Gail risk alone, showing an AUC of 0.557 (95% CI = 0.537 to 0.575). Although this statistically significant improvement was modest, in this dataset, the Gail model itself had an AUC that was only 5.7% greater than that expected by chance.
Although ROC curves are useful in some contexts, they have been criticized for several reasons: 1) they summarize classification information across the full range of sensitivities and specificities (in most clinical contexts, only a subset of these sensitivities and specificities are relevant); 2) they do not provide information about the actual risks predicted by the model; 3) they do not provide information about the proportion of individuals with particularly high- or low-risk values; and 4) the area under the ROC curve, which is the probability that a predicted risk for an individual with an event is higher than for an individual without an event, has minimal direct clinical relevance (38). Therefore, we used reclassification tables (34,35) to calculate NRI (36) as a more helpful measure of the potential impact of the Gail × SNP risk score in assessing invasive breast cancer risk (27). This analysis demonstrated a statistically significant improvement in classification (NRI = 0.085; P = 1.0 × 10−5). Importantly, NRI can be substantially improved by focusing SNP genotyping on those individuals who are predicted to be at intermediate risk by the Gail score, as these women are most likely to be reclassified after the addition of SNP risk. For example, limiting SNP testing to women with Gail 5-year risks between 1.5% and 2.0% results in a substantially larger NRI of 0.195. Taking this information into account, future efforts should rigorously evaluate the clinical utility of targeted strategies to incorporate the combined risk score into the clinical decision-making process in the context of both breast cancer primary prevention and screening (17−20).
NRI is a relatively new statistic but has gained increasing acceptance as an important part of the evaluation of new biomarkers and risk stratifiers (38,39). The statistic has also been criticized, for sensitivity to model calibration (40,41) and dependence on arbitrary cut points (42). The use of NRI presupposes that classification performance is important, and in that context, sensitivity to calibration could be seen as a feature, not a limitation. In any case, the Gail model is embedded within our combined risk model, so the calibration discrepancy of the Gail model is also present in the combined model. We obtained a similar NRI using recalibrated Gail risk estimates, and we found that the NRI statistic is robust to moderate changes in the cut points.
We also evaluated whether it might be possible to obtain better test performance by focusing on a subset of women at particularly high risk—those with previous breast biopsies (43,44). Here, classification improved for only 2.8% of case patients but for 14.8% of control subjects (P = 4.4 × 10−5). Although this improvement suggests that the combined Gail × SNP risk score might assist in identifying a subset of women with previous biopsies who might not need as aggressive risk reduction and surveillance efforts as their biopsy history suggests, these results should be interpreted with caution and will require further validation in other datasets with available histopathology from the previous biopsies.
This study has several strengths. First, we have taken a rigorous approach to identifying the SNPs to include in the panel; only including those that have been reproducibly associated with breast cancer risk and for which consistent risk estimates have been reported in the literature (5−8). Second, we have genotyped individuals from a large prospectively recruited cohort with meticulous data collection and rigorous ascertainment of relevant breast cancer outcomes. Third, we have used literature-based genetic risk estimates and have combined them in a straightforward fashion to form risk predictors. Importantly, we did not train our predictors on the WHI Clinical Trial data and only used the trial samples to assess their performance.
Our study has a few limitations that include the composition of the WHI Clinical Trial cohort, which limits inferential scope to white non-Hispanic postmenopausal women, and the clinical characteristics of the women therein. For example, individuals within the WHI Clinical Trial received hormone replacement therapy at a higher frequency than women currently do at present. Importantly, the age-matching design inherent to this study does remove one of the Gail model variables and contributes both to the relatively low AUC seen for the Gail model in our analysis and to its poor calibration. In addition, the absence of pathology records for previous breast biopsies in the WHI Clinical Trial required us to estimate individual Gail risks by coding atypical hyperplasia status as unknown for women with prior breast biopsies. Although the incidence of atypical hyperplasia is sufficiently low that it seems unlikely to have affected the analyses of the entire nested case–control study, it is unclear to what extent this may have affected the analyses focusing on the subset of women with one or more previous biopsies. Additionally, case–control sampling means we cannot evaluate calibration of absolute event rates; we can only effectively test the slope of the relationship between expected and observed risk and not the intercept. Therefore, although the WHI Clinical Trial cohort is sufficient to support the validity of the SNP risk and Gail × SNP risk models in predicting breast cancer risk, there is a need for further assessment of the clinical validity of the combined Gail × SNP risk model, especially in population-based cohorts.
