Abstract
Purpose
We have previously shown that VHL regulates ubiquitylation and Proline P1465 hydroxylation of the large subunit of RNA polymerase II, Rpb1, in human RCC cell lines. Here, our goal was to determine the impact of this VHL function and the status of P1465 hydroxylation in human RCC tumors.
Experimental Design
Primary human tumors and matched normal kidney samples were probed for expression levels of the large subunit of RNA polymerase II (Rpb1), Rpb1 hydroxylated on P1465 (Rpb1(OH)), Rpb1 phosphorylated on Ser5 (Rpb1(S5P)), and proline hydroxylases PHD1, PHD2, and PHD3. Results from RCC tumors were categorized according to the status of VHL gene. Mechanistic analysis was performed in orthotopic xenograft model using 786-O RCC cells with wild-type VHL and knockdown of PHD2, characterized by high levels of Rpb1(OH) and PHD1.
Results
Levels of Rpb1(OH), PHD1, and PHD2 were significantly higher in RCC tumors as compared with normal kidneys. RCC tumors with wild-type VHL had higher levels of Rpb1(OH) and PHD1 and lower levels of PHD2 than tumors with VHL gene alterations. Levels of Rpb1(OH) significantly correlated with levels of PHD1 in tumors and in normal kidneys. Knockdown of PHD2 in 786-O VHL(+) cells resulted in a more malignant phenotype in orthotopic xenografts and higher expression of specific cell cycle regulators (CDC25A, CDK2, CCNA2), as compared with VHL(−) RCC cells.
Conclusions
Elevated PHD1 concomitant with decreased PHD2 are causatively related to Rpb1 hydroxylation and oncogenesis in human RCC tumors with wild-type VHL gene. Thus, P1465-hydroxylated Rpb1 and PHD1 represent attractive drug targets for new RCC treatments.
Keywords: proline hydroxylase, von Hippel-Lindau tumor suppressor, renal clear cell carcinoma, RNA Polymerase
Introduction
Renal clear cell carcinoma (RCC) is the most prevalent and malignant histological type of kidney cancer. The majority of RCC (60%–80%) is associated with intragenic mutations or hypermethylation of the von Hippel-Lindau tumor suppressor gene (VHL) and loss of heterozygosity at the VHL locus (3p26). In VHL-deficient tumors, activation of metabolic, angiogenic, and survival pathways resulting from VHL loss is an early and critical event in RCC tumorigenesis. In cases of RCC with the wild-type VHL gene, oncogenesis is not well understood. That is, it is not clear which activities of VHL are retained in those tumors.
VHL is a substrate-recognition molecule of an E3 ubiquitin ligase complex, recognizing hydroxylation of proline within the LxxLAP motif, which leads to ubiquitylation of the alpha subunits of hypoxia-inducible transcription factor (HIF) (1). This hydroxylation is mediated by two of the O2-, Fe(II)-, and oxyglutarate-regulated Egln9-type proline hydroxylases (PHDs), PHD2 and PHD3 (2). The function of the third enzyme from this group, PHD1, is less well understood but it is known to hydroxylate conserved LQYLAP motifs of IKKβ, and by doing so, inhibit activation of the NFκB transcription factor (3).
Importantly, we have discovered that VHL mediates ubiquitylation of the large subunit of RNA Polymerase II, Rpb1, in a manner dependent on the hydroxylation of proline 1465 (P1465) within the LGQLAP motif (4–8). P1465 is located near the C-terminal domain (CTD) of Rpb1 and is hydroxylated in response to oxidative stress, in a VHL- and PHD1-dependent manner. In contrast, PHD2 has an inhibitory effect on P1465 hydroxylation and its knockdown in VHL(+) cells induces high levels of hydroxylated and ubiquitylated Rpb1 (6). The hydroxylation and ubiquitylation of Rpb1 is also associated with phosphorylation of Rpb1 on serine 5 residues of the CTD. Most importantly, overexpression of wild-type Rpb1, but not Rpb1 mutated on P1465 (P1465A), in VHL(+) RCC cells stimulates the formation of tumors by orthotopic xenografts in nude mice. These data support an oncogenic role for P1465 hydroxylation in RCC (6).
The current study was undertaken to assess the clinical relevance of VHL- and PHD1-dependent P1465 hydroxylation of Rpb1. We determined that levels of P1465-hydroxylated Rpb1 were significantly elevated in a large population of human RCC tumors, as compared with normal kidneys, and that this parameter correlated strongly with levels of PHD1, supporting a role for PHD1 as the main hydroxylase of Rpb1. Importantly, we also demonstrated that distinct patterns of Rpb1(OH), PHD1, and PHD2 expression levels correlated with VHL gene status in these tumors and that recapitulation of these expression patterns in cultured cells led to increased malignancy in an orthotopic xenograft model and higher levels of protein expression for CDC25A, CDK2 and CCNA2. In summary, this study points to a new VHL-dependent mechanism in RCC oncogenesis and suggests that hydroxylation of Rpb1 and PHD1 activity could present novel therapeutic targets for the treatment of RCC.
Materials and Methods
Collection of human RCC tumor and kidney samples
Fresh-frozen samples of renal clear cell carcinomas (n=128) and matched normal kidneys (n=114) were purchased from NDRI (Philadelphia, PA) (50%), or obtained from the departments of pathology at the University of Cincinnati (35%) or Yale University (15%). In addition, four kidney-tumor pairs of papillary carcinoma samples and three such pairs of chromophobe cancer were also obtained. All samples were anonymous and therefore exempted from the need for an IRB protocol. Cryostat-cut sections of tumors were stained by hematoxylin and eosin to assure that the samples were from tumor regions consisting of more than 90% tumor cells. Two adjacent fragments of the tumor, not bigger than 3 to 5 mm in each dimension, were collected for DNA and protein analyses. While DNA was obtained from every tumor, sufficient tissue extract for analysis of all proteins of interest were obtained from 112 tumor and 95 kidney specimens.
