Abstract
Elevated expression of the Nedd9/HEF1/Cas-L scaffolding protein promotes tumor cell invasion and metastasis in multiple cancer cell types. Conversely, generation of mammary tumors in the MMTV-polyoma virus middle T (PyVT) genetic model is delayed by a Nedd9−/− genotype. These activities arise from the role of Nedd9 in assembling complexes and supporting activity of cancer signaling proteins including FAK, SRC, SHC, and AKT, and would support evaluation of Nedd9 expression as an unambiguous biomarker for tumor aggressiveness. However, we here show that despite the initial delay in tumor growth, cells derived from MMTV-PyVT;Nedd9−/− tumors are characteristically hyperaggressive versus MMTV-PyVT;Nedd9+/+ cells in anchorage-independent growth, growth on 3D matrix produced by tumor-associated fibroblasts, and in formation of tumors after mammary orthotopic reinjection, and of lung metastases after tail vein injection. This reversal suggests the specific selection of MMTV-PyVT;Nedd9−/− cells for growth in an in vivo microenvironment. Indeed, MMTV-PyVT;Nedd9−/− cells have increased cell cycle, centrosomal, and mitotic defects, phenotypes compatible with the increased selection of these cells for aggressive growth. Intriguingly, in spite of their aggressive phenotype, MMTV-PyVT;Nedd9−/− cells persistently have low levels of SRC activation and are hypersensitive to the SRC kinase inhibitor dasatinib. These studies identify Nedd9 as a complex modulator of different aspects of mammary tumor growth.
Introduction
The Nedd9/HEF1/Cas-L non-catalytic scaffolding protein coordinates signaling cascades that control migration and invasion (1-3), cell cycle transition through M-phase (4-6), and apoptosis (7). Upregulation of Nedd9 is associated with enhanced invasion and metastasis in human and murine tumors and cell lines including melanoma, lung, breast, and brain (reviewed in (8)). Reciprocally, we recently reported that a Nedd9−/− genotype increased the latency of tumors formed in the aggressive MMTV-polyoma virus middle T antigen (PyVT) mouse mammary tumor model (9). Nedd9 is a central component of integrin-dependent signaling cascades that activate the FAK and Src kinases to promote cell migration, and also involved in lateral communication through Shc and other proteins to the Ras signaling cascades (8, 9). In MMTV-PyVT;Nedd9−/− tumors, activation of FAK, Src, AKT, and ERK were typically significantly reduced, as was the migratory and invasive behavior of isolated cell lines (9). Taken in sum, these data support the straightforward interpretation that Nedd9 is a tumor-promoting factor, and that high levels of Nedd9 in tumors correlate with poor prognosis and treatment resistance. As such, this would support adaptation of Nedd9 expression as a biomarker for aggressive disease, as has been proposed (10).
However, some observations argue that Nedd9 may have a more complex role in tumor formation, invasion, and metastasis. For example, while the majority of studies have shown a positive effect of Nedd9 on regulation of cell migration, two works have reported Nedd9 can negatively regulate migration of MCF10A normal breast epithelial cells, while mRNA profiling experiments suggest Nedd9 expression negatively correlates with the metastasis of MDA-MB-231 breast cancer cells to bone (11, 12). This may reflect cell type-specific differences in Nedd9 function, or alternatively, indicate that cellular response to altered expression of Nedd9 is dynamic at different stages of tumor growth. In potential support of the latter interpretation, we have previously shown that Nedd9 directly binds and regulates activity of the Aurora-A kinase (5, 6, 13). Because of its role in timing mitotic progression, Aurora-A levels and activity are highly regulated within a narrow range, with both overexpression and depletion causing centrosomal and mitotic defects, and contributing to genomic instability (14). In cell culture experiments, we have determined that exogenous overexpression or siRNA depletion of Nedd9 results in similar cell cycle phenotypes, based on aberrant regulation of Aurora-A (5).
These data led us to hypothesize that although absence of Nedd9 might initially delay tumor appearance, it may also increase in vivo selection pressures affecting tumor growth, thus reconciling contradictory observations. In this study, we have analyzed early passage cells derived from the mammary tumors of MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ mice to test this idea. Results of this study strongly support the idea that constitutive absence of Nedd9 in tumors induces defects in cell cycle associated with reduced function of Aurora-A and a more aggressive tumor phenotype. Intriguingly, our data also indicate that the absence of Nedd9 causes a persistent deficiency in Src activation, and influences the susceptibility of mammary cancer cell lines to growth inhibition by dasatinib, a drug targeting Src kinases. These studies in sum should significantly inform the use of Nedd9 as a biomarker for clinical response.
