Abstract
Excessive bone destruction is a major cause of morbidity in myeloma patients. However, the biologic mechanisms involved in the pathogenesis of myeloma-induced bone disease are not fully understood. Heparanase, an enzyme that cleaves the heparan sulfate chains of proteoglycans, is upregulated in a variety of human tumors, including multiple myeloma. We previously demonstrated that heparanase promotes robust myeloma tumor growth and supports spontaneous metastasis of tumor cells to bone. In the present study, we demonstrate, for the first time, that the expression of heparanase by myeloma tumor cells remarkably enhances bone destruction locally within the tumor microenvironment. In addition, enhanced heparanase expression in the primary tumor also stimulated systemic osteoclastogenesis and osteolysis, thus mimicking the systemic osteoporosis often seen in myeloma patients. These effects occur, at least in part, as the result of a significant elevation in the expression and secretion of RANKL by heparanase-expressing myeloma cells. Moreover, analysis of bone marrow biopsies from myeloma patients reveals a positive correlation between the level of expression of heparanase and RANKL. Together these discoveries reveal a novel and key role for heparanase in promoting tumor osteolysis and demonstrate that RANKL is central to the mechanism of heparanase-mediated osteolysis in myeloma.
Keywords: Heparanase, osteolysis, multiple myeloma, RANKL, tumor microenvironment
Introduction
Multiple myeloma is a hematologic B cell malignancy and is the most frequent cancer to involve the skeleton, with up to 90% of patients developing destructive bone lesions (1). Myeloma-induced bone disease is associated with bone pain, fractures, and hypercalcemia which affect quality of life and lead to increased morbidity in myeloma patients. Numerous studies have shown that bone disease in myeloma is caused by elevated bone resorption, which is stimulated by the activation of osteoclasts and the inhibition of osteoblasts (2-4). However, the specific components and mechanisms responsible for driving the elevated bone resorption have not been fully determined.
Heparanase-1 (heparanase, HPSE) is the only known enzymatically active endoglycosidase that degrades heparan sulfate at specific intrachain sites (5, 6). Expression of heparanase is rare in normal tissues, but it is upregulated in many human tumors, including myeloma (7-13). Upregulation of heparanase has been correlated with increases in lymph node metastasis, distant metastasis, microvessel density, and reductions in post-operation survival of several types of cancer – all of which provide strong clinical support for the pro-metastatic and pro-angiogenic features of the enzyme (11, 14-17). Upregulation of heparanase expression is associated with poor prognosis in myeloma (18) and we previously demonstrated that elevated expression of heparanase in myeloma cells dramatically enhances their growth, angiogenesis, and metastasis to bone (19-22). However, regarding the role of heparanase in regulating bone turnover, there are conflicting reports in the literature. Expression of heparanase by breast cancer cells implanted in the mammary fat pad of mice enhances systemic bone turnover (23, 24). A limited descriptive study using a murine model of prostate cancer suggested a role for heparanase in enhancing osteolysis (25). However, in this study heparanase also increased tumor growth and it was not determined if the increased osteolysis was due to heparanase or simply due to an increase in the tumor size. In contrast to the proposed role of heparanase in osteolysis, it was discovered that mice transgenic for heparanase exhibited a marked increase in bone density as compared to controls, suggesting that heparanase may promote bone formation (26). To address the role of heparanase in tumor osteolysis we utilized two murine models of myeloma as well as biopsies from myeloma patients. We discovered that the expression of heparanase by tumor cells induces significant osteolysis both locally and systemically and that this was not solely a function of increased tumor size. Furthermore, we found that elevated levels of heparanase lead to the upregulation of the expression of receptor activator of nuclear factor κ B ligand (RANKL) in myeloma cells with a resulting increase in osteoclastogenesis, thus defining a mechanism whereby heparanase expression can drive the destruction of bone in myeloma. These novel findings point to heparanase as a key regulator of myeloma bone disease that is upstream of RANKL and further validate heparanase as a viable target for osteolytic cancers.
Methods and materials
Cells and reagents
CAG myeloma cell line was established at the Myeloma Institute for Research and Therapy (Little Rock, AR) as described previously (27). CAG cells with modified levels of heparanase expression have been previously extensively characterized (19, 20, 28) and include i) heparanase-low (HPSE-low) cells prepared by transfection with empty vector; ii) heparanase high (HPSE-high) cells prepared by transfection with vector containing the cDNA for human heparanase (Although HPSE-high cells express a 4 fold-higher level of heparanase than do the HPSE-low cells, the elevated levels of enzyme activity present in HPSE-high cells is in the same range as that present in the bone marrow of many myeloma patients (21). This indicates that HPSE-high cells very closely mimic the level of heparanase activity present in many myeloma patients and thus represent an appropriate model for examining the effects of heparanase on myeloma tumors); iii) cells transduced with viral vectors containing a control shRNA sequence; and iv) HPSE k/d cells transduced with viral vectors containing a shRNA sequence to knockdown heparanase expression. U266 cells were obtained from ATCC, (Manassas, VA) and MM.1S human myeloma cells were generously provided by Dr. Steve T. Rosen and Dr. Nancy Krett (Northwestern University). During the course of this study the CAG cells were confirmed as myeloma cells by their expression of CD138 and kappa immunoglobulin light chain.