DNA extraction and analysis of VHL gene status
DNA was extracted with DNAsol following standard manufacturer’s protocols (MRC, Cincinnati, OH). For sequence analysis of the VHL gene, DNA for each of the three coding exons was amplified by PCR by using previously published primers (9). Amplicon 1 included nucleotides 195 to 586 (exon 1 is from nucleotide 214 to 553), amplicon 2 included nucleotides 4846 to 5053 (exon 2 is from nucleotide 4880 to 5002), and amplicon 3 included nucleotides 8124 to 8373 (exon 3 is from nucleotides 8153 to 8331). Amplicons were purified by using the Millipore Montage-PCR Filter Unit (Fisher Scientific, Pittsburgh, PA) and then sequenced. The methylation status of the VHL promoter was examined by sodium bisulfite modification and methylation-specific PCR. Genomic DNA (0.3 to 0.5 μg) from tumor tissues was subjected to bisulfite modification via the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA). Methylation-specific PCR was performed by using specific primers designed to amplify either methylated or unmethylated VHL promoter sequences, as described previously (10).
Preparation of tumor protein extracts and western blot analysis
Total tissue lysates were prepared by using a previously described protocol (6). The protein extracts from all tissues were stored as concentrated stocks of known protein concentration. Before PAGE analysis of tumor and kidney samples, stocks were diluted, and protein concentration was measured in all samples again and determined to be in the range of 4 to 7 μg/μl per sample. Aliquots of all samples, corresponding to 15 or 50 μg of protein, were denatured by boiling in SDS buffer and then processed by PAGE. For immunoblotting, all blots (between 7 and 12) were cut, and strips containing proteins in the appropriate size ranges were processed at the same time in the same solutions for each antibody and developed together on the same film. Blots were probed, as we described before (6), with the following antibodies: H14, which detects Rpb1 phosphorylated on serine 5 residues; 8WG16, which detect all forms of Rpb1 (Covance, Princeton, NJ); anti-pVHL (BD Pharmingen, San Jose, CA); anti-PHD1/2/3 (Novus, Littleton, CO); anti-CDC25A (Santa Cruz, Santa Cruz, CA)and anti-GAPDH (Abcam, Cambridge, MA). The antibody against hydroxylated proline within the Rpb1 peptide was previously characterized by our laboratory (6). Secondary antibodies were obtained from Sigma (St. Louis, MD), Cell Signaling (Danvers, MA), or Amersham (Piscataway, NJ). The relative level of the proteins of interest was determined by scanning the blots with an Epson Perfection 4990 Photo (Epson, Japan) scanner and measuring the optical density of protein bands using Image Quant 5.2 software. Care was taken to insure that all the scanned bands were in the linear range of the optical density. To compensate for systematic technical differences between gels, we ran a set of four to six dilutions of the same standard extract from 786-O VHL(−) cells on each gel with specimen extracts. For measurements of pVHL levels in the kidney extracts, the standard extracts were from 786-O VHL(+) cells. The amount of a protein in each band, represented by optical density, was normalized to the standards in the linear range run on each gel and to the level of GAPDH accepted as a loading control (Fig. S1). All samples were analyzed by PAGE and immunoblotting, and all indicated proteins were quantified in at least two independent experiments. All the outliers identified in the statistical analysis were additionally rerun and re-quantified as described above.
Data analysis
Only tumors and kidneys for which we were able to obtain measurements for all proteins of interest were used for statistical evaluation (n=95 for kidneys, n=112 for tumors). The normalized values were log2 transformed to facilitate robust, outlier-insensitive statistical and discriminatory analyses. Standard box-and-whisker plots were used to compare distributions of the normalized abundance measure for individual proteins. Log2 levels of each protein were analyzed by weighted least squares regression and t-test to investigate differences between mean levels in kidneys and tumors. For each protein, both paired and unpaired analyses were performed with essentially the same result. Correlations between variables were assessed using regression analysis as well as by Pearson and Spearman rank correlation coefficients. The ability to discriminate between VHL-positive (WT) and VHL-negative (DEL) tumors was further assessed by using standard decision trees, C4.5 and CART, as implemented in Weka and R packages respectively (11–13). Accuracy, generalization, and consistency of decision trees was estimated by using multiple (100) runs of 5-fold and 10-fold cross-validation using Weka, and 5 runs of 10-fold cross-validation using the rpart function in R with explicit separation of training and control sets to ensure that feature selection was done with training subsets only. The selection of top features, optimal values of decision cutoffs, and the resulting strata, were largely insensitive to the choice of data subsets, yielding an average classification accuracy of about 80% in cross-validation.
Cell line experiments
Human RCC cell lines, pools of 786-O VHL(−) or VHL(+), with and without knockdown of PHD2, were developed and cultured as described before (3–7). Preparation of the lysates, western blot analysis of cellular extracts, and injections of cells into the kidneys of nude mice were performed as described before (6). Xenograft tumors, along with the invaded kidney, were collected at 12 weeks post-injection and weighed. Sections were prepared for immunocytochemistry, which was performed using antibodies against endothelial cell marker Cd31 (BD Pharmingen) or proliferation marker Ki67 (NeoMarkers, Fremont, CA), followed by secondary biotinylated antibody (M.O.M. kit, Vector Laboratories, Burlingame, CA) and ABC reagent (Vector Laboratories). Positive staining was detected with DAB substrate (Sigma) and sections were counterstained with hematoxylin. Fresh tumors were homogenized in TRI-Reagent for subsequent RNA extraction and analysis; frozen tumors were used for preparation of total or nuclear/chromatin enriched lysates (6) for immunoblotting. Antibodies against CDC25a, CDC25b, CDK2, and Cyclin A were purchased from Cell Signaling. All image quantification was performed as described above. For analysis of mRNA levels, equal fractions of RNA were employed for first-strand cDNA synthesis using a SuperScriptR III First Strand Synthesis Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Sequences of primers used for amplification of VEGF-A, -B, -C and Glut-1 are shown in Table S1. Amplifications were performed using a standard built-in manufacturer’s protocol with the ABI7900HT thermocycler (Applied Biosystems, Carlsbad, CA). Averaged data are expressed as mean ± standard error of the mean (SEM) of all independent experiments. The statistical analysis of the values was performed using one-way ANOVA with post-tests (*P<0.5; **P<0.001; ***P<0.001). All procedures involving animals were approved by the IACUC of the University of Cincinnati and followed guidelines provided by NIH.
Results and Discussion
Correlation of Rpb1 hydroxylation with prolyl hydroxylase levels and VHL gene status in human RCC tumor specimens
We analyzed a total of 128 human RCC tumors for VHL gene status. 36% of these tumors (46/128) had the wild-type VHL gene, 41% (52/128) had frameshift and nonsense mutations, 10% (13/128) were hypermethylated, and 13% (17/128) of tumors had missense point mutations (Table S2 and Table S3). 112 tumor and 95 kidney samples were used for the analysis of normalized protein levels as they showed measurable levels of all proteins of interest in the extracts (Table S4).
