Abstract
Microfluidic image cytometry (MIC) has been developed to study phenotypes of various hPSC lines by screening several chemically defined serum/feeder-free conditions. A chemically defined hPSC culture was established using 20 ng mL−1 of bFGF on 20 μg mL−1 of Matrigel to grow hPSCs over a week in an undifferentiated state. Following hPSC culture, we conducted quantitative MIC to perform a single cell profiling of simultaneously detected protein expression (OCT4 and SSEA1). Using clustering analysis, we were able to systematically compare the characteristics of various hPSC lines in different conditions.
Human pluripotent stem cells (hPSCs) such as human embryonic stem cells (hESCs)1 and human induced pluripotent stem cells (hiPSCs)2–6 exhibit unique characteristics and may provide great opportunities for cell-based therapy and regenerative medicine. These characteristics include unlimited propagation capacity in the undifferentiated stage with a normal euploid karyotype and the ability to differentiate into all cell types in the human body.
Typically, hPSC culture conditions contain serum such as KnockOut serum replacement (KSR) and feeders such as mouse embryonic fibroblasts (MEFs). Although these conditions can successfully maintain pluripotency of hPSCs, these animal products could cause xenogenic contamination and immunorejection in patients after transplantation of hPSCs, posing a major challenge to the use of hPSCs in cell-based therapy applications. Additionally, these factors are undefined and some are proprietarily formulated, forming an obstacle in being able to systematically study the regulation of stem cell biology. Therefore, it is essential to develop serum/feeder-free culture methods for hPSCs in order to define culture elements and later apply them to effective therapeutic use.
Currently, there is an ongoing trend towards establishing chemically defined conditions for hPSC culture. Several chemically defined culture systems have been introduced to maintain hESCs in combination with (i) growth factors/cytokines (e.g., basic fibroblast growth factor (bFGF), nodal, transforming growth factor-β1 (TGF-β1), activin A and insulin-like growth factor-1 (IGF-1) analog (heregulin-1β)) and (ii) supplements (e.g., GABA, pipecolic acid and lithium chloride,7,8 and N2/B279) on extracellular matrices (ECM) such as Matrigel or other ECM components. A chemically defined culture system with serum/feeder-free conditions is ideal since it excludes the unknown factors and enhances the reproducibility and robustness of hPSC propagation. Thus, to facilitate practical applications involving hPSCs, optimal chemically defined culture conditions must be established that will not only maintain phenotypically and karyotypically stable cells for extended periods but will also retain the ability for directed and reproducible differentiation.
Until now, even with the conventional culturing methods, controlling hPSC fate (e.g., self-renewal, differentiation, apoptosis and quiescence) has been challenging and underlying mechanisms are mostly unidentified. However, recent studies have uncovered some extrinsic factors that can influence state stability of hPSCs and contribute to fate decisions.10 These extrinsic factors include various soluble factors, cell-cell interactions, and ECM, which are key components of the hPSC microenvironment by definition (Fig. 1a).11 Additionally, soluble factors such as growth factors added to the culture or secreted by stem cells are often potent in their effects on cell fate.12 Indeed, undifferentiated hPSCs are highly sensitive to the soluble growth factors that are usually contained in these media. However, the effects of various defined media for maintaining self-renewal states over extended periods have not been fully studied and optimally defined culture conditions have yet to be further refined. Therefore, screening chemically defined media (CDM) to evaluate the influence of these factors will also be essential for acquiring more qualified and defined culture methods that support self-renewal of hPSCs.
Fig. 1.
(a) The extrinsic factors such as soluble growth factors, cell-cell interactions, and ECM play an important role in controlling stem cell fate in the microenvironment. (b) Schematic illustration of a microfluidic hPSC array for hPSC culture and phenotype assay. (c) Microfluidic image cytometry (MIC) was conducted followed by segmentation and quantification analysis.