Methods
Cell lines and cell growth measurements
The protocol for derivation of mammary tumor cell lines from MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ mice was described in (9). Basal proliferation rates for all cell lines were measured by seeding 1 × 105 cells into 0.1% gelatin-coated T25 flasks, and then propagating them for up to 96 hours, after which cells were trypsinized and counted using Trypan Blue exclusion analysis at times indicated. To contrast cell proliferation on tissue culture plastic, or on matrix produced by NIH3T3 cells or tumor associated fibroblasts (TAFs) to simulate an in vivo microenvironment, matrix was prepared and measurements made using protocols described in detail in (15, 16). Typically, cell lines were plated at a density of 50000 cells/well in 12 well plates directly onto tissue culture plastic, NIH3T3-derived 3D matrix, or TAF-derived 3D matrix. Cells were grown for 24h, 48h, 72h, and 96h then treated with 10% (v/v) Alamar Blue solution (Invitrogen), and fluorescence measured by platereader. For drug treatment experiments, cells were seeded into 48 or 96 well plates. After 24 hours, vehicle (DMSO, 0.1%) or drugs (dasatinib and erlotinib, obtained from the Fox Chase Cancer Center pharmacy; and C1368 (Sigma)), were added to medium. After 72 hours, cell viability was assessed by Alamar Blue assay. All assays were performed a minimum of three times in triplicate. Cell cycle compartmentalization was measured using a Guava Personal Cell Analysis-96 (PCA-96) System (Guava Technologies, Hayward, CA). Soft agar experiments were performed using standard techniques, as in (17). For rescue experiments, cells were transfected with the plasmids pEGFP-C1 vector (Clontech, Mountain View, CA) containing Nedd9 and pCDNA3.1-mRFP (Invitrogen, Carlsbad, CA) containing AurA, to overexpress GFP-Nedd9 and RFP-AurA, respectively.
Orthotopic and tail vein injections
Care of mice and injection protocols were approved by the Fox Chase Cancer Center Institutional Animal Care and Use Committee and followed the National Institutes of Health guidelines. For orthotopic injection, 1 × 106 cells in 200 μl PBS were injected (bilateral inguinal) into the fourth mammary fat pad of SCID mice (5 per cell line). Mice were palpated twice weekly for tumor onset. Tumors were measured by caliper beginning 6 days after injection, and volume calculated as width × length × 0.4. The mice were euthanized by methoxy-fluorane (Metofane) inhalation when the longest dimension of the largest tumor reached 2 cm or alternatively, if mice exhibited signs of illness or distress. For each mouse, the tumor and lungs were excised, divided in half, and processed either for Western analysis or pathology. Xenografted tumors and both lungs were fixed in 10% phosphate buffered formaldehyde for 24 hours, embedded in paraffin, sectioned and stained with hematoxylin-eosin. Three sections of each lung separated by 1 mm were evaluated for metastases. Metastases were counted by a pathologist (AKS), using a Nikon Eclipse 50i microscope. The surface area of the lungs determined with a planimetric software (Image Pro Pus, Media Cybernetics, Bethesda MD). Metastases were expressed as number of metastases/cm2.
For tail vein injections into SCID mice, 0.35×106 cells suspended in 200 μl PBS were injected per mouse (5 mice per cell line). Mice were monitored daily, for signs of developing tumor burden such as weight loss, reduced mobility, hunched posture, and ruffled fur in SCID mice. All mice were sacrificed at the end of the week 3, when two mice showed signs of breathing problems. For each mouse, the lungs were excised, divided in half, and processed either for Western analysis or pathology.
Biochemical analysis
For Western analysis, tumor sections histologically confirmed to contain >90% tumor tissue were harvested, homogenized, and lysed in PBS-TDS buffer (1x PBS, 1% Triton X-100, 0.1% SDS, 20% glycerol) containing complete protease and phosphatase inhibitor cocktail (Roche Diagnostic). Whole cell lysates from the MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ cell lines were prepared using CelLytic™ MT Mammalian Tissue Lysis/Extraction Reagent (Sigma). Primary antibodies targeted Nedd9 (2G9, (5), diluted 1:1000), p130Cas (Santa Cruz Biotechnology, 1:1000), Aurora (BD Biosciences, 1:1000), phospho-Aurora A T288 (Cell Signaling, 1:500) phospho-FAK-Y397 (1:2000), ShcA-Y317 (1:1000), and Src-Y418 (1:1000), FAK (1:2000), Src (1:1000), and ShcA (1:1000) (Abcam), phospho-ERK1/2-T202/Y204 (1:500), AKT-S475 (1:1000) and AKT-T308 (1:500), and ERK1/2 (1:1000) and AKT (1:1000) (Cell Signaling), and phospho-Lyn-Y507 (1:500) and Lyn (1:1000) (Cell Signaling), and β-actin (1:20000) (Sigma). Secondary horseradish peroxidase–conjugated antibodies were from Pierce Biotechnology. Proteins were visualized using the West-Pico system (Pierce). Image analysis was done using NIH ImageJ (NIH), with signal intensity normalized to β-actin.
For in vitro kinase assays, cell lysates prepared from MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+cell lines were immunoprecipitated using anti-Aurora A antibody (BD Transduction Laboratories) and incubated with γ-32P(ATP) and histone H3 (Upstate) substrate and standard methods were followed for a kinase assay as described in (5). Aliquots of the reaction mix were used for SDS-PAGE and Western analysis to confirm levels of AurA or T288-AurA (using T288 phospho-specific antibody (1:500), Cell Signaling).