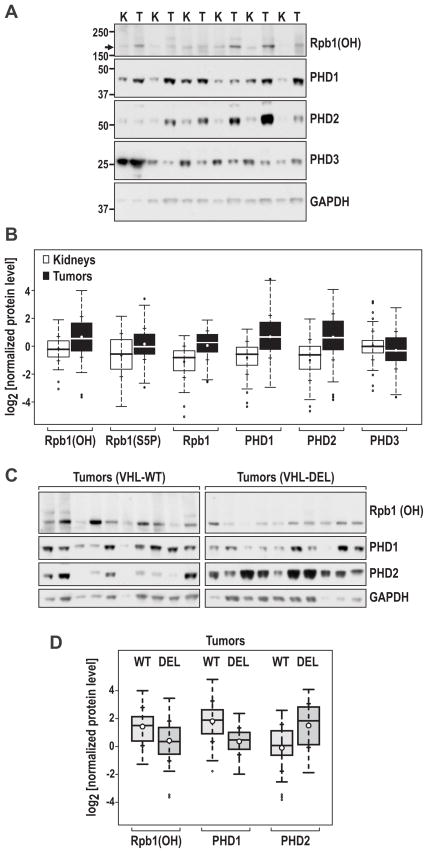
Initial analysis revealed that levels of all three forms of Rpb1, P1465-hydroxylated, Serine-5-phosphorylated, and total, were increased in tumors as compared with normal kidneys (Fig. 1A and 1B). Similarly, levels of both PHD1 and PHD2 were significantly increased in tumors as compared with kidneys; however, the PHD3 levels were decreased at borderline significance (Fig. 1A and B). For each protein level, the tumor mean was higher than the kidney mean, with a similar level of significance, using both unpaired and paired analyses. These results are likely to be specific for RCC tumors, as analysis of a small number of tumor-kidney pairs of papillary and chromophobe renal cancer did not indicate any trends corresponding to the differences in Rpb1(OH), PHD1, and PHD2 measured in RCC tumors (Fig. S2). However, to verify this finding, a larger sample of non-RCC tumors will need to be analyzed in the future.
Figure 1. VHL-regulated induction of Rpb(OH), PHD1, and PHD2 in human RCC tumors.
A Examples of western blots of matched kidney-tumor pairs to detect the indicated proteins. B. Quantification of the indicated normalized protein levels (log2) of the total unpaired kidney and tumor samples using box-and-whisker plots. The boxes represent lower and upper quartiles separated by the median (thick horizontal line), and the whiskers extend up to the minimum and maximum values, excluding points that are considered to be outliers, i.e., those outside the 1.5 interquartile range from the box (outliers are marked as circles). Means and standard deviations of each distribution are indicated by dots and crosses on the whiskers, respectively. All differences were statistically significant at p<0.001, except for PHD3, where the difference was statistically significant at p< 0.04. C. Representative western blots for indicated proteins in the example set of 10 tumors with VHL-WT and 10 tumors with VHL-DEL. D. Box-and-whisker plots of Rpb1(OH), PHD1, and PHD2 levels in RCC tumors stratified by VHL gene status (WT vs. DEL). The differences in the levels of all three proteins were statistically significant at p<0.001.
Stratification of tumors by VHL gene status revealed significantly larger increases in Rpb1(OH) and PHD1, but relatively smaller increases in PHD2 as compared with normal kidneys in tumors with the wild-type VHL gene (VHL-WT, Fig. 1C and 1D). On the other hand, relatively lower levels of Rpb1(OH) and PHD1 (although still above levels measured in normal kidneys) and significantly higher levels of PHD2 were observed in tumors with lost VHL gene (VHL-DEL, Fig. 1C and 1D). Histograms of the distributions of the levels of Rpb1(OH), PHD1 and PHD2 in VHL-WT and VHL-DEL tumors further support the idea that VHL-WT and -DEL tumors can be separated relatively well based on the levels of these three proteins, although the separation is most clear for PHD1 (Fig. S3). The levels of other forms of Rpb1 and PHD3 did not differ in tumors on the basis of VHL gene status (data not shown). Thus, these data indicate that, while high levels of Rpb1(OH) contribute to RCC oncogenesis independently of the status of the VHL gene, there are VHL-dependent mechanisms that specifically allow for higher levels of Rpb1(OH) and a potentially higher contribution of hydroxylated Rpb1 to the oncogenesis in tumors with an intact VHL gene.
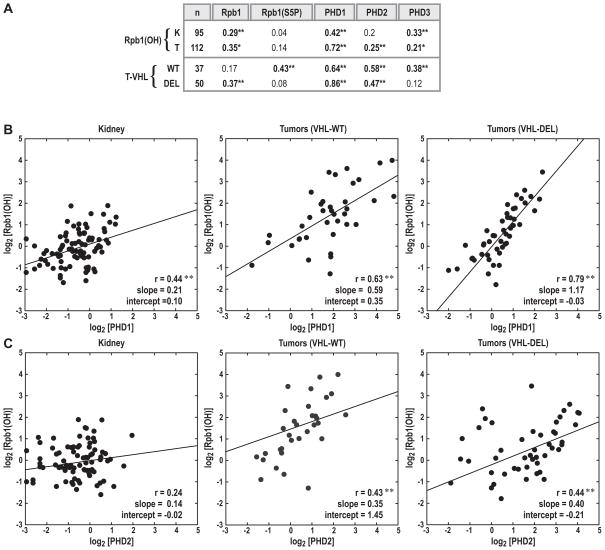
Our previous experimental work demonstrated that PHD1 and PHD3 hydroxylated Rpb1, with PHD1 being the main hydroxylase (6). In contrast, PHD2 had an inhibitory effect on hydroxylation as its knockdown resulted in increased levels of Rpb1(OH) due to the activity of PHD3 (6). Here we sought to assess the correlations between Rpb1(OH) and PHDs in different groups of samples by using regression analysis. Pearson and Spearman rank correlation coefficients were computed, yielding very similar results (Fig. 2). In normal kidneys, consistent with our previous findings, levels of Rpb1(OH) correlated with levels of PHD1 and PHD3, but not with levels of PHD2 (Fig. 2). Similar analysis of the tumor samples revealed a much stronger correlation between levels of Rpb1(OH) and PHD1, compared with that observed for these two proteins in kidney, and a similar correlation to that in kidneys with regard to Rpb1(OH) and PHD3 levels. However, a significant correlation between Rpb1(OH) levels and PHD2 was also found (Fig. 2). When tumors were stratified by VHL gene status, in the VHL-WT tumors, Rpb1(OH) levels correlated with the levels of all three PHDs, similar to the unstratified population of tumors. However, in the case of VHL-DEL tumors, levels of Rpb1(OH) correlated only with levels of PHD1 and PHD2, with a particularly high correlation with PHD1 in this group of tumors (Fig. 2A, and 2B), but the correlation with PHD3 level was lost. This strong correlation between Rpb1(OH) and PHD1 in VHL-DEL tumors (Pearson correlation coefficient of 0.79) indicates that PHD1 alone explains approximately 65% of variance in Rpb1(OH) levels, further supporting PHD1 as the major hydroxylase of Rpb1 in these tumors. Thus, even though the levels of both Rpb1(OH) and PHD1 were lower in this group of tumors, as compared with VHL-WT tumors, the