Still, there are several other parameters that must be addressed in the study of hPSCs. First, although hPSCs can self-renew indefinitely, it is known that there is enormous variation between different PSC lines with regard to expression of pluripotency and differentiation markers.13 This is likely due to that fact that hES cell lines have been derived from embryos with different characteristics and further isolated by different procedures.14 In the case of hiPSCs, there is also variation due to (i) the factors used for reprogramming, (ii) the methods to deliver these factors, (iii) the source of the original cell lines, (iv) the expression levels of delivered factors, (v) the culture conditions for obtained hiPSCs and (vi) the methods to identify obtained hPSCs.15 Second, various commercially available hPSC defined culture media and ECM7–9,16–18 contain different components that may cause variable effects depending on the cell line and culture periods. Thus, taking into consideration all of these parameters as influential factors, there is also a need to systematically compare the differences between hPSC lines in order to comprehend their fundamental biology.
However, there are some disadvantages in conventional experimental settings for hPSCs. Especially, when screening the characteristics among various hPSC lines, conventional analyses such as flow cytometry, microarray or RT-PCR require large amounts of cells, resulting in high costs in maintenance.19 On the other hand, the introduction of microfluidics can allow major advances in stem cell research. While there are tremendous efforts to compare the similarities and differences of various hPSC characteristics worldwide,13 it is highly important to establish a standardized hPSC culture condition, which causes less deviation and uncertainty. In microfluidics, miniaturization of cell culture platforms not only allows us to observe cellular behavior on the scale found in living systems but also provides a means to engineer miniaturized cell culture platforms that are more in vivo-like than conventional dish cultures,20,21 thereby fostering robust, reproducible and uniform culture conditions. Additionally, with the ability to manipulate the fluid flow precisely, microfluidics can make excellent perfusion cell-culture devices, which are powerful tools to control the soluble and mechanical parameters of the cell culture environment.22 These aspects are extremely essential since hPSCs interact strongly with their microenvironmental factors, which can directly influence the fate decisions. Furthermore, microfluidic technology can be integrated with a variety of biological assays and is compatible with Micro Electro Mechanical System (MEMS) technology for further applications including electrophoresis and cell sorting.19 A microfluidic device is made out of polydimethylsiloxane (PDMS), which is an elastomeric material utilizing the process of soft lithography for fabrication. Its beneficial features for stem cell biology include biocompatibility, gas permeability and durability. It is also safe and easy to handle within general laboratories performing biological research. Additionally, with its scalability and automation, it has more potential for clinical applications. Ultimately, since microfluidic devices can perform standard tissue culture in a more rapid, controllable and reproducible fashion with considerably low costs in a high-throughput fashion,23,24 microfluidic technology is well-suited for evaluating multiple hPSC culture conditions and simultaneously observing their responses.
Previously, Villa-Diaz et al. and we have reported maintaining hESCs in conventional KSR/MEF conditions inside a hESC-μChip.25,26 However, to date, it has not yet been reported that hPSCs, especially hiPSCs, can be cultured in chemically defined culture conditions and quantitatively studied in a microfluidic device. More importantly, there has not been a systematic comparison of the similarities or differences of each of these hPSCs cultured in various chemically defined culture conditions.
Therefore, we have developed a microfluidic hPSC array (Fig. 1b) to perform hPSC culture and phenotype assay. Subsequently, microfluidic image cytometry (MIC) was conducted (Fig. 1c) followed by segmentation and quantification analysis. In this study, we have reported (i) a microfluidic platform to optimize ECM and various CDM for evaluating optimal culture conditions in hPSCs, combined with (ii) a systematic and quantitative analysis and small-scale screening of the hPSCs cultured in various CDM using multi-parallel detected protein expressions. Using this array, we have also performed (iii) a side-by-side comparison of the hPSC phenotypic responses across available stem cell lines and CDM. This analysis allowed for examination of the cell fate of a single hPSC in a hPSC colony in each condition and demonstrated the sensitivity and effectiveness of our microfluidic hPSC array for use in quantification of multiple stem cell culture parameters.