A recent study (29) has performed a similar evaluation of breast cancer risk models incorporating SNP information, which combined case patients and control subjects from four cohort studies and one case–control study (including the WHI Observational Study), and refit Gail and SNP model parameters rather than comparing prespecified models. It reported a difference in AUC of 0.038 from adding SNP information to the best clinical risk model compared with 0.037 in our study. Although NRI itself was not reported, it can be calculated from the reported data for quintiles of risk, and the resulting NRI of 0.141 matches our result. These results suggest that our reported differences in model performance are somewhat robust to differences in underlying data and model-fitting methodologies.
The use of improved risk models, such as the one described in our study, may benefit the public health if shown to have clinical utility when combined with optimal individualized screening and risk reduction strategies. We agree with previous assessments that the benefits are likely to be modest (26,29), although our work points to several strategies that may give better results. A previous evaluation of potential utility considered an unselective strategy for SNP testing broadly applied to women regardless of Gail risk (26). Our results suggest that, because utility is sensitive to how a test is targeted, it may be wise to focus further evaluation of the application of SNP genotyping for breast cancer risk on women at intermediate risk as measured by the Gail model. Such a strategy clearly boosts classification performance in this study. Use of broad highly multiplexed multi-disease SNP panels would also improve cost-effectiveness by reducing the marginal cost of testing. Future research should assess performance in population-based cohorts and ultimately address whether classification improvement can be translated into improved prevention and/or screening outcomes in the clinic.
Funding
National Heart Lung and Blood Institute (NHLBI, contract number HHSN268200764314C to R.L.P. and M.P.); National Cancer Institute (PO1 CA53996 to R.L.P. and M.P.) at the National Institutes of Health, Department of Health and Human Services. The Women's Health Initiative was supported by the NHLBI.
Supplementary Material
Footnotes
M. E. Mealiffe, R. P. Stokowski, B. K. Rhees, and D. A. Hinds were employees of Perlegen Sciences during this work, and these analyses were relevant to development of a diagnostic product by Perlegen.
Present address: Clinical Utility Consulting, LLC, Palo Alto, CA (M. E. Mealiffe).
Present address: Tandem Diagnostics, Inc, San Jose, CA (R. P. Stokowski).
Present address: Artemis Health, Inc, San Carlos, CA (B. K. Rhees).
Present address: 23andMe, Inc, Mountain View, CA (D. A. Hinds).
We thank Ellen Beasley, Allison Kurian, Kevin Hughes, Anne-Renee Hartman, and Bryan Walser for helpful comments and discussions. The authors thank Women's Health Initiative (WHI) investigators and staff for their dedication and the study participants for making the WHI possible. A listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.Robson M, Offit K. Clinical practice. Management of an inherited predisposition to breast cancer. N Engl J Med. 2007;357(2):154–162. doi: 10.1056/NEJMcp071286. [DOI] [PubMed] [Google Scholar]
- 4.Pharoah PDP, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BAJ. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31(1):33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 5.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 8.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(6):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 12.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 13.Nam RK, Zhang WW, Trachtenberg J, et al. Utility of incorporating genetic variants for the early detection of prostate cancer. Clin Cancer Res. 2009;15(5):1787–1793. doi: 10.1158/1078-0432.CCR-08-1593. [DOI] [PubMed] [Google Scholar]
- 14.Weersma RK, Stokkers PC, Cleynen I, et al. Confirmation of multiple Crohn's disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104(3):630–638. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- 15.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358(12):1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 16.Domchek SM, Eisen A, Calzone K, Stopfer J, Blackwood A, Weber BL. Application of breast cancer risk prediction models in clinical practice. J Clin Oncol. 2003;21(4):593–601. doi: 10.1200/JCO.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Mahoney MC, Bevers T, Linos E, Willett WC. Opportunities and strategies for breast cancer prevention through risk reduction. CA Cancer J Clin. 2008;58(6):347–371. doi: 10.3322/CA.2008.0016. [DOI] [PubMed] [Google Scholar]