correlation between these two factors was much stronger. Overall, these data support our previous experimental evidence that PHD1 is the main and direct hydroxylase for Rpb1, while PHD3 plays an essential auxiliary role, particularly under conditions of relatively low levels of PHD2, as in VHL-WT tumors. Observation that high levels of PHD2, as measured in VHL-DEL tumors, coincide with lost correlations between Rpb1(OH) and PHD3, and overall lower Rpb1 hydroxylation as compared with VHL-WT tumors, also supports our previous experimental data (6). Here we propose that PHD2 may not hydroxylate Rpb1 directly, but that it primarily influences its hydroxylation by PHD1 and PHD3 via protein-protein interactions (Fig. S4). Previous biochemical analysis of the PHD protein complexes demonstrated that PHD3 can heterodimerize with the other two PHDs (14). Thus, one potential model is that higher levels of PHD2, as in VHL-DEL tumors, titrate PHD3 away from Rpb1, leaving PHD1 as the only hydroxylase and resulting in an overall lower, but primarily PHD1-mediated, Rpb1 hydroxylation in VHL-DEL tumors as compared with VHL-WT tumors (Fig. S4).
Figure 2. Levels of Rpb1(OH) show the strongest and most uniform correlation with levels of PHD1 in all investigated populations of kidneys and tumors.
The analysis involved parametric and non-parametric methods of correlation using Log2 values of normalized protein levels. A. Table showing Spearman correlation coefficients between levels of indicated proteins in kidneys (K) and tumors (T), or for the subpopulation of tumors with VHL WT or DEL. ** p<0.01; * p<0.05. Regression analysis of Rpb1(OH) levels as a function of PHD1 (B) and PHD2 (C) levels in kidney and tumor samples stratified by VHL gene status. Each dot represents a sample and the solid line represents the linear regression fit, with the Pearson correlation coefficients (r) as well as the slope and intercept of the fitted line shown in the lower right corner of each box. ** p<0.01
Importantly, we found an overall positive correlation between Rpb1(OH) and total Rpb1 levels in normal kidneys and tumors, but not with Rpb1 phosphorylated on Ser5. The only exceptions were VHL-WT tumors, where levels of Rpb1(OH) were positively correlated with levels of Rpb1 phosphorylated on Ser5 (Fig. 2A). This observation is consistent with our cell-culture observation that hydroxylation and phosphorylation of Rpb1 were co-regulated in VHL(+) RCC cells (6). We did not detect any meaningful correlations between protein levels in subsets of samples stratified according to RCC tumor grade (data not shown).
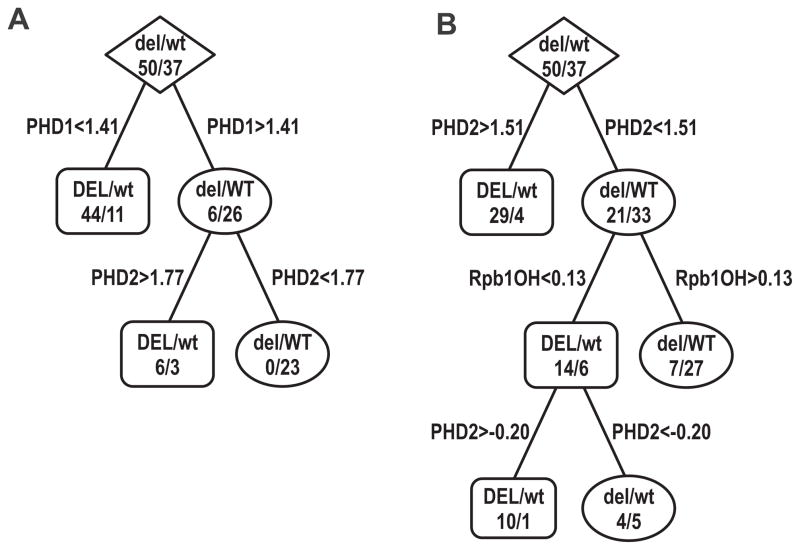
The above univariate analysis suggested that relatively good separation between the types of tumors could be achieved by using levels of PHD1, or to a lesser degree by levels of PHD2 or Rpb1(OH); however, there was some overlap between VHL-WT and -DEL tumors (Fig. S3). Thus, we used recursive partitioning with C4.5 and CART decision trees to independently determine if the stratification of tumors on the basis of distinct expression patterns of the proteins investigated in this study could also group tumors according to VHL status in a multivariate approach. We performed multiple runs of cross-validation with randomly selected subsets of the data used for training and the remaining data in each run used only for validation. We found that levels of PHD1, Rpb1(OH), and PHD2 changed in a coordinate fashion dependent on VHL status (Fig. 3A). In particular, according to CART-optimized decision tree analysis, low levels of PHD1 (<1.41 in normalized log2 transformed values) were sufficient to classify most (44 out of 50) RCC tumors with VHL-DEL status, while misclassifying 11 WT tumors (Fig. 3A). Importantly, 31 out of these 44 tumors also had relatively low levels of Rpb1(OH), defined as the median Rpb1(OH) expression among all analyzed tumors. In contrast, VHL-WT tumors were characterized by high levels of PHD1 (>1.41) and low levels of PHD2 (<1.77) (Fig. 3A). The majority of VHL-WT samples and none of the VHL-DEL samples satisfied these conditions. Consistent with the separation of tumors observed in univariate analysis (Fig. S3), PHD1 was always the top feature with an average decision cutoff of 1.41 and standard deviation of 0.23 in a set of 50 CART decision trees optimized using different subsets of the data in 5 runs of 10-fold cross-validation. We also performed discriminatory analysis without taking PHD1 into account. In this case, a high PHD2 level (>1.51) was sufficient to identify most of the VHL-DEL tumors, while the rest of the VHL-DEL tumors were characterized by low Rpb1(OH) (Fig. 3B). In contrast, VHL-WT tumors were characterized by relatively high Rpb1(OH) in conjunction with low PHD2 levels (Fig. 3B). Stratification of the samples was largely insensitive to sampling of data subsets, i.e., in an alternative set of 50 decision trees, PHD2 was always the top feature, with a mean cutoff value of 1.52 and a standard deviation of 0.07. Thus, this analysis further supports the conclusion that high levels of PHD1 and Rpb1(OH) with low levels of PHD2 occur in VHL-WT tumors, whereas low levels of PHD1 and Rpb1(OH) with high levels of PHD2 occur in VHL-DEL tumors.