For fabrication of a PDMS-based microfluidic hPSC array, we used the process of soft lithography (ESI Fig. S1a †). The PDMS was mounted and assembled on a glass slide. During the hPSC culture assays, this array was set on an inverted microscope stage for routine monitoring of hPSCs. Our PDMS-based microfluidic hPSC array was comprised of 24 cell culture chambers (700 μm (W) × 900 μm (L) × 100 μm (H), Total volume 630 nL). For on-chip cell culture, each chamber was used for the static culture conditions. Using an electrical pipette (0.5–12.5 μL, Thermo Fisher Scientific) capable of handling precise volume and flow rates, four μL of solution containing hPSCs or reagents were filled into the tip. The tip was gently inserted into the inlet of a microfluidic hPSC culture array and solution was dispensed at 6 μL sec−1 with accurate piston movement (ESI Fig. S1b†). A few hours after hPSC loading, the medium was changed every 12 h (see also in ESI Methods and Fig. S2†). In this array, each chamber can perform immunocytochemical analysis under discrete hPSC culture conditions to determine the levels of protein expression (see also in ESI Methods†). For cell line study, we examined 5 lines including (i) OCT4-enhanced green fluorescent protein (EGFP) knock-in HSF1 cell line (HSF1-OCT4-EGFP),25,27 (ii) hESC lines (HSF1 and H1) and (iii) hiPSC lines (iPSA1 and iPSB2, ESI Fig. S3†). The OCT4-EGFP cell line is unique in that it allows live cell monitoring of its pluripotency status in real-time. We therefore used it to optimize the defined culture conditions. Other cell lines were used to further make comparisons between their protein expressions.
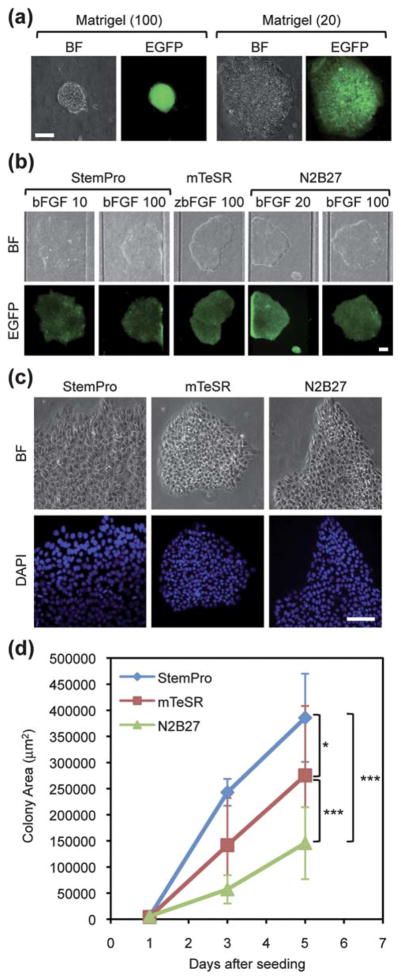
For the purpose of establishing optimal culture conditions in a microfluidic hPSC array, we began with examining the optimal concentration of ECM by using MEF-conditioned medium (CM). We chose to use hESC qualified Matrigel, (see also in ESI Methods†) since this is commonly used for feeder-free hPSC culture in current stem cell research. As mentioned, we used HSF1-OCT4-EGFP cell lines to monitor the morphology of hPSC colonies and EGFP expression levels during culturing periods. The results showed that HSF1-OCT4-EGFP colonies were unable to attach, spread out and grow well on the substrate coated with 100 μg mL−1 (Fig. 2a). On the other hand, the HSF1-OCT4-EGFP colonies extended well and maintained their growth in an undifferentiated state for 7 days with 20 μg mL−1. Thus, we determined that 20 μg mL−1 of Matrigel was an optimal ECM condition for hPSC culture.
Fig. 2.
The establishment of serum/feeder-free chemically defined hPSC culture conditions in a microfluidic hPSC array. Bright field (BF) and fluorescence images of hPSCs are shown on the top and bottom, respectively. (a) Optimization of Matrigel coating conditions. HSF1-OCT4-EGFP cells were cultured on ECM with two concentrations (20 or 100 μg mL−1) using MEF-CM. Scale bar represents 50 μm. (b) Screening of CDM (StemPro, mTeSR and N2B27) with different concentrations of bFGF using HSF1-OCT4-EGFP cells. HSF1-OCT4-EGFP cells were cultured on the glass slide coated with an optimal Matrigel concentration (20 μg mL−1). Scale bar represents 100 μm. (c) Morphologically different HSF1-OCT4-EGFP colonies cultured in three CDM. (d) Quantitative comparison of the growth curves of HSF1-OCT4-EGFP cells cultured in three CDM. Each dot represents mean ± S.D. (*p < 0.05, ***p < 0.001). Scale bar represents 50 μm.