- 18.Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57(2):75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 19.Visvanathan K, Chlebowski RT, Hurley P, et al. American Society of Clinical Oncology clinical practice guideline update on the use of pharmacologic interventions including tamoxifen, raloxifene, and aromatase inhibition for breast cancer risk reduction. J Clin Oncol. 2009;27(19):3235–3258. doi: 10.1200/JCO.2008.20.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinsinger LA, Harris R, Lewis C, Woddell M. Chemoprevention of breast cancer: a review of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2002;137(1):59–69. doi: 10.7326/0003-4819-137-1-200207020-00017. [DOI] [PubMed] [Google Scholar]
- 21.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 22.Elmore JG, Fletcher SW. The risk of cancer risk prediction: “What is my risk of getting breast cancer.”. J Natl Cancer Inst. 2006;98(23):1673–1675. doi: 10.1093/jnci/djj501. [DOI] [PubMed] [Google Scholar]
- 23.Chlebowski RT, Anderson GL, Lane DS, et al. Predicting risk of breast cancer in postmenopausal women by hormone receptor status. J Natl Cancer Inst. 2007;99(22):1695–1705. doi: 10.1093/jnci/djm224. [DOI] [PubMed] [Google Scholar]
- 24.Armstrong K, Quistberg DA, Micco E, et al. Prescription of tamoxifen for breast cancer prevention by primary care physicians. Arch Intern Med. 2006;166(20):2260–2265. doi: 10.1001/archinte.166.20.2260. [DOI] [PubMed] [Google Scholar]
- 25.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pepe MS, Janes HE. Gauging the performance of SNPs, biomarkers, and clinical factors for predicting risk of breast cancer. J Natl Cancer Inst. 2008;100(14):978–979. doi: 10.1093/jnci/djn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pharoah PDP, Antoniou AC, Easton DF, Ponder BAJ. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 29.Wacholder S, Hartge P, Prentice R. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prentice RL, Anderson GL. The Women's Health Initiative: lessons learned. Annu Rev Public Health. 2007;29:131–150. doi: 10.1146/annurev.publhealth.29.020907.090947. [DOI] [PubMed] [Google Scholar]
- 31.Hosmer DW, Lemeshow S. Applied Logistic Regression. New York, NY: Wiley; 1989. Section 5.2.2. [Google Scholar]
- 32.Prentice RL, Pyke R. Logistic disease incidence models and case control studies. Biometrika. 1979;66(4):403–411. [Google Scholar]
- 33.Janes H, Longston G, Pepe MS. Accommodating covariates in receiver operating characteristic analysis. Stata J. 2009;9(1):17–39. [PMC free article] [PubMed] [Google Scholar]
- 34.Cook NR, Buring JE, Ridker PM. The effect of including C-reactive protein in cardiovascular risk prediction models for women. Ann Intern Med. 2006;145(1):21–29. doi: 10.7326/0003-4819-145-1-200607040-00128. [DOI] [PubMed] [Google Scholar]
- 35.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115(7):928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 36.Pencina MJ, D’Agostino Sr RB, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2007;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 37.Cook NR. Comments on ‘Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond’. Stat Med. 2008;27(2):191–195. doi: 10.1002/sim.2987. [DOI] [PubMed] [Google Scholar]
- 38.Janes H, Pepe MS, Gu W. Assessing the value of risk predictions by using risk stratification tables. Ann Intern Med. 2008;149(10):751–760. doi: 10.7326/0003-4819-149-10-200811180-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGeechan K, Macaskill P, Irwig L, Liew G, Wong TY. Assessing new biomarkers and predictive models for use in clinical practice: a clinician's guide. Arch Intern Med. 2008;168(21):2304–2310. doi: 10.1001/archinte.168.21.2304. [DOI] [PubMed] [Google Scholar]
- 40.Pencina MJ, D’Agostino Sr RB, D’Agostino RB, Jr, Vasan RS. Comments on ‘Integrated discrimination and net reclassification improvements—Practical advice’. Stat Med. 2008;27(2):207–212. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 41.Gail MH. Response: Re: discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;101(24):1732. [Google Scholar]
- 42.Vickers AJ, Elkin EB, Steyerberg E. Net reclassification improvement and decision theory. Stat Med. 2009;28(3):525–536. doi: 10.1002/sim.3087. [DOI] [PubMed] [Google Scholar]
- 43.Hartmann LC, Sellers TA, Frost MH, et al. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 44.Ashbeck EL, Rosenberg RD, Stauber PM, Key CR. Benign breast biopsy diagnosis and subsequent risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(3):467–472. doi: 10.1158/1055-9965.EPI-06-0394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.