Figure 3. CART-optimized decision trees for the discrimination and stratification of RCC tumors with the wild-type VHL gene (WT) vs. tumors with a nonfunctional VHL gene (DEL).
A. The level of PHD1 was the strongest discriminating factor for the entire population of RCCs, and was consistently found to be so in multiple runs of cross-validation. B. Exclusion of PHD1 from the analysis revealed that the levels of PHD2 were consistently the top discriminating feature in multiple runs of cross-validation with the CART and C4.5 algorithms. Levels of Rpb1(OH) became the second most discriminating feature, with lower values occurring in VHL-DEL tumors and higher values in VHL-WT tumors.
Overall, these data emphasize the role of VHL in modulation of Rpb1 hydroxylation by a set of three prolyl hydroxylases and further support previous experimental evidence that a high level of Rpb1(OH) is associated with RCC oncogenesis, particularly in VHL-WT tumors.
Identification and characterization of a cell-line model system that recapitulates high Rpb1(OH)/PHD1 and low PHD2 levels in the presence of wild-type VHL
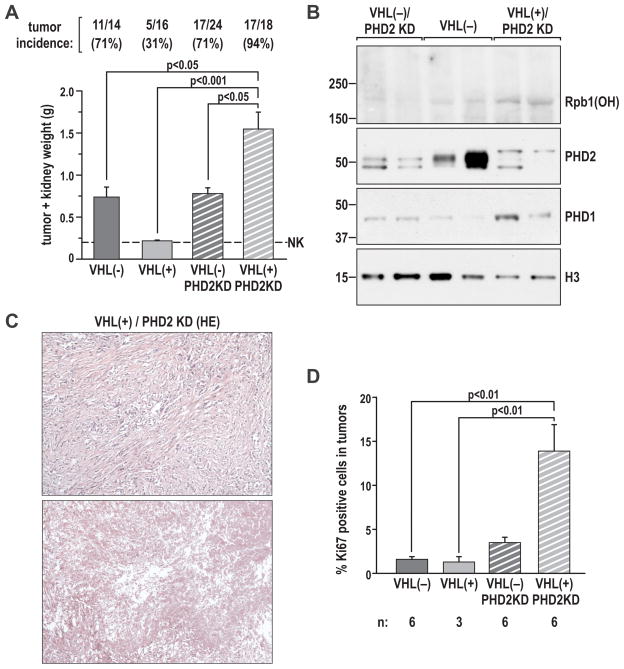
In view of our intriguing findings showing that VHL-WT tumors are characterized by relatively high levels of Rpb1(OH) and PHD1 and low levels of PHD2 as compared to VHL-DEL tumors (Fig. S4A), we wanted to determine if similar coexisting features could be recapitulated in a model using RCC cells, and, if so, how they would affect tumor formation. In our previous work we observed that efficient knockdown of PHD2 (PHD2KD) in 786-O VHL(+) RCC cells resulted in a robust upregulation of constitutive levels of Rpb1 hydroxylated on P1465 and phosphorylated on Ser5, which correlated with some enrichment for PHD1 and PHD3 in the chromatin fraction (6). Thus, we hypothesized that these cells may represent an experimental system to model the aforementioned group of VHL(+) tumors. To investigate their tumorigenic potential, we injected 786-O VHL(+)/PHD2KD cells into the kidneys of nude mice. We found that these cells formed large and highly malignant tumors in nearly 100% of cases, which was not only significantly more than 786-O VHL(+) RCC cells, which form very small tumors in about 20% to 30% of mice, but also more than VHL(−) cells (Fig. 4A). These VHL(+)/PHD2KD tumors had, as expected, high levels of Rpb1(OH) and PHD1 (Fig. 4B), showed a sarcomatoid malignant phenotype and large areas of necrosis (Fig. 4C), and were characterized by high levels of the proliferation marker Ki67 (Fig. 4D). In contrast, a comparable knockdown of PHD2 in VHL(−) RCC cells was without any effect on Rpb1 hydroxylation (Fig. 4B and (6)), or formation of tumors in nude mice as compared to VHL(−) cells (Fig. 4A). These data indicate that, in the presence of VHL, downregulation of PHD2 promotes tumor formation with a highly malignant phenotype, and it is possible that the upregulation of Rpb1(OH) plays an important role in this process.
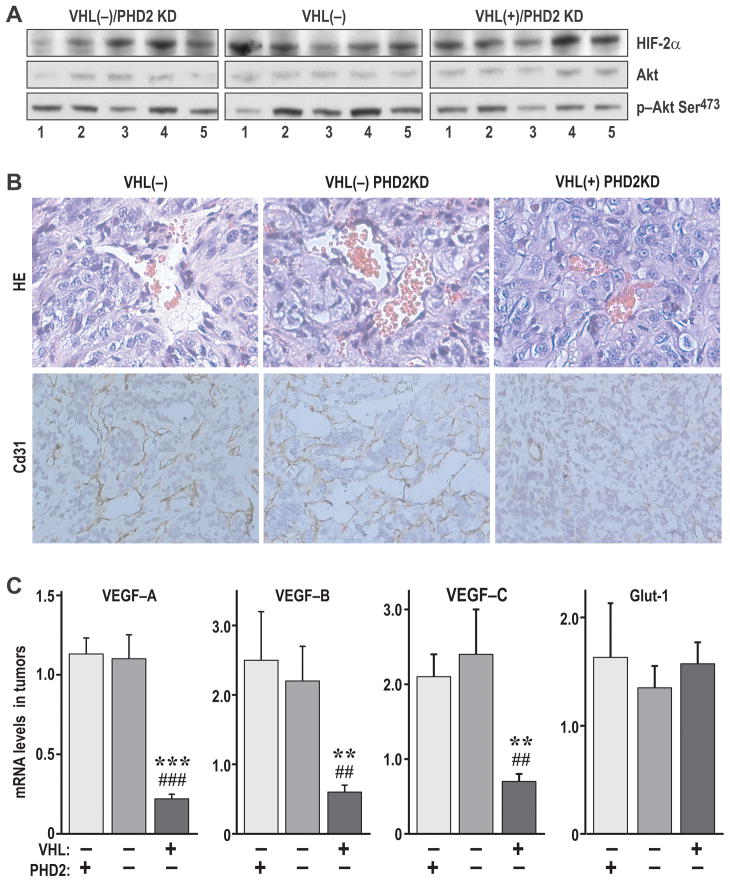
Figure 4. Knockdown of PHD2 in VHL(+) but not VHL(−) RCC tumors induces formation and growth of RCC tumors in orthotopic xenografts.