Next, using an optimized ECM condition, we then screened CDM for optimal culture conditions. We tested three CDM (StemPro,16 mTeSR7,8,32 and N2B279), which had been published to support undifferentiated growth of hPSCs with defined components. Each medium was also supplemented with varying bFGF concentrations and the morphology of hPSC colonies and EGFP expression levels were then monitored over 5 days (Fig. 2b). After five days in culture, HSF1-OCT4-EGFP cells were able to form colonies and express EGFP driven by an OCT4 promoter in all three CDM conditions. We also found that bFGF concentrations did not influence cell viability and pluripotency of hPSCs. In a previous study, we used 100 ng mL−1 of bFGF with KSR/MEF conditions.15 However, at this time, we observed that 20 ng mL−1 bFGF in feeder-free chemically defined hPSC culture conditions was sufficient to grow undifferentiated hPSCs. Here, we confirmed that all three chemically defined conditions were able to sustain the growth of hPSCs with undifferentiated states using optimized ECMs and accordingly established optimal defined culture conditions in a microfluidic hPSC array. Interestingly, within optimal conditions we now found a variation in physical and biochemical characteristics in hPSCs cultured with different media. We then compared the effects of these media on various phenotypes across the cell lines including morphology, growth rates and expression level of pluripotency protein markers. A recent study showed that colony morphology was an important parameter to determine characteristics of hPSCs and molecular phenotype and differentiation potential could vary within morphologically different hPSC colonies.28 According to the results of DAPI nuclear staining, we found that HSF1-OCT4-EGFP cells cultured in StemPro formed colonies with sharp pointed edges (Fig. 2c). These cells also had a tendency to form a relatively larger nuclear size than those cultured in the other CDM. Additionally, HSF1-OCT4-EGFP cells cultured in mTeSR represented more dense and tight colonies. We then conducted growth assays to examine the average growth rate of colonies cultured in each medium by measuring the surface area of HSF1-OCT4-EGFP colonies (Fig. 2d). We found that although all the conditions were able to support self-renewal of hPSCs and maintain pluripotency marker protein expression over four days, the growth rate of colonies differed depending on culturing media. Among three CDM, HSF1-OCT4-EGFP cultured in Stem-Pro showed the fastest growth rate. Compared to N2B27, StemPro and mTeSR conditions showed 2.65 and 1.85-fold changes in their average colony size, respectively. We speculated that the components heregulin-1β and activin A were responsible for promoting proliferation of HSF1-OCT4-EGFP cells in StemPro.16
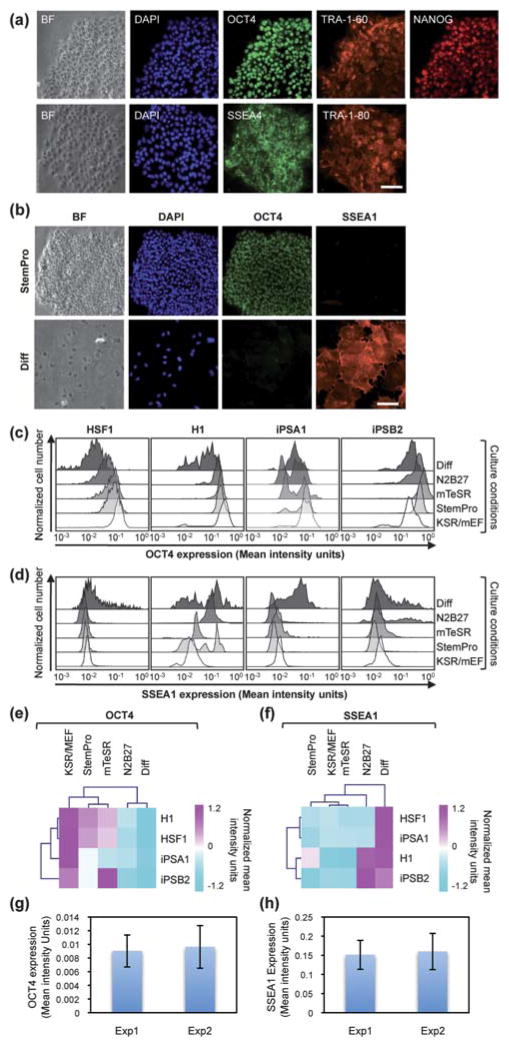
To further characterize the effects of these media on a collection of hPSC lines, we performed immunocytochemistry to evaluate expression of pluripotency markers in hES and hiPS cells quantitatively (Fig. 3 and ESI Fig. S2†). The pluripotent markers we used were OCT4, NANOG, SSEA4, TRA-1-60 and TRA-1-80. Here, we introduced one more condition where we induced differentiation by adding 10% fetal bovine serum to DMEM medium as a negative control. The differentiation marker we used was SSEA1. As the results indicated, all of the hPSCs cultured in chemically defined conditions uniformly expressed these pluripotency markers (Fig. 3a). After confirming