Immunofluorescence
Epifluorescence microscopy was performed using an inverted Nikon TE300 microscope (Melville, NY). All images were acquired as 12-bit images with a Spot RT monochrome camera (Diagnostic Instruments, Sterling Heights, MI). Secondary antibodies labeled with Alexa-488, Alexa-568, and DAPI to stain DNA, were from Molecular Probes/Invitrogen. To measure cell spreading, cells were fixed in 4% paraformaldehyde for 10 min, permeabilized in 0.2% Triton X-100 for 5 min and blocked with 3% BSA in PBS. Anti-paxillin monoclonal antibodies (mAb) (BD Transduction laboratories) were used to mark focal adhesions and outline cells. Cell area measurements were made for ~100 cells/experiment using MetaMorph or MetaVue software (Molecular Devices, Universal Imaging, Downingtown, PA) software to score pixels within cell perimeters.
To count centrosomes and analyze mitotic spindles, cells were fixed with 4% paraformaldehyde followed by methanol, permeabilized with 1% Triton-X100 in PBS, blocked in 1x PBS, 3% BSA, and incubated with antibodies to γ-tubulin (Sigma, 1:200) and α-tubulin (Sigma, 1:800) to visualize centrosomes and mitotic spindles, respectively.
Statistical analysis
We used Kaplan-Meier curves for survival outcomes. For figures, we used generalized linear models with restricted cubic splines estimated via Generalized Estimating Equations. We also used semiparametric regressions and generalized linear mixed effects models. Approaches are similar to those detailed in (9). Analyses were conducted in R (R Foundation for Statistical Computing, Vienna, Austria) and STATA 10 (STATACorp, College Station, Texas).
Results
Lack of Nedd9 increases the aggressiveness of selected MMTV-PyVT mammary tumor cell lines
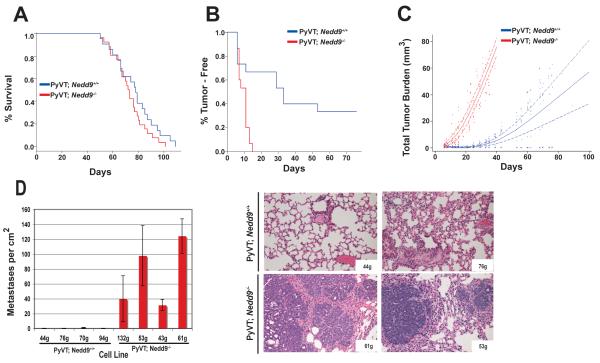
We previously compared mammary tumor development in MMTV-PyVT;Nedd9+/+ and MMTV-PyVT;Nedd9−/− mice (9). This work indicated tumor development was very significantly delayed in mice with an MMTV-PyVT;Nedd9−/− genotype (p=0.0001). Based on this observation and additional analysis indicating reduced activation of FAK, SRC, AKT, and ERK1/2 in MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ tumor lysates (9), and reduced cell migration by mammary tumor-derived cell lines lacking Nedd9−/−, we concluded Nedd9 acted positively in promoting tumor growth. However, continuing analysis of the data from the study indicated that while initial appearance of MMTV-PyVT;Nedd9−/− tumors was delayed, once tumors had attained palpable mass, the average time until euthanasia was required because of tumor size was similar to that of MMTV-PyVT;Nedd9+/+ animals (average 129 versus 133 days, median 128 versus 134 days) (Figure 1A).
Figure 1. Comparison of in vivo growth of MMTV-PyVT;Nedd9+/+ versus MMTV-PyVT;Nedd9−/− primary tumors and derivative cell lines.
A. Kaplan-Meier curve showing insignificant increase in time from appearance of tumors to maximal growth before euthanasia in MMTV-PyVT;Nedd9−/− (n=26) versus MMTV-PyVT;Nedd9+/+ (n=21) mice (p=0.181). B. Independent cell lines derived from MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ primary mammary tumors (3 per genotype) were injected orthotopically into the mammary pads of SCID mice (n=5/cell line; 30 mice total). Data shown represent significant difference in time till appearance of tumors (p=0.001). C. Significant decrease in total tumor burden in SCID mice with tumors derived from MMTV-PyVT;Nedd9+/+ versus MMTV-PyVT;Nedd9−/− cell lines (p=<0.001). D. MMTV-PyVT;Nedd9−/− cell lines more effectively form lung metastases following tail vein injection. Left, quantification of lung metastases/cm2, averaged from 5 mice for each of 8 cell line, 21 days after tail vein injection. Difference between genotypic groups is highly significant, P<0.0001. Right, hematoxylin and eosin stained lungs derived from SCID mice 3 weeks after tail vein injection of representative MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ cell lines, taken at magnification 10x.
This adaptation suggested that absence of Nedd9 might select for compensatory changes that increase tumor aggression. To test this idea, we used 3 MMTV-PyVT;Nedd9+/+ and 3 MMTV-PyVT;Nedd9−/− tumor-derived cell lines for orthotopic reinjection into mammary fat pads, and again monitored growth of tumors (Figures 1B, 1C). All three MMTV-PyVT;Nedd9−/− cell lines formed palpable tumors within 14 days (median, 11 days), reaching maximal size within 40 days after appearance. In contrast, two of the three MMTV-PyVT;Nedd9+/+ cell lines yielded mammary tumors that were detected within 55 days after injection (median, 33 days), and grew more slowly, while the third cell line failed to form any tumors over a four month period (Figure 1C). In a complementary analysis, we used 4 cell lines of each genotype to perform tail vein injections followed by measurement of metastases in the lung after 3 weeks. Strikingly, four out of four injected MMTV-PyVT;Nedd9−/− cell lines generated a large number of metastases in 20/20 injected animals (with an average of 73 metastases/cm2). In contrast, a single micrometastasis was detected in 1/20 mice injected with one of the four MMTV-PyVT;Nedd9+/+ cell lines assessed, while 19/20 mice had none, at the same time point (Figure 1D). These data clearly supported the idea that absence of Nedd9 ultimately selected for more aggressive tumor growth in vivo.
MMTV-PyVT;Nedd9−/− cells have a consistent growth advantage on 3D matrices and in soft agar
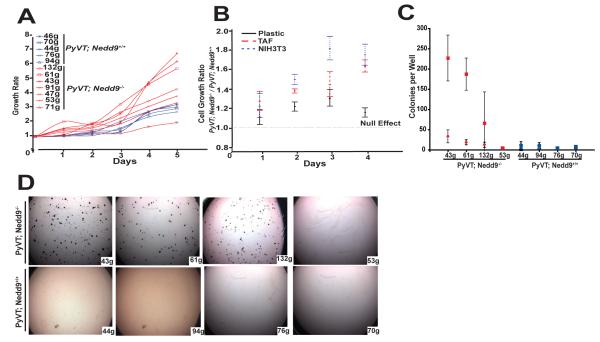
We had previously determined that MMTV-PyVT;Nedd9−/− cell lines were less migratory, and more sensitive to detachment-induced anoikis, than MMTV-PyVT;Nedd9+/+ cell lines (9). Given these observations, the most plausible explanation for the increased aggressiveness of the MMTV-PyVT;Nedd9−/− cells in vivo would be if they had undergone specific selection for proliferation in a tumor microenvironment. To begin assess this possibility, we first compared the coefficient of variation in growth rate in a larger panel of cell lines derived from the two genotypes (Figure 2A). This analysis indicated that among a group of 12 cell lines, the range of doubling times of cells cultured on plastic was significantly greater with the MMTV-PyVT;Nedd9−/− genotype (p<0.001), with some growing very rapidly and others growing relatively slowly.
Figure 2. Comparative growth of MMTV-PyVT;Nedd9+/+ versus MMTV-PyVT;Nedd9−/− cell lines in vitro.
A. Doubling time of 7 MMTV-PyVT;Nedd9−/− versus 5 MMTV-PyVT;Nedd9+/+ cell lines grown on tissue culture plastic indicates greater overall variability in growth rates associated with absence of Nedd9 (<0.0001). B. Ratio of growth rate of 3 MMTV-PyVT;Nedd9−/− versus 3 MMTV-PyVT;Nedd9+/+ cell lines grown on tissue culture plastic versus matrix derived from NIH3T3 cells or TAFs indicates significant difference at 90 hours(for each, p<0.001). C. Colony formation on soft agar, 3 weeks after plating of 2500 cells/well. Experiments were performed in quadruplicate, at least 3 times independently. Squares, large colonies (>150 pixels); diamonds, small colonies (< 150 pixels). Difference between genotypic groups is highly significant, P=0.001.
Second, growth of cells on three-dimensional (3D) matrices secreted by either immortalized fibroblast cell lines, or by primary fibroblasts that have been “primed” to support the growth of tumor cells (i.e., tumor associated fibroblasts, TAFs), can simulate a stromal tumor microenvironment (18). We compared proliferation rates of a set of MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines cultured on tissue culture plastic, on 3D matrix prepared from NIH3T3 cells, and on 3D matrix generated from tumor-associated fibroblasts (TAFs) (Figure 2B). Strikingly, although there was no overall statistically significant difference between the two genotypic groups cultured on plastic (as both fast and slow-growing MMTV-PyVT;Nedd9−/− lines were assessed), all MMTV-PyVT;Nedd9−/− cell lines grew much faster on NIH3T3- or TAF-derived matrix (p <0.001).
Third, we also compared growth of MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines cultured in soft agar, to assess fully anchorage-independent growth. In this case, 3/4 MMTV-PyVT;Nedd9−/− cell lines efficiently formed large colonies in soft agar, while 0/4 MMTV-PyVT;Nedd9+/+ cell lines formed large colonies (Figure 2C). The increased growth rate of MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cells on 3D matrices, in soft agar, and in vivo, coupled with the greater growth variability of MMTV-PyVT;Nedd9−/− cell lines on tissue culture plastic, suggested that MMTV-PyVT;Nedd9−/− cell lines may have been more subject to selective pressures for growth in the in vivo microenvironment, particularly as the effects were seen in early (<6) passage populations of cells recovered from tumors.
MMTV-PyVT;Nedd9−/− cells have more cell cycle, spindle, and centrosome abnormalities than MMTV-PyVT;Nedd9+/+ cells
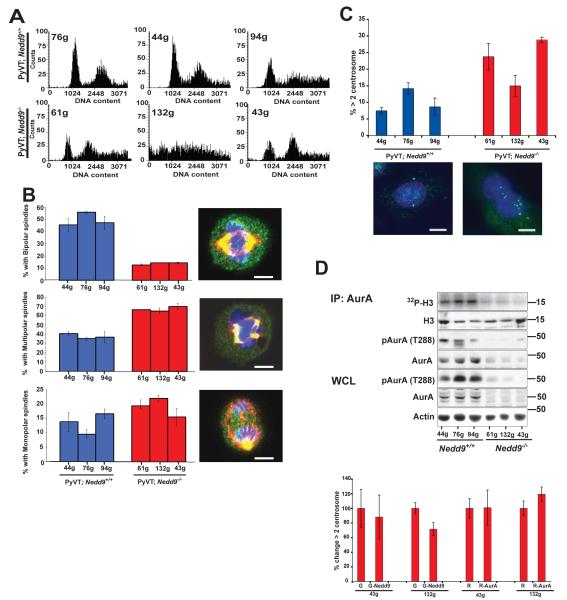
To begin to establish the basis for the increased variability in the growth of MMTV-PyVT;Nedd9−/−cells, we directly assessed whether these cells had signs of increased genomic instability and aneuploidy. Indeed, flow cytometric analysis (Figure 3A and not shown) indicated a greater proportion of >4N cells and more abnormal profiles in cells with the MMTV-PyVT;Nedd9−/− genotype. These cells typically had a much greater number of abnormal mitoses than MMTV-PyVT;Nedd9+/+ lines, including both monopolar (19% versus 13%, p=0.002) and multipolar (67% versus 38%, p=0.001) spindles (Figures 3B), with consistent increases observed in all cell lines of the MMTV-PyVT;Nedd9−/− genotype. Commensurate with this observation, a higher proportion of these cells had supernumerary centrosomes (Figure 3C).
Figure 3. Cell cycle abnormalities in MMTV-PyVT;Nedd9+/+ versus MMTV-PyVT;Nedd9−/− cell lines.
A. FACS analysis of asynchronously growing MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines indicates more cells with 4N or greater DNA content. B. Mitotic profiles in asynchronously growing cells demonstrate fewer normal bipolar spindles (p=<0.001) and more multipolar (p=0.001) and monopolar (p=0.002) spindles in MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines. >50 mitoses were counted/cell line in each of 3 independent experiments. Representative cells are visualized with DAPI (blue), α-tubulin (red) and γ-tubulin (green). Scale bar, 5 μn. C. Increased frequency of supernumerary centrosomes in MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines (p=<0.001). >100 cells were counted/cell line in each of 3 independent experiments. Images indicate representative cell with normal centrosome count (left) and supernumerary centrosomes (right), visualized with DAPI (blue) and γ-tubulin (green). Scale bar, 10 μm. D, IP:AurA, immunoprecipitation (IP) of total AurA from cell lines followed by in vitro kinase assay with phosphorylated histone H3 as substrate visualized by 32P-ATP, and with immunoprecipitated, autophosphorylated (T288) AurA, total AurA, and H3 substrate visualized by Western analysis. WCL, Western analysis of T288-phospho-AurA and total AurA in whole cell lysates (WCL). Bottom, change in proportion of MMTV-PyVT;Nedd9−/− cells with >2 centrosomes following expression of GFP (G) versus GFP-Nedd9 (G-Nedd9) or RFP (R) versus RFP-AurA (R-AurA) is insignificant (p>0.1).
We have previously demonstrated that Nedd9 binds and controls the activation of the mitotic kinase Aurora-A (AurA) (5, 6). Defects in AurA activation are often associated with centrosomal abnormalities and aneuploidy in human cancers. Intriguingly, we found that the levels of AurA kinase activity were sharply reduced in MMTV-PyVT;Nedd9−/−cells (Figure 3D, top). Further, and unexpectedly, this activity drop correlated with a reduction in the levels of overall AurA kinase in this genetic background, rather than a proportional reduction in AurA activity (Figure 3D, middle). A Nedd9-dependent decrease in AurA might provide an initiating event for the genomic instability manifested by the MMTV-PyVT;Nedd9−/−cells. However, overexpression of neither GFP-Nedd9 nor RFP-AurA in these cell lines reverses the centrosomal abnormalities, implying that at minimum, other changes in the regulatory machinery had become critical (Figure 3D, bottom).
The MMTV-PyVT;Nedd9−/− genotype is sensitized to dasatinib
These data in sum demonstrated that absence of Nedd9 resulted in a very different profile of mammary tumor cell growth, and signs of genomic instability associated with loss of Aurora-A. We had previously shown that Src and FAK had consistently reduced activity in the tumors and derivative cell lines of MMTV-PyVT;Nedd9−/− mice, while SHC, ERK, and AKT have statistically significant reduction activity in primary MMTV-PyVT;Nedd9−/− tumors, but greater tumor to tumor variability (9). We therefore assessed whether mammary tumor cells positive or negative for Nedd9 displayed different profile for response to drugs targeting proteins with which Nedd9 functionally interacts.
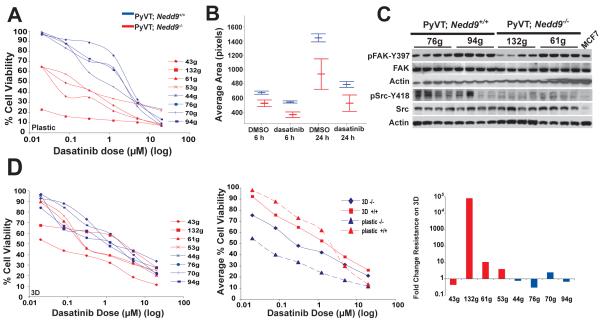
Strikingly, MMTV-PyVT;Nedd9−/− cell lines were much more sensitive to dasatinib, an inhibitor of the Src kinases, then were MMTV-PyVT;Nedd9+/+ cells (Figure 4A). This correlated with not only a significantly decreased basal ability of MMTV-PyVT;Nedd9−/− cell lines to spread, but enhanced inhibition of spreading following plating in low doses of dasatinib, indicating an inhibition of cytoskeletal rearrangment (Figure 4B), and the depressed activation of FAK we had previously noted in these cell lines (9). Re-analysis of FAK and SRC activation in lysates from the primary tumors arising from orthotopic reinjection (Figure 1B) indicated that activation of SRC remained consistently significantly depressed in tumors of the MMTV-PyVT;Nedd9−/− genotype, although FAK activation was now similar in tumors of both genotypes (Figure 4C).
Figure 4. Response to dasatinib predicted by Nedd9 status and compensated by 3D TAF matrix.
A. IC50 curve calculations for 4 dasatinib-treated MMTV-PyVT;Nedd9−/− versus 4 MMTV-PyVT;Nedd9+/+ cell lines. Cell lines analyzed in quadruplicate, in 3 independent experiments. B. Comparative rate of attachment of MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines plated in dasatinib. Dasatinib-dependent decrease in spreading differs significantly at 6 (p=0.001) and 24 (p=0.016) hours. At 24 hours, proportionately fewer MMTV-PyVT;Nedd9−/− cells remain attached to plate (not shown). C. Immunoblots of tumor lysates prepared from tumors generated by mammary orthotopic reinjection of the cell line indicated at top. Each lane represents an independent tumor arising from the noted cell line. D. 3D TAF matrix rescues dasatinib sensitivity of MMTV-PyVT;Nedd9−/− cell lines. Left panel, experiment as in A, in 8 cell lines plated on 3D matrix generated from TAFs. Center, IC50 curves represented as averages, for genotypes and conditions indicated. Taking 1uM as an example, differences in IC50 between genotypes are significant on plastic (P=0.002) and on 3D TAF matrix (P=0.033). Right, fold difference in IC50 values for cell lines indicated in plating on TAF matrix versus tissue culture plastic.
These data suggest MMTV-PyVT;Nedd9−/− cells might be specifically vulnerable to additional dasatinib-based targeting of Src because of predisposing prior reduction in pathway activity arising from absence of the SRC partner Nedd9. If so, and given the prior data suggesting MMTV-PyVT;Nedd9−/− cells were more adapted to extrinsic microenvironment, 3D matrix cues might be expected to rescue cell growth. Strikingly, plating of cell lines on 3D TAF matrix produced markedly different results between MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ cell lines (compare Figure 4D left and center panels with 4A). Although the MMTV-PyVT;Nedd9−/− lines remained more sensitive to dasatinib, overall IC50 values were significantly increased in 3 of 4 lines (Figure 4D, right panel). In contrast, MMTV-PyVT;Nedd9+/+ cell lines as a group had little responsiveness to 3D TAF matrix, with 3 of 4 lines instead becoming more sensitive to dasatinib.
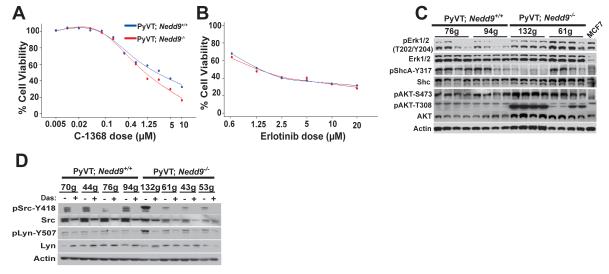
We also compared the responsiveness of MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ cells to C1368, which targets the Aurora-A kinase (Figure 5A), and erlotinib, which targets EGFR-dependent signaling (Figure 5B). In neither case was a specific difference observed, suggesting the sensitization we observed was specific for SRC. Interestingly, although activity of the EGFR effectors ERK, SHC, and AKT had also been depressed in primary tumors, as with FAK, this was not observed in tumors arising from orthotopic reinjection (Figure 5C), where levels were comparable or higher in the MMTV-PyVT;Nedd9−/− cells, and some tumors showed evidence of recovery of Aurora-A levels (not shown).
Figure 5. Nedd9 status does not affect response to inhibitors of Aurora-A or EGFR.
A. Experiment as in 4A with the Aurora-A inhibitor C1368, for 2 cell lines of each genotype, and resulting data averaged (IC50 1.11 μM versus 2.21 μM; p value is not significant). Similar results were obtained with a second Aurora-A inhibitor, PHA 680632; results not shown. B. Experiment as in A with the EGFR inhibitor erlotinib (IC50 1.29 μM versus 1.37μM; p value is not significant) indicates no cell genotype-dependent difference in IC50 value. C. Immunoblots of tumor lysates as in 4C. D. Expression and activation (reflected by levels of autophosphorylated forms) of Src and Lyn in MMTV-PyVT;Nedd9−/− versus MMTV-PyVT;Nedd9+/+ cell lines treated for 2 hours with 200 nM dasatinib.
Finally, although SRC is often considered the primary physiological target of dasatinib in solid tumors (19), this drug can affect other relevant cellular kinases including SRC-family kinases such as LYN (20), but also c-Kit (19), EGFR (19), Ephrin (21), and others. Given the close physical interactions between Nedd9 and Src kinases (1-3), one possibility was that tumors developing in the absence of Nedd9 might affect the basal expression or activity of Srcrelated kinases. We determined that Src and Lyn were similarly expressed in MMTV-PyVT;Nedd9−/− and MMTV-PyVT;Nedd9+/+ cells (Figure 5D), and also established that the activation of these kinases was similarly inhibited by dasatinib. Expression and activity of a number of other dasatinib targets (including EGFR and c-Kit) were also unaffected by Nedd9 genotype (not shown).
Discussion
The variant proliferation rates, increased percentage of cells characterized by supernumerary centrosomes, and anomalous cell cycle compartmentalization in MMTV-PyVT;Nedd9−/− tumor-derived cell lines suggests that in each case, the Nedd9−/− genotype has provided a selection for mutations in the tumor population. The most direct explanation for this selection lies in deficient expression and activation of Aurora-A in Nedd9−/− cells. Aurora-A activation is required for mitotic entry and transit, and we have shown that siRNA depletion of Nedd9 impairs Aurora-A activation and mitotic progression, leading to the accumulation of aneuploid binucleate cells (4, 5). Our present data demonstrate that genetic loss of Nedd9 results in a similar phenotype, in which context it is remarkable that the consequences of a Nedd9−/− genotype are predominantly restricted to effects on tumorigenesis. We suggest this may reflect a specific requirement for Nedd9 scaffolding function in sustaining the activity and expression of some of its partner proteins in the constitutive growth environment of a tumor. By contrast, the transient activation of focal adhesion-associated proteins such as Src or FAK in a normally growing or quiescent cellular context may not require Nedd9 interactions, but be supported by the Nedd9 paralog BCAR1/p130Cas (discussed in (22)); while Aurora-A activation may be supported by its other partners, such as Ajuba (23) and Tpx2 (24). The fact that very significant Nedd9 upregulation has been observed in a large subset of aggressive tumors from the lung, breast, and brain ((10, 25, 26); discussed in (22)) and head and neck (27), while BCAR1, Ajuba, FAK, and Tpx2 have not been reported to be thus overexpressed, is compatible with such a model.
Identifying effective biomarkers for cancer prognosis and drug responsiveness is of great importance in improving the clinical management of cancer. While earlier studies focused on Nedd9 upregulation in tumors suggest that high Nedd9 levels are specifically associated with poor prognosis and metastasis, the work presented here and in (9) indicate a more nuanced role for Nedd9 in mammary tumor biology, in which both elevated or absent levels of Nedd9 may be associated with aggressive tumor phenotypes. This argues against the straightforward consideration of high Nedd9 levels in tumors as a predictive biomarker. However, the observation that absence of Nedd9 greatly sensitizes cells to the Src-family targeting agent dasatinib is particularly significant, given the increasing use of dasatinib in the clinic for treatment of breast (28) and other cancers. MMTV-PyVT;Nedd9−/− cells maintain persistently low levels of active Src, suggesting the “double hit” of dasatinib and a MMTV-PyVT;Nedd9−/− genotype is particularly deleterious to cell survival signaling networks. In this context, the modulation of sensitivity by the 3D matrix produced by TAFs suggests that in vivo, specific niches of tumor cells may be protected, while other populations may be more vulnerable. Conversely, the subset of metastatic tumors overexpressing Nedd9 (8, 10, 25, 26, 29) may be particularly resistant to Src-targeting agents, and independent of microenvironment-associated survival cues.
MMTV-PyVT;Nedd9−/− cells do not have increased sensitivity to EGFR- or Aurora-A targeting agents. In contrast to the situation with Src, the cells have overcome the inhibition of the EGFR effectors AKT and ERK observed in primary tumor lysates. While levels of Aurora-A remain depressed in these tumors, much of the actual cell killing associated with Aurora-inhibiting small molecules has been proposed to be associated with inhibition of Aurora-B, rather than Aurora-A kinase (30), compatible with our observation that reductions in Aurora-A expression do not correlate with increased sensitivity to Aurora-targeting inhibitors. Based on these data, we suggest that Nedd9 may be an excellent candidate to examine as a modulator of response to dasatinib and potentially other Src kinase inhibitors in ongoing clinical development of this and other Src-targeting agents.
Acknowledgments
We are grateful to Sharon Howard, Anthony Lerro, and Jackie Valvardi for technical help with cell line generation and animal work. We thank Sachiko Seo for the original gift of Nedd9−/− mice, which allowed derivation of the cell lines. We thank Nicolas G. Day for assistance with soft agar and xenograft assays, and graphical rendering of data. We thank Ilya Serebriiskii, Edna Cukierman, and Natalia Skobeleva for guidance in some experiments and analysis, and Denise Connolly and Joy Little for critical comments on the manuscript. This work was supported by NIH R01-CA63366 and R01-CA113342; W81XWH-07-1-0676 from the Army Materiel Command; and Pennsylvania Tobacco Settlement funding (to EAG); by the Israel Cancer Association and Stanley Abersur Research Foundation (to MW); and by NIH core grant CA-06927 and the Pew Charitable Trust (to Fox Chase Cancer Center). EI was supported in part by the American Associates, Ben-Gurion University of the Negev.
References
- 1.Fashena SJ, Einarson MB, O’Neill GM, Patriotis CP, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115:99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 2.van Seventer GA, Salman HJ, Law SF, et al. Focal adhesion kinase regulates beta1 integrin dependent migration through an HEF1 effector pathway. Eur J Imm. 2001;31:1417–27. doi: 10.1002/1521-4141(200105)31:5<1417::AID-IMMU1417>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Elia AEH, Law SF, Golemis EA, Farley J, Wang T. A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member, HEF1. EMBO J. 2000;19:6759–69. doi: 10.1093/emboj/19.24.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dadke D, Jarnik M, Pugacheva EN, Singh MK, Golemis EA. Deregulation of HEF1 impairs M-phase progression by disrupting the RhoA activation cycle. Mol Biol Cell. 2006;17:1204–17. doi: 10.1091/mbc.E05-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–46. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugacheva EN, Golemis EA. HEF1-aurora A interactions: points of dialog between the cell cycle and cell attachment signaling networks. Cell Cycle. 2006;5:384–91. doi: 10.4161/cc.5.4.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neill GM, Golemis EA. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol Cell Biol. 2001;21:5094–108. doi: 10.1128/MCB.21.15.5094-5108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975–9. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumchenko E, Singh MK, Plotnikova OV, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198–206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Simpson KJ, Selfors LM, Bui J, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–38. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 12.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–63. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 15.Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 2008;27:573–85. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castello-Cros R, Cukierman E. Stromagenesis during tumorigenesis: characterization of tumor-associated fibroblasts and stroma-derived 3D matrices. Methods Mol Biol. 2009;522:275–305. doi: 10.1007/978-1-59745-413-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skobeleva N, Menon S, Weber L, Golemis EA, Khazak V. In vitro and in vivo synergy of MCP compounds with mitogen-activated protein kinase pathway- and microtubule-targeting inhibitors. Mol Cancer Ther. 2007;6:898–906. doi: 10.1158/1535-7163.MCT-06-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol. 2005;167:475–88. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardo LJ, Lee FY, Chen P, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–61. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 20.Choi YL, Bocanegra M, Kwon MJ, et al. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res. 70:2296–306. doi: 10.1158/0008-5472.CAN-09-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang F, Reeves K, Han X, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res. 2007;67:2226–38. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
- 22.Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirota T, Kunitoku N, Sasayama T, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–98. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 24.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–62. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 25.Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–32. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 26.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 27.Lucas JT, Jr., Salimath BP, Slomiany MG, Rosenzweig SA. Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent. Oncogene. 2010 doi: 10.1038/onc.2010.185. Epub May 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RS, Dering J, Ginther C, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/“triple-negative“ breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–26. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 29.Bui LC, Tomkiewicz C, Chevallier A, et al. Nedd9/Hef1/Cas-L mediates the effects of environmental pollutants on cell migration and plasticity. Oncogene. 2009;28:3642–51. doi: 10.1038/onc.2009.224. [DOI] [PubMed] [Google Scholar]
- 30.Keen N, Taylor S. Mitotic drivers--inhibitors of the Aurora B Kinase. Cancer Metastasis Rev. 2009;28:185–95. doi: 10.1007/s10555-009-9184-9. [DOI] [PubMed] [Google Scholar]