A. Incidence and weight of tumors formed in nude mice by 786-O cell lines with the indicated modifications. The number of mice with tumors, as compared to the total number of injected mice, is shown above the graph. The graph shows the average values of tumor weights (including the kidney) calculated as mean±SEM for all mice that had tumors. The overall significance of the differences among the different groups, calculated by non-parametric ANOVA, was P<0.0001. The P values for relevant post-tests are provided in the graph. NK, average weight of a normal mouse kidney. B. Representative western blot of two tumors from each group to detect Rpb1(OH), PHD1 and PHD2. Nuclear extracts enriched for the chromatin fraction from tumor tissues were used. The blot shows increases in Rpb1(OH) and PHD1 in tumors formed by cells with PHD2 knockdown, similar to what has previously been shown in the corresponding cell lines (6). Samples from the PHD2KD tumors show double bands, which represent either mouse PHD2 or incompletely knocked down human PHD2. C. H&E staining of representative sections of a tumor without necrosis (top) and one with a large area of necrosis (bottom). D. Quantification of the proliferation index (by Ki67 staining) in the indicated types of tumors.
Knockdown of PHD2 also induces accumulation of HIF-2α, and overexpression of HIF-2α mutated on its proline hydroxylation sites promotes formation of tumors by VHL(+) RCC cells, which normally have suppressed ability to form tumors as compared to isogenic VHL(−) cells (15,16). While knockdown of PHD2 resulted in the expected accumulation of HIF-2α in VHL(+) cells, this accumulation was comparable to that measured in VHL(−) and VHL(−)/PHD2KD cells (Fig. S5). Consistent with this, we did not detect any differences in HIF-2α protein levels among tumors formed by VHL(−), VHL(−)/PHD2KD, or VHL(+)/PHD2KD cells (Fig. 5A). Most importantly, we found that the VHL(+)/PHD2KD tumors were significantly less well vascularized as compared with VHL(−)/PHD2KD or VHL(−) tumors (Fig. 5B). Moreover, despite similar levels of HIF-2α protein expression, mRNAs for all three VEGFs (A,B,C) were significantly lower, as compared with VHL(−) or VHL(−)/PHD2KD tumors, even though the mRNA for another HIF target, GLUT1, was expressed at the same level in all the tumor types (Fig. 5C). These data indicate that, while accumulated HIF may contribute similarly to the regulation of metabolism in all three investigated cell lines, the HIF-induced angiogenic phenotype does not play a leading role in the enhanced growth of VHL(+)/PHD2KD tumors as compared with that of VHL(−) or VHL(−)/PHD2KD tumors. Similarly, as shown in Fig. 5A, we did not measure any differences in the levels of total or phosphorylated Akt among the tumors formed by the three different cell lines, an indication that this pathway was not induced by PHD2 knockdown, and does not play a role in the excessive growth of tumors formed by VHL(+)/PHD2KD cells as compared with tumors formed by VHL(−) or VHL(−)/PHD2KD cells. Note that the material obtained from very small VHL(+) tumors, when formed, was insufficient for any molecular analysis.
Figure 5. Growth of VHL(+)/PHDKD RCC tumors in orthotopic xenografts does not result from increased HIF activity or angiogenesis.
A. Western blot analysis of the indicated proteins in total extracts obtained from individual tumors formed by VHL(−), VHL(−)/PHD2KD, and VHL(+)PHD2KD cells. HIF-2α protein levels are very similar in tumors derived from each cell line. Likewise, levels of Akt and phosphorylated Akt do not differ among these tumors. Note that these are the same tumors as shown in Fig. 6A. All tumor extracts were run on the same gel and the image was cut to better visualize the different groups of tumors. B. Immunocytochemistry for cd31 on representative sections from the indicated tumors indicates lower angiogenesis in VHL(+)/PHD2KD tumors as compared with VHL(−) or VHL(−)/PHD2KD tumors. C. RT-PCR of several targets of HIF: VEGF A, B, C; and GLUT-1. GLUT-1 levels are similar in all three groups of tumors, corresponding to the similar levels of HIF-2α. However, levels of all three VEGFs were significantly reduced in VHL(+)/PHD2KD mice as compared with the VHL(−) (*) or VHL(−)/PHD2KD (#) tumors. P values were computed as described for Fig. 4A. These results indicate that factors other then HIF regulate expression of VEGF in these tumors. Moreover, low levels of VEGFs correspond to poor vascularization and a high level of necrosis in these tumors.
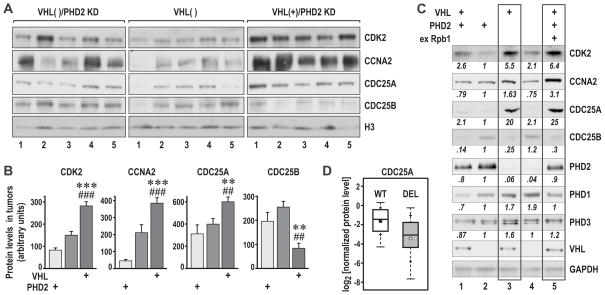
Because the main feature of the VHL(+)/PHD2KD tumors was highly augmented cell proliferation, we investigated the effects of PHD2KD on expression levels of several cell-cycle regulators. In that respect, we found that tumors formed by VHL(+)/PHD2KD cells showed higher levels of cyclin A (CCNA2), CDK2, and CDC25A, but lower levels of CDC25B, as compared to tumors formed by VHL(−) cells (Fig. 6A and 6B). In contrast, tumors formed by VHL(−)/PHD2KD cells, showed smaller increases in the levels of CCNA2 and CDK2, and no change in CDC25A or CDC25B expression as compared to VHL(−) tumors (Fig. 6A and 6B).