their pluripotency, we further quantified OCT4 and SSEA1 expression at the single-cell level based on immunofluorescence imaging (Fig. 3b). Image cytometry is an image-based measurement that allows quantitative analysis of these marker expressions at the single cell level by using software such as CellProfiler, which can generate flow-cytometry-like data. Single-cell based immunofluorescent histograms presented a variable distribution in OCT4 and SSEA1 expression (Fig. 3c,d, respectively) after each cell line in each condition was co-stained with OCT4 and SSEA1 and analyzed in a single cell. In general, this visually expressed how heterogeneous/homogeneous each colony was within the undifferentiated and differentiated conditions. As an illustration, when a histogram of protein expression showed the broad distribution, populations were more heterogeneous and vice versa. The histograms of OCT4 expression in HSF1, H1 and iPSB2 cultured in KSR/MEF and three CDM conditions had a homogenous distribution with the higher level of OCT4 expression compared to Differentiation condition. The iPSA1 cells cultured in mTeSR and N2B27 had two subpopulations with both high and low OCT4 expression. The histograms for SSEA1 expressionin HSF1, iPSA1 and iPSB2 cultured in KSR/MEF and three CDM conditions exhibited a homogeneous distribution with the low SSEA1 expression level, indicating that most of the hPSC populations remained undifferentiated. On the other hand, all the cell lines in Differentiation condition exhibited the broad distribution of the histogram for SSEA1 expression, revealing various responsiveness and sensitivity of the highly heterogeneous cells upon the serum inducement. This could also be attributed to the weak and/or short-time period of treatment, but still enabled visual dynamics of the protein expression during the differentiation process. Additionally, while the H1 cells cultured in mTeSR and KSR/MEF conditions had low SSEA1 expression, some populations in StemPro and N2B27 conditions showed relatively high SSEA1 regardless of the fact that they simultaneously expressed high OCT4.
Fig. 3.
Evaluation of pluripotency/differentiation marker expression in a microfluidic hPSC array using MIC. (a) Bright-field (BF) images, DAPI nuclear fluorescence images and other fluorescence images of HSF1 cells cultured in StemPro immunostained against pluripotent markers (OCT4, NANOG, SSEA4, TRA-1-60 and TRA-1-80). Scale bar represents 50 μm. (b) BF images, DAPI nuclear fluorescence images, OCT4 and SSEA1 fluorescence images of HSF1 cells cultured in StemPro or Differentiation condition (Diff). Scale bar represents 50 μm. (c,d) Single-cell based immunofluorescent histograms of (c) OCT4 expression and (d) SSEA1 expression in individual HSF1, H1, iPSA1 and iPSB2 cells cultured in KSR/MEF, StemPro, mTeSR, N2B27 and Differentiation condition. (e,f) Heat maps based on the quantified (e) OCT4 expression and (f) SSEA1 expression. The protein expression level was normalized among samples of H1, HSF1, iPSA1 and iPSB2 cultured in KSR/MEF, StemPro, mTeSR, N2B27 and Differentiation condition and analyzed by Euclidean distance hierarchical clustering to categorize similar groups together. Each row represents marker expression in each cell line. Each column represents a particular condition. (g,h) Evaluation of MIC in terms of the reproducibility. The quantification of (g) OCT4 fluorescence intensity and (h) SSEA1 fluorescence intensity between the two experiments is shown.