Recombinant heparanase (rHPSE) and heparanase antibodies were kindly provided by Dr. Israel Vlodavsky (Technion, Haifa, Israel). Human kappa antibody and kappa ELISA kit were purchased from Bethyl Laboratories (Montgomery, TX); TRAP staining kit and β- actin antibody were from Sigma (St Louis, MO); Human TRAP 5b ELISA kit was from Immunodiagnostic Systems (Fountain Hills, AZ); Human RANKL and OPG antibodies were from R&D Systems (Minneapolis, MN); and human RANKL and OPG ELISA kits were from ALPCO (Salem, NH).
In vivo models of myeloma
5- to 6-week-old male CB.17 scid/scid mice obtained from Harlan Sprague Dawley (Indianapolis, IN) were used in the studies. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee.
SCID-hu model
The SCID-hu model has been previously described (19, 29, 30). Briefly, human femora (Advanced Bioscience Resources, Inc., Alameda, CA) were cut into halves and implanted subcutaneously, one on each side of the dorsum of the SCID mice. 6-8 weeks after implantation, 105 CAG HPSE-low or HPSE-high cells were injected directly into the cut end of one human bone graft in each SCID-hu mouse. Murine sera were collected bi-weekly. After mice were euthanized, tumor-injected human bones and non-injected contralateral human bones were collected and evaluated for the extent of bone resorption. Final tumor burden in each mouse was determined by measurement of human kappa light chain in mouse serum harvested just prior to euthanasia.
SCID-tibia model
105 CAG HPSE-low or HPSE-high cells were injected into the right-side tibia of SCID mice. Murine sera were collected every two weeks. After mice were euthanized, tumor-injected tibia and non-injected contralateral tibia were evaluated for the extent of bone resorption.
Evaluation of bone resorption and osteoclast number
Implanted human bones (SCID-hu model) or murine tibia (SCID-tibia model) were fixed in 10% neutral-buffered formalin, bone microarchitecture and density was assessed by microCT (Scanco Medical AG, Bassersdorf, Switzerland) as described previously (31). After imaging, the bones were decalcified with 10% EDTA, embedded in paraffin and sectioned. Bone Sections were stained using a TRAP staining kit (19). Osteoclast number was determined via counting of TRAP positive multinucleated osteoclasts (3 or more nuclei per cell) adjacent to the bone surface as described previously (23). In addition, the levels of human TRAP 5b in murine sera of SCID-hu mice were measured using a human TRAP 5b ELISA kit.
Immunohistochemistry
Human kappa, heparanase, RANKL and OPG were stained on paraffin-embedded tissue sections harvested from animal models following the manufacturer’s recommendation (19, 28).
Paraffin-embedded bone marrow core biopsy specimens of myeloma patients, obtained from the Department of Pathology at UAB, were also stained for heparanase, RANKL and OPG. The experimental procedures and protocols were approved by the UAB Institutional Review Board. Scoring for staining densities was determined in a blinded fashion by two different readers including a board certified hematopathologist as described previously (21).
Real-Time PCR
RNA was extracted from cultured cells using Qiagen RNeasy columns (Qiagen, Valencia, CA) and equal amounts were reverse transcribed using RevertAid First Strand cDNA synthesis kit (Fermentas, Hanover, MD). For real- time PCR 2X IQ™ SYBR® green supermix (Bio-Rad), along with 25ng of cDNA from each sample and gene specific primers were used. The primers and the cycle parameters for real-time PCR analysis of human RANKL (32) and the internal control 18S ribosomal RNA (33) have been published previously. The PCR cycle at which the fluorescence exceeded a set threshold (CT), was determined by the iCycler software and data were analyzed according to the comparative CT method (34). Semiquantitative results represent the n-fold difference of normalized human RANKL transcript levels.
Western blotting
Equal amounts of protein from cell or tumor tissue extract were subjected to 4-12% gradient SDS-PAGE (BioRad) and transferred to nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany) (19). They were then probed with anti-human RANKL or OPG or β- actin antibodies and were visualized by an enhanced chemiluminescence system (Amersham Biosciences, Buckinghasmshire, UK).