Figure 6. Analysis of proliferation markers in RCC tumors formed by VHL(−) or VHL(+)/PHD2KD cells.
A. Western blot analysis of the indicated proteins in sets of tumors formed by VHL(−), VHL(−)/PHD2KD, and VHL(+)PHD2KD cells as in Fig. 5A. All tumors were run on the same gel and the image was cut to better visualize individual groups of tumors. B. Quantification of the normalized data. Comparison between VHL(+)PHD2KD and VHL(−) is marked by (*), and between VHL(+)PHD2KD and VHL(−)/PHD2KD by (#). C. Western blot analysis of the indicated proteins in cellular extracts from the indicated original cell lines used in the xenograft experiment. Rpb1, exogenously expressed Rpb1. Boxes mark lanes comparing indicated proteins in VHL(+) 786-O cells with knockdown of PHD2 or overexpression of exogenous Rpb1. D. Box-and-whisker plots of CDC25A levels in RCC tumors stratified by VHL gene status (WT vs. DEL). The difference in the levels was statistically significant at p<0.0001.
Further analysis of RCC cell lines confirmed observations from the orthotopic xenografts (Fig. 6C). In particular, 786-O VHL(+) RCC cells expressed higher levels of CDK2 and CDC25A as compared to VHL(−) cells (Fig. 6C, compare lanes 1 and 2). In contrast, 786-O VHL(−) cells expressed higher levels of CDC25B. Knockdown of PHD2 in 786-O VHL(+) cells further induced expression of CDK2, CDC25A, and CCNA2, an effect that was also seen upon overexpression of wild-type Rpb1 in VHL(+) cells (compare lanes 1, 3, and 5 in Fig. 6C). Knockdown of PHD2 in VHL(−) RCC cells had only a small effect on expression of CDK2, but was without effect on CCNA2 or CDC25A. Also note that knockdown of PHD2 in both VHL(−) and VHL(+) RCC cells resulted in increased expression of PHD1, and in VHL(+) RCC cells, PHD3. These data show that the reduced level of PHD2 in VHL(+) RCC, which is associated with increased hydroxylation of Rpb1(6), has effects similar to those of Rpb1 overexpression, both resulting in increased expression of three cell-cycle regulators: CDK2, CCNA2 and CDC25A (Fig. 6C, lanes 3 and 5), and augmented tumor growth by VHL(+)/PHD2KD cells (Fig. 4) or by VHL(+)/Rpb1 cells (6). This VHL-dependent switch from expression of CDC25B to CDC25A, and the increase in CDC25A expression under conditions of augmented Rpb1 hydroxylation on P1465 are particularly intriguing, implicating Rpb1 hydroxylation in the stimulation of CDC25A and/or inhibition of CDC25B expression.
CDC25 phosphatases regulate key transitions between cell cycle phases. Predominantly, CDC25A functions to accelerate G1-S transition but also plays a role in G2-M transition, while CDC25B is required for entry into mitosis. Endogenous CDC25 phosphatases are overexpressed in several human cancers; they contribute to the growth of different tumors and are targets for anticancer therapies (17). Because the induction of CDC25A was particularly strongly associated with elevated levels of Rpb1(OH) in RCC cells, we analyzed CDC25A expression in VHL-WT and VHL-DEL human RCC tumors. Protein levels of CDC25A were significantly higher in VHL-WT tumors as compared to VHL-DEL tumors (Fig. 6D and Fig. S6). Moreover, there was a significant correlation between the levels of CDC25A and Rpb1(OH), however, only in the VHL-DEL tumors, which had overall lower levels of CDC25A and Rpb1(OH), but not in the VHL-WT tumors, which had higher levels of both proteins (Fig. S7). This result is reminiscent of the stronger correlation in the levels of Rpb1(OH) and PHD1 in VHL-DEL as compared to VHL-WT tumors (Fig. 2), and indicates that in the VHL-WT tumors, mechanisms additional to Rpb1(OH) positively regulate the expression of CDC25A. In contrast, in the VHL-DEL tumors, the levels of CDC25A depend more on the levels of hydroxylated Rpb1. At this point, we do not yet understand how Rpb1(OH) regulates expression of CDC25A. One possibility is that hydroxylation of Rpb1 may directly affect transcription and/or translation of CDC25A. Another possibility is that this regulation is indirect, and hydroxylation affects other cell cycle regulatory mechanisms which in turn affect expression of CDC25A. Importantly, this is the first insight into the molecular mechanism by which elevated hydroxylation of Rpb1 exercises pro-oncogenic activities in RCC tumors. This is also the first demonstration of higher expression of CDC25A in VHL-WT vs. VHL-DEL RCC tumors.
Overall, these results contribute to an enhanced understanding of RCC tumors that have the wild-type VHL gene. These tumors have not been extensively investigated. However, several reports point to a more malignant character and poorer survival of patients without VHL gene alterations (18–21), although these tumors do not have elevated levels of HIFs and are less vascularized as compared to tumors expressing high levels of HIFs (22). Similarly, RCC tumors associated with genetic VHL disease, where VHL loss follows the classic Knudsen two-hit model for tumor suppressors, have a much better prognosis and lower metastatic potential, despite occurrence much earlier in life and multifocal disease (23). Our data demonstrate that the presence of VHL enables hydroxylation of Rpb1 and expression of certain proliferation genes, such as CDC25A, and plays an oncogenic role rather than that of a tumor suppressor. Rpb1 hydroxylated on P1465 likely exercises different transcriptional activities as compared with the non-hydroxylated form, resulting in different patterns of gene expression that are potentially associated with more aggressive oncogenesis. This is in accordance with our previously published data showing different patterns of proteins induced in VHL(+)/PHD2KD vs. VHL(−) RCC cells (8). Moreover, VHL has been reported to have growth-promoting effects during development (24) and during formation of teratomas or fibrosarcomas by VHL(+/−) vs. VHL(−/−) cells (25,26). It remains to be determined if any of these effects of VHL are mediated through regulation of Rpb1 hydroxylation.