For systematic analysis, we further conducted Euclidean distance hierarchical clustering29 based on the mean values of OCT4 and SSEA1 expression and generated the heat maps, which represented values of data in two-dimensional maps as colors (Fig. 3e,f, respectively. ESI Fig. S2), to compare the distinct protein expression resulting from the various cell lines and CDM. Hierarchical clustering generates a hierarchy of sample groups represented by a dendrogram (a tree-like diagram). To determine the similarities of two groups, we used Euclidean distance calculated with the eqn (1),
| (1) |
if p and q are the two points (here, samples) in Euclidean n-space for finding nearest neighbors of sample groups. The heat maps generated based on Euclidean distance hierarchical clustering are commonly used for, for instance, microarray analysis to reduce data dimension, categorize samples, and show a multidimensional data set in 2-D maps in colors. In terms of the different media treatments for 4 days, according to the heat map, hPSCs cultured in StemPro and mTeSR expressed high OCT4 and low SSEA1 across all the cell lines. The heat map also showed a similar expression pattern across all the cell lines in the OCT4 level between hPSCs cultured in StemPro and mTeSR conditions. In contrast, hPSCs cultured in N2B27 condition rendered relatively lower OCT4 expression across the cell lines, exhibiting its tendency to direct differentiation during the culturing periods. Therefore, N2B27 condition was categorized as similar to the hPSCs cultured in Differentiation condition based on the clustering. In the case of SSEA1 expression, all cell lines cultured in Differentiation condition showed strong SSEA1 expression. Additionally, we observed that some populations of H1 and iPSB2 cells cultured in N2B27 condition also expressed relatively high SSEA1. In terms of cell lines within the same condition, each cell line responded differently and resulted in various phenotypes. The iPSB2 line especially appeared to have different OCT4 and SSEA1 expression compared to the other three lines. However, there seems to be no clear trend in cell lines concluded based on the level of pluripotent marker expression.
Finally, we evaluated the robustness of MIC to confirm the fidelity of our study (Fig. 3g,h). Two microfluidic chips that cultured H1 cells in Stem Pro (Exp1 and Exp2) were randomly chosen and both OCT4 (Fig. 3g) and SSEA1 (Fig. 3h) expression were quantified. Between the two chips, there were no significant differences in OCT4 or SSEA1 fluorescence intensity value therefore we concluded that our microfluidic hPSC array in conjunction with MIC was precise and reproducible.
Conclusions
We developed a simple microfluidic platform to optimize ECM, screen CDM and establish the optimal chemically defined culture system for both human ESCs and iPSCs. By using this microfluidic platform, we were also able to study hPSC phenotypic response by comparing the effects of various CDM and hPSC lines. Although we cannot ignore the fact that PDMS may absorb molecules from solution due to their characteristics (e.g., highly porous and hydrophobic material)30 and release them during culturing periods, according to the results, not only this microfluidic platform can effectively maintain pluripotency of hPSCs over a week in CDM with 20 ng mL−1 of bFGF but all the results were consistent and reproducible across the hPSC lines. Also, our concentration of bFGF was the original concentration7–9,16 found in other studies with the conventional macro-scale settings. Thus, we considered this PDMS effect was negligible. Additionally, we found that the condition with StemPro medium on the ECM of 20 μg mL−1 of Matrigel for culturing hPSCs generally provides high OCT4 and low SSEA1 expression across the cell lines including hiPSCs. In this work, we have demonstrated that a microfluidic hPSC array can achieve robust and reproducible hPSC cultures on a simple setting when combined with highly quantitative single-cell profiling methods. Furthermore, not only can this array be utilized for real-time live cell monitoring of hPSCs, but this platform can also perform small-scale screening with multi-parallel detection system, using a small amount of samples and reagents that are roughly 3 orders of magnitude less than the conventional 6 well plates. According to the results, we found that culturing in various CDM resulted in different phenotypes in each hPSC line including morphology, growth rate and pluripotent marker expression. We speculated that over culturing periods, heterogeneous cell populations within a single colony showed varied growth factor responsiveness and protein expressions by intricately interplaying with the microenvironmental factors at the single cell level. In general, the final phenotype in a single cell relies on the current state of the cell and the microenvironment that is composed of the extrinsic factors such as soluble factors in media and output signals of hPSCs.11,31 Here, by presenting the detail phenotypic analysis, we have also demonstrated the ability of our device to study the heterogeneity of hPSCs and the interaction of different hPSC lines with the microenvironment, which will have an overall effect in governing stem cell fate. With a carefully selected set of markers (e.g. pluripotency, apoptosis, differentiation and cell cycle), this tool can be applied to conduct more phenotype studies when combined with signaling cascades transduced by extrinsic factors using its multiplexity to determine the hPSC molecular signatures. Because of these unique features, we envision that this microfluidic platform will be beneficial to investigate stem cell biology in a wide range of biomedical settings and applications in regenerative medicine.