Enzyme-linked immunosorbent assay (ELISA)
Human immunoglobulin kappa light chain in murine sera was measured using human kappa ELISA kit (29). RANKL levels in conditioned media of HPSE-low and HPSE-high cells, as well as U266 and MM.1S myeloma cells treated with rHPSE were measured using human RANKL ELISA kit. Though heparanase transfection does not affect cell proliferation in vitro (19, 28) we seeded an equal number of cells at the beginning of each experiment and counted them again when the conditioned medium was collected. RANKL levels in the media were then normalized to cell number.
Osteoclast differentiation assay
The effect of medium conditioned by HPSE-low and HPSE-high CAG cells on osteoclastogenesis was determined as previously described (23). Medium was harvested 48 hours after cell plating, diluted 50% with αMEM, and added to the cultures of human osteoclast precursors. Macrophage colony-stimulating factor (mCSF; 25 ng/mL) was present in all groups. rRANKL (25 ng/mL) was used as a positive control. Cell cultures were maintained at 37°C and half of the medium in each well was replaced with fresh medium thrice weekly. After 10 days of culture, the cells were fixed with 10% formalin. TRAP staining was utilized for quantification of multinucleated osteoclasts (23).
Statistical analysis
Statistical significance between each two experimental groups was analyzed by Student’s t test, and the correlations between heparanase and RANKL expression and between heparanase and OPG expression in MM patients’ samples were assessed using Spearman correlation coefficient. P<0.05 was considered statistically significant.
Results
Heparanase dramatically enhances osteolysis locally and systemically in animal models of multiple myeloma
To explore the effect of heparanase expression in myeloma related bone disease, two animal models were utilized: the SCID-hu model in which myeloma tumor cells are grown within human bone implanted in mice, and the SCID-tibia model in which tumor cells are injected directly into the tibia of SCID mice. In SCID-hu mice, although tumors were allowed to grow to a larger size in the HPSE-low group than in the HPSE-high group (as shown on x-ray image and as determined by human kappa light chain analysis, Fig. 1A, left panel), bones injected with HPSE-low cells retained a normal spongy appearance with only minor osteolysis (arrows mark osteolytic areas, Fig. 1A, middle panel). In contrast, bones injected with HPSE-high cells showed massive destruction. Thus, the elevated bone destruction in the animals bearing tumors formed from HPSE-high cells was not simply due to high tumor burden. Rather, it points to a key role for heparanase in the osteolytic phenotype. In addition to the human bone that was injected with tumor cells, there was a second human bone implanted in each animal that did not receive injection of tumor cells. Importantly, in the animals bearing tumors formed by HPSE-high cells, extensive osteolysis also appeared in this uninjected bone. In contrast, uninjected bones from animals bearing HPSE-low tumor cells exhibited no evidence of osteolysis (Fig. 1A, right panel). Immunohistochemical staining for human kappa light chain, which specifically detects myeloma cells, revealed no detectable tumor cells in these uninjected contralateral bones (data not shown). Together these results indicate that heparanase dramatically enhances osteolysis within the tumors growing in bone and can also stimulate osteolysis in distal bones prior to the arrival of tumor cells at those sites.
Figure 1. Elevated expression of heparanase by myeloma cells dramatically enhances osteolysis locally and systemically in animal models.
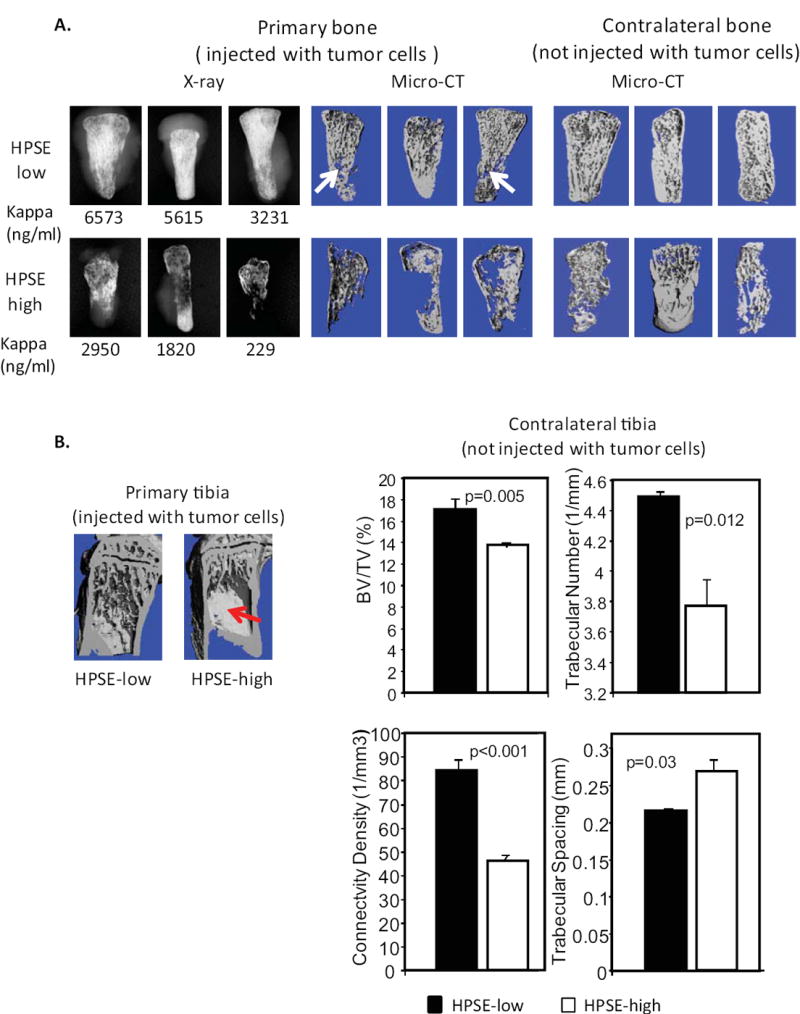
A. Imaging of human bones harvested from the SCID-hu model (7 mice per group). Left panel: X-ray image showing three most highly degraded bones from each group that were injected with either HPSE-low or HPSE-high cells. Numbers below the images are the ng/ml of human kappa light chain present in the murine serum, which reflects the whole animal tumor burden. Middle panel: MicroCT imaging of the same bones shown in the x-ray. Right panel: human bones contralateral to those shown in the left panel. B. Left panel: MicroCT imaging of a primary murine tibiae injected with tumor cells (HPSE-low and HPSE-high) Significant bone destruction is observed only in the tibia bearing HPSE-high tumor cells (red arrow, note loss of trabecular architecture) (5 mice per group). Right panel: MicroCT microarchitectural analyses of contralateral tibiae of mice injected with either HPSE-low or HPSE-high tumor cells. Significant decreases in bone volume/total volume (BV/TV), trabecular number, connectivity density, and increased trabecular spacing are observed n animals bearing HPSE-high tumors, as compared to animals bearing HPSE-low tumors.
To confirm these findings in a different animal model, we implanted tumor cells directly within the tibia of mice (SCID-tibia model). HPSE-high cells again induced severe bone destruction, whereas little damage was observed in bones injected with HPSE-low cells (Fig. 1B, left panel). MicroCT analyses of the contralateral non-injected tibia demonstrated that in mice bearing HPSE-high tumors, the bone volume, trabecular number and connectivity density were all significantly decreased, while trabecular spacing was increased compared to mice bearing HPSE-low tumors (Fig.1B, right panel). Immunostaining for human kappa light chain confirmed the absence of metastasized tumor cells in the distal tibias.
The enhanced bone destruction in both injected and contralateral bones in two animal models suggest that heparanase activates both local and systemic upregulation of osteoclastogenesis in vivo. To test this idea, the number of osteoclasts in bones harvested from SCID-hu animals was assessed by TRAP staining. In addition, we also measured levels of human TRAP 5b (a soluble marker that reflects the level of human bone resorption (23)) in the serum of SCID-hu mice bearing tumors. The number of TRAP positive cells (osteoclasts) in both the injected bone and the contralateral bone were significantly higher in animals injected with HPSE-high cells compared to animals with HPSE-low cells (Fig. 2A and 2B). Similarly, levels of human TRAP 5b were also markedly elevated in the serum of SCID-hu mice bearing HPSE-high tumors (Fig. 2C), whereas murine soluble TRAP levels were unchanged. These results demonstrate that the expression of heparanase by the myeloma tumor cells promotes bone resorption by driving an increase in both local and systemic osteoclastogenesis.
Figure 2. Osteoclastogenesis is enhanced in mice bearing HPSE-high tumors.
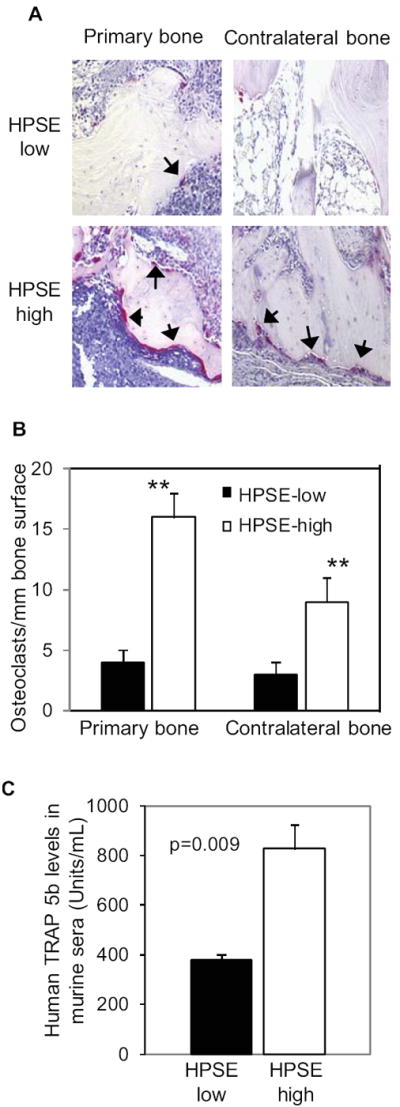
A. TRAP staining was performed on the primary and contralateral bones harvested from SCID-hu mice. The purple-red stained cells are osteoclasts (indicated by arrows; original magnification 200x). B. Numbers of TRAP-positive osteoclasts in human bones harvest from the SCID-hu model were determined. ** p<0.01 for HPSE-high compared with HPSE-low. C. TRAP 5b present in the murine sera is significantly higher in animals bearing HPSE-high tumors compared with HPSE-low tumors.
RANKL expression is enhanced in myeloma tumors formed by cells expressing high levels of heparanase
Because RANKL is a primary stimulator of osteoclastogenesis, we compared the level of RANKL expression in HPSE-low and HPSE-high tumors. Immunostaining of human bones injected with tumor cells (SCID-hu mice) revealed that the HPSE-high tumors had much higher RANKL levels than the tumors formed by HPSE-low cells (Fig. 3A). In addition, western blots confirmed a higher level of RANKL protein was present in subcutaneous tumors formed by the HPSE-high cells as compared to HPSE-low cells (Fig. 3B and 3C). This demonstrates that RANKL expression is enhanced in HPSE-high tumors even if they are not growing within the bone marrow microenvironment. Interestingly, levels of osteoprotegerin (OPG, the RANKL competitive inhibitor) were not significantly altered in HPSE-high tumors (Fig. 3B and 3C).
Figure 3. RANKL expression is elevated in tumors formed by HPSE-high cells.
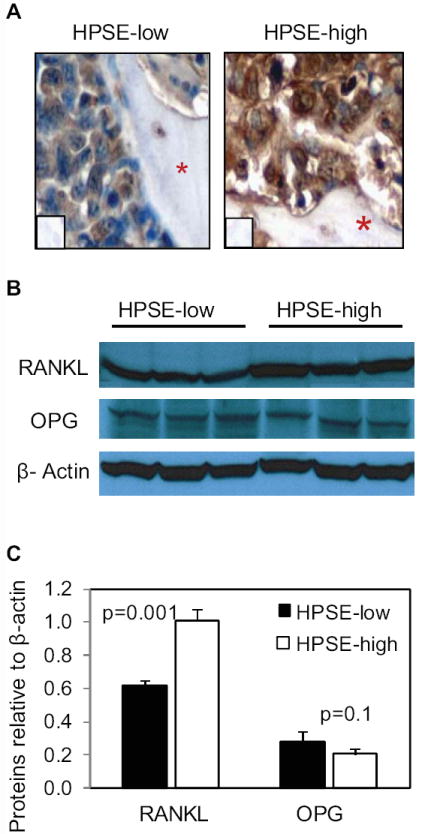
A. Human RANKL staining within human bones from SCID-hu mice bearing tumors formed by HPSE-low or HPSE-high cells. The red asterisks denote bone and inserts are controls lacking addition of primary antibody (original magnification 400x). B. Primary myeloma tumors formed by HPSE-low or HPSE-high cells growing subcutaneously in SCID mice were extracted with detergent and western blotting was performed for detection of RANKL, OPG, and β-actin. Each lane of the blot represents tumor from a different animal. C. The bands from western blots were quantified relative to β-actin using ImageJ software. The bars represent means ± SD.
Heparanase enhances RANKL expression and secretion in myeloma cells
Real-time PCR and western blots of the extracts from HPSE-low and HPSE-high cells growing in vitro confirmed that RANKL expression was increased in HPSE-high cells at both mRNA and protein levels, compared to HPSE-low cells (Fig. 4). Furthermore, RANKL expression was decreased in CAG cells following knockdown of heparanase. Note that levels of RANKL expressed by the HPSE-high CAG cells are even higher than those expressed by Saos-2 cells. This is important since Saos-2 cells are known to express high levels of RANKL and often serve as a positive control for RANKL assays (35). Thus, the heparanase-expressing CAG cells are making substantial amounts of RANKL as indicated by the immunohistochemistry results shown in Fig. 3.
Figure 4. Heparanase upregulates RANKL expression and secretion in myeloma cells.
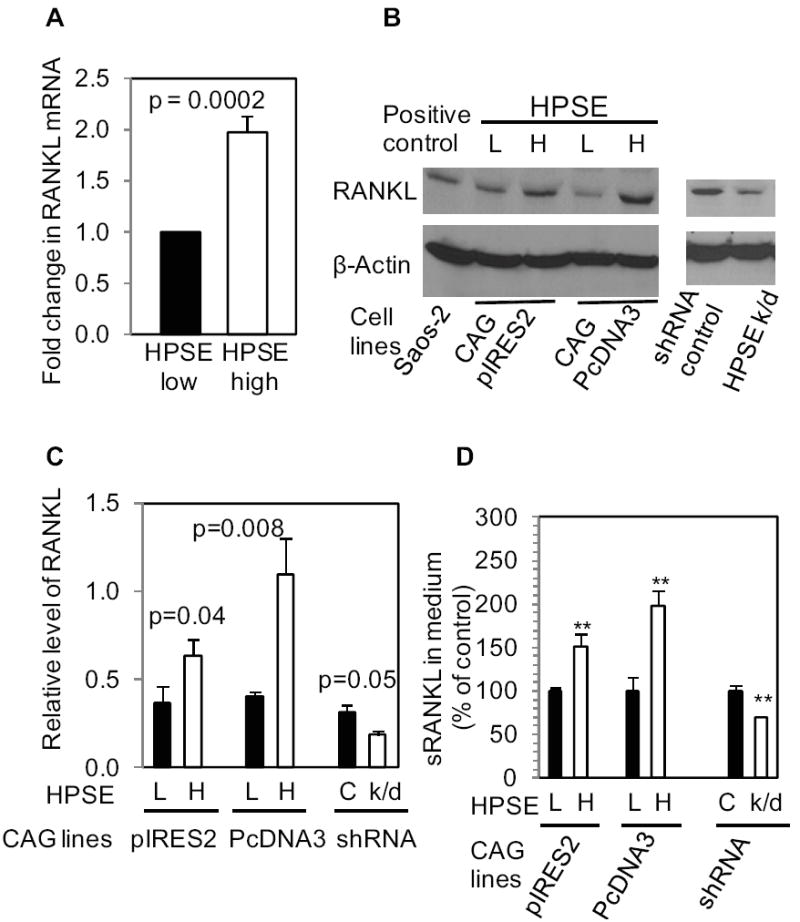
A. Real-time PCR for human RANKL. The graph is representative of three individual experiments performed in triplicate. B. CAG cells from two different heparanase transfections using different vectors (pIRES2 or pcDNA3) or from heparanase knock-down (HPSE k/d) cells were lysed and probed for human RANKL by Western blotting((HPSE-low (L), HPSE-high (H), HPSE knock-down (k/d)). C. Quantification of the RANKL from Western blots. Each bar graph represents an average reading number of RANKL/β-actin from 3 independent experiments. D. RANKL levels in conditioned medium were determined by ELISA. The bar graph shows the changes in RANKL levels in HPSE-high cells or HPSE k/d cells as the percentage of control. The results (representative of 3 independent experiments) are means ± SD, **p < 0.01 compared to controls.
Analysis of medium conditioned by CAG cells demonstrated that RANKL secretion was elevated in the medium of HPSE-high cells and reduced in the medium of HPSE-knockdown cells, compared to the corresponding controls (Fig. 4D). OPG was undetectable in either the cells or the conditioned medium, confirming that myeloma cells do not express OPG, which is consistent with previously published data (2, 36). In addition, IL-8, a chemokine known to promote osteoclastogenesis, was not detectable by ELISA of cell extracts or condition medium of either CAG HPSE-low or HPSE-high cells (data not shown).
To ensure that the heparanase induced upregulation of RANKL in CAG cells was not cell type-specific or a non-specific consequence of cell transfections or viral infection, we also determined the effects of heparanase on myeloma cell lines U266 and MM.1S by treating these cells with recombinant human heparanase (rHPSE). Previous studies have shown that rHPSE when added exogenously to cells is taken up by the cells and remains biologically active (20, 28, 37). As shown in Fig. 5, after the addition of rHPSE, the expression and secretion of RANKL was significantly increased in both U266 and MM.1S cells, confirming findings with the CAG myeloma cells.
Figure 5. Recombinant heparanase (rHPSE) induces RANKL expression and secretion in myeloma cells.
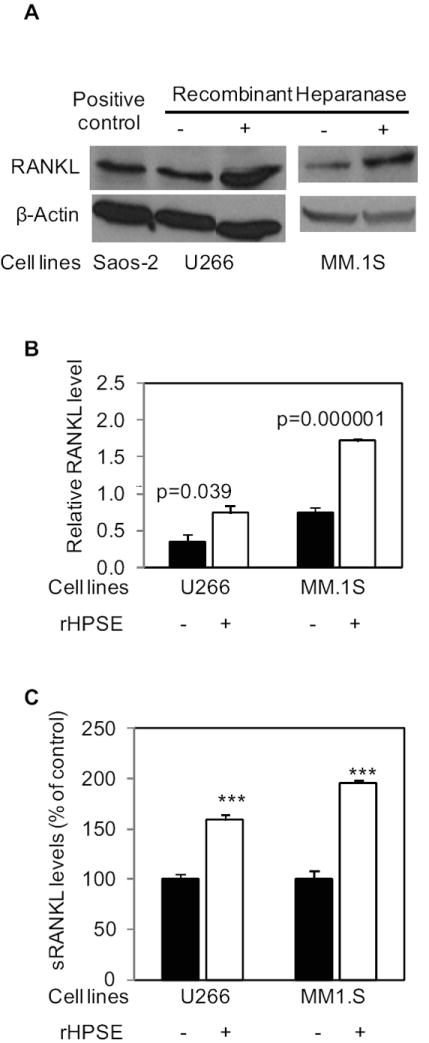
Equal numbers of human myeloma U266 or MM.1S cells were grown in complete medium in the absence or presence of rHPSE (100 ng/ml, added every 12 hours) for 48 h. The cells were lysed for western blotting, and the conditioned media were harvested for ELISA. A. Western blots were probed with antibody to human RANKL or β-actin. Saos-2 cell lysate was used as a positive control for human RANKL. B. RANKL bands from western blots were quantified using ImageJ and normalized to the corresponding β-actin bands. Each bar graph represents an average quantitative result of two independent experiments with two western blots. C. RANKL ELISA of conditioned media. The graph shows the percentage increase in RANKL present in medium conditioned by cells treated with rHPSE compared to control treated cells. The results are means ± SD from 2 independent experiments, *** p<0.001 to the controls.
Conditioned medium from HPSE-high cells induces osteoclastogenesis via RANKL
The finding that osteolysis is enhanced in tumor free bones that are contralateral to bones injected with tumor cells in both SCID-hu and SCID-tibia animals (Fig. 1) suggests that osteoclastogenesis is being enhanced by a factor or factors released into the systemic circulation by HPSE-high cells. To test this idea, in vitro osteoclastogenesis assays were utilized to measure the osteoclastogenic activity of conditioned medium from HPSE-high and HPSE-low CAG myeloma cells. The results revealed that the medium conditioned by HPSE-high cells significantly enhanced osteoclastogenesis in vitro as compared to medium from HPSE-low cells (Fig. 6). Moreover this enhancement was significantly inhibited by addition of OPG, thus confirming that the enhanced osteoclastogenic activity in the medium is mediated by RANKL (Fig. 6).
Figure 6. Medium conditioned by CAG HPSE-high cells induces osteoclastogenesis via increased RANKL.
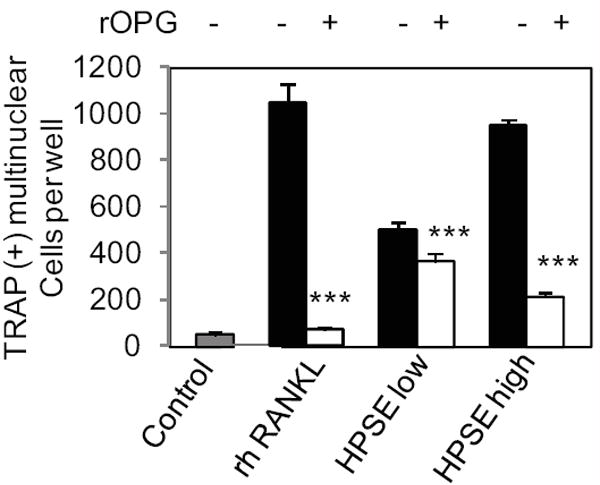
Human PBMCs were cultured in conditioned medium from CAG cells expressing either low or high levels of heparanase (HPSE-low or HPSE-high) in the presence or absence of OPG. Addition of recombinant human (rh) RANKL was used as a positive control. The negative control was cells cultivated only with DMEM and mCSF. *** p< 0.001 compared to “no OPG” group.
The level of RANKL expression correlates with the level of heparanase expression in tumor cells growing in the bone marrow of myeloma patients
Immunohistochemical staining of heparanase and RANKL was performed on 20 bone marrow core biopsy specimens of myeloma patients. A significant positive correlation (p<0.0001) between expression of heparanase and RANKL by the myeloma tumor cells was observed (Fig 7 and table 1 shown in Supplemental material). Interestingly, the subcellular localization of heparanase and RANKL within the bone marrow myeloma cells also correlated significantly (p<0.0001). Variable expression of heparanase and RANKL was also noted on bone marrow cells including osteoblasts (data not shown). As expected, the expression of OPG was detected in bone marrow cells but not in myeloma cells. However, there was no significant correlation between the expression of heparanase and OPG within the bone marrow (p=0.22).
Figure 7. Myeloma tumor cell expression of heparanase correlates with the level of RANKL in the bone marrow of myeloma patients.
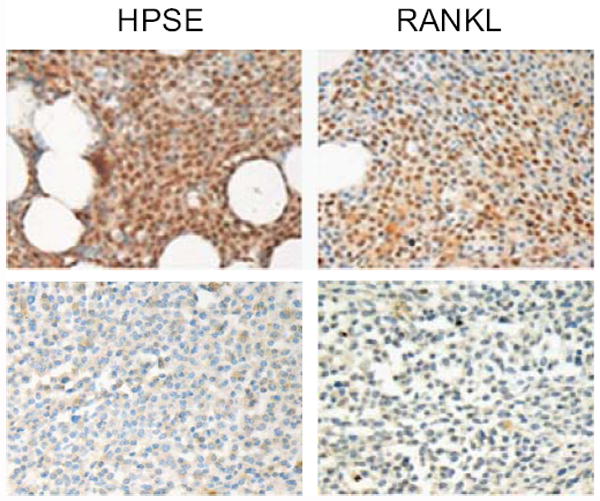
Heparanase and RANKL staining were performed on 20 of bone marrow biopsy specimens from myeloma patients. Myeloma cells expressing high level of heparanase show elevated RANKL expression (original magnification 400x).
Discussion
Although several studies have implicated heparanase in playing a role in bone turnover, definitive studies examining the role of heparanase in osteolytic cancers have not been reported (23-25). We demonstrate in the present work that when tumors are growing in bone and expressing high levels of heparanase, there is a marked increase in both systemic and local osteolysis as compared to controls. This increased osteolysis is not the result of an increase in the tumor size mediated by the heparanase. Rather, it is due, at least in part, to an increase in expression and secretion of RANKL leading to enhanced osteoclastogenesis. Evidence of a role for RANKL in heparanase driven osteolysis includes the detection of high levels of RANKL within tumors formed by heparanase-high CAG myeloma cells and the fact that elevated RANKL in medium conditioned by heparanase-high CAG cells drives osteoclastogenesis in vitro. Enhanced osteoclastogenesis in animals bearing heparanase-high tumors was confirmed by the presence of elevated levels of TRAP5b and by the presence of elevated numbers of osteoclasts both at the injected tumor site (primary tumor) and in distal bones where no tumor cells were injected or detected. In fact, the measurements of osteoclast number per bone surface are likely underrepresented, because in the heparanase-high CAG tumors, bone destruction is rampant and little bone (with resident osteoclasts) remain, thus contributing to the wide variation in osteoclast numbers observed (although the difference is still significant).
Multiple signaling pathways are involved in RANKL expression and we do not yet know how the expression of heparanase upregulates RANKL levels. We have previously examined the activation status of several signaling mediators—including Src, p38 MAPK, and ERK—in HPSE-low and HPSE-high cells. We found that levels of phosphorylated Src and phosphorylated p38 were not affected by an elevation in heparanase expression; but ERK activation was significantly greater in the HPSE-high cells (28). Thus, this upregulation of ERK signaling may be involved in heparanase-induced RANKL expression in myeloma cells. This and other possibilities await further investigation.
Furthermore, the analysis of primary human myeloma bone marrow biopsies confirmed that patients having high levels of heparanase expression also express higher levels of RANKL than do patients expressing low levels of heparanase. These clinical correlations are important since osteolytic bone disease is found in over 80% of newly diagnosed myeloma patients, is the most debilitating manifestation of this cancer and is a strong indicator of poor overall survival in myeloma (38, 39). Moreover, as a consequence of tumor osteolysis, factors may be released that drive tumor growth and further stimulate disease progression (3). The discovery of a role for heparanase in the cascade of events leading to osteolysis underscores its important role in regulating the tumor microenvironment and contributes to our understanding of the contribution of heparanase with poor prognosis in myeloma (18).
Our finding that heparanase enhances RANKL expression is consistent with the elevation of RANKL and systemic osteolysis observed in many myeloma patients. The hyperstimulation