Our data support the need for the development of inhibitors specific for the individual PHDs, with the expectation that a specific inhibitor of PHD1 could be used in the treatment of RCC carrying the WT VHL gene.
Statement of Translational Relevance.
Renal clear cell carcinoma (RCC) is the most frequent and malignant form of kidney cancer. Most RCC is associated with loss of the von Hippel-Lindau tumor suppressor (VHL), while in 20% to 30% of tumors the wild-typeVHLgene is maintained. We previously reported that VHL regulates coordinated ubiquitylation and proline P1465 hydroxylation of the large subunit of RNA Polymerase II, Rpb1, in RCC cell lines. Here we determined that tumors with wild-type VHL and tumors withVHLgene loss have distinct patterns of P1465 hydroxylation and protein levels for two proline hydroxylases, PHD1 and PHD2. We also found that P1465 hydroxylation is significantly increased in human RCC tumors as compared with normal kidneys, and that PHD1 serves as the main hydroxylase for Rpb1. Thus, PHD1 and P1465-hydroxylated Rpb1 may represent drug targets for new RCC treatments.
Supplementary Material
Acknowledgments
This work was supported in part by the following grants: NCI CA122346, DoD W81XWH-07-02-0026 (to M.F.C-K) and P30-ES006096 to the Center for Environmental Genetics at UC. We thank Karen Winstead for the collection of human RCC tumors at UC, Zhengyuan Shan for the histological preparation of tumor sections, G. Doerman for preparing the figures, and Dr. M. Daston for editorial assistance. Part of the work presented in the manuscript has been submitted as patent application #107 38-83.
References
- 1.Kaelin WG., Jr Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–82. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 2.Fraisl P, Aragonés J, Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Disc. 2009;8:139–52. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 3.Cummins EP, Berra E, Comerford KM, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappa B activity. Proc Natl Acad Sci USA. 2006;103:18154–59. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuznetsova AV, Meller J, Schnell PO, et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA. 2003;100:2706–2711. doi: 10.1073/pnas.0436037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czyzyk-Krzeska MF, Meller J. Von Hippel-Lindau tumor suppressor complex: not only HIF’s executioner. Trends Mol Med. 2004;10:146–49. doi: 10.1016/j.molmed.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Mikhaylova O, Ignacak ML, Barankiewicz TJ, et al. The von Hippel-Lindau tumor suppressor protein and Egl 9-type proline hydroxylases regulate the large subunit of RNA Polymerase II in response to oxidative stress. Mol Cell Biol. 2008;28:2701–17. doi: 10.1128/MCB.01231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ignacak ML, Harbaugh SV, Dayyat E, et al. Intermittent hypoxia regulates RNA polymerase II in hippocampus and prefrontal cortex. Neuroscience. 2009;158:1436–45. doi: 10.1016/j.neuroscience.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haffey WD, Mikhaylova O, Meller J, Yi Y, Greis KD, Czyzyk-Krzeska MF. iTRAQ proteomic identification of pVHL-dependent and -independent targets of Egln1 prolyl hydroxylase knockdown in renal carcinoma cells. Adv Enzyme Reg. 2009;49:121–32. doi: 10.1016/j.advenzreg.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Houwelingen KP, van Dijk BAC, Hulsbergen-van de Kaa CA, et al. Prevalence of von Hippel-Lindau gene mutations in sporadic renal cell carcinoma: results from the Netherlands cohort study. BMC Cancer. 2005;5:57. doi: 10.1186/1471-2407-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banks RE, Tirukonda P, Taylor C, et al. Genetic and Epigenetic Analysis of von Hippel-Lindau (VHL) gene Alterations and Relationship with Clinical Variables in Sporadic Renal Cancer. Cancer Res. 2006;66:2000–11. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 11.Therneau TM, Atkinson B. R port by Brian Ripley, rpart: Recursive Partitioning. 2009 http://CRAN.R-project.org/package=rpart.
- 12.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update. SIGKDD Explorations. 2009;11:1. [Google Scholar]
- 13.Quinlan JR. C4.5: Programs for Machine Learning. Morgan Kaufmann Publishers; 1993. [Google Scholar]
- 14.Nakayama K, Gazdoiu S, Abraham R, Pan ZQ, Ronai Z. Hypoxia-induced assembly of prolyl hydroxylase PHD3 into complexes: implications for its activity and susceptibility for degradation by the E3 ligase Siah2. Biochem J. 2007;401:217–26. doi: 10.1042/BJ20061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 16.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2-alpha is sufficient to suppress pVHL-defective tumor growth. Plos Biol. 2003:439–44. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutros R, Lobjois V, Ducommun B. CDC25 phoasphatases in cancer cells: key players? Good targets? Nature Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 18.Parker AS, Cheville JC, Lohse CM, Igel T, Leibovich BC, Blute ML. Loss of expression of von Hippel-Lindau tumor suppressor protein associated with improved survival in patients with early-stage clear cell renal cell carcinoma. Urology. 2005;65:1090–95. doi: 10.1016/j.urology.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 19.Yao M, Yoshida M, Kishida T, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J Natl Cancer Inst. 2002;94:1569–75. doi: 10.1093/jnci/94.20.1569. [DOI] [PubMed] [Google Scholar]
- 20.Patard JJ, Fergelot P, Karakiewicz PI, et al. CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer. 2008;123:395–400. doi: 10.1002/ijc.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patard JJ, Rioux-Leclercq N, Masson D, et al. Absence of VHL gene alteration and high VEGF expression are associated with tumour aggressiveness and poor survival of renal-cell carcinoma. Br J Cancer. 2009;101:1417–24. doi: 10.1038/sj.bjc.6605298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordan JD, Lal P, Dondeti VR, et al. HIF-α effects on c-myc distinguish two subtypes of sporadic VHL-deficient celar cell carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann HP, Bender BU, Berger DP, et al. Prevalence, morphology and biology of renal cell carcinoma in von Hippel-Lindau disease compared to sporadic renal cell carcinoma. J Urol. 1998;160:1248–54. [PubMed] [Google Scholar]
- 24.Gnarra JR, Ward JM, Porter FD, et al. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci USA. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack FA, Rathmell WK, Arsham AM, et al. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell. 2003;3:75–88. doi: 10.1016/s1535-6108(02)00240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack FA, Patel JH, Biju MP, Haase VH, Simon MC. Decreased growth of Vhl−/−fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol Cell Biol. 2005;25:4565–4578. doi: 10.1128/MCB.25.11.4565-4578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.