Supplementary Material
Acknowledgments
This work was supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at the Institute of Molecular Medicine at University of California, Los Angeles. H.R.T., M.A.T., A.D.P., A.T.C., and O.N.W. were supported by California Institute of Regenerative Medicine. O.N.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Electronic supplementary information (ESI) available: Fabrication of microfluidic hPSC array, generation of hiPSC (i.e., hiPSA1 and hiPSB2), microscopy settings and image processing are available. See DOI: 10.1039/b922884e
Contributor Information
Ken-ichiro Kamei, Email: kkamei@mednet.ucla.edu.
Hsian-Rong Tseng, Email: hrtseng@mednet.ucla.edu.
Notes and references
- 1.Thomson JA. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane J, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson J. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park I, Zhao R, West J, Yabuuchi A, Huo H, Ince T, Lerou P, Lensch MW, Daley G. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig T, Levenstein M, Jones J, Berggren W, Mitchen E, Frane J, Crandall L, Daigh C, Conard K, Piekarczyk M, Llanas R, Thomson J. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig T, Bergendahl V, Levenstein M, Yu J, Probasco MD, Thomson J. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 9.Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, Ding S. Proc Natl Acad Sci U S A. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enver T, Pera M, Peterson C, Andrews PW. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murry CE, Keller G. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O’Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman LM, Carpenter MK. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 15.Maherali N, Hochedlinger K. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Schulz TC, Sherrer ES, Dauphin DS, Shin S, Nelson AM, Ware CB, Zhan M, Song C, Chen X, Brimble SN, Mclean A, Galeano MJ, Uhl EW, D’Amour KA, Chesnut JD, Rao MS, Blau CA, Robins AJ. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furue MK, Na J, Jackson JP, Okamoto T, Jones M, Baker D, Hata R, Moore HD, Sato JD, Andrews PW. Proc Natl Acad Sci U S A. 2008;105:13409–13414. doi: 10.1073/pnas.0806136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu R, Peck R, Li D, Feng X, Ludwig T, Thomson J. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 19.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 20.van Noort D, Ong SM, Zhang C, Zhang S, Arooz T, Yu H. Biotechnol Prog. 2009;25:52–60. doi: 10.1002/btpr.171. [DOI] [PubMed] [Google Scholar]
- 21.Bhatia SN, Balis UJ, Yarmush ML, Toner M. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 22.Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Chem Biol. 2003;10:123–130. doi: 10.1016/s1074-5521(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 23.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe D. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 24.Warrick J, Meyvantsson I, Ju J, Beebe DJ. Lab Chip. 2007;7:316–321. doi: 10.1039/b613350a. [DOI] [PubMed] [Google Scholar]
- 25.Kamei K, Guo S, Yu Z, Takahashi H, Gschweng E, Suh C, Wang X, Tang J, Mclaughlin J, Witte ON, Lee K, Tseng H. Lab Chip. 2009;9:555–563. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]
- 26.Villa-Diaz LG, Torisawa Y, Uchida T, Ding J, Nogueira-de-Souza NC, O’Shea KS, Takayama S, Smith GD. Lab Chip. 2009;9:1749–1755. doi: 10.1039/b820380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwaka TP, Thomson JA. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 28.Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 29.Quackenbush J. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 30.Toepke MW, Beebe DJ. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 31.Solanki A, Kim JD, Lee KB. Nanomedicine. 2008;3:567–578. doi: 10.2217/17435889.3.4.567. [DOI] [PubMed] [Google Scholar]
- 32.The mTeSR medium is commercially available in the original condition with 100 ng mL−1 of zebrafish bFGF (zbFGF) to begin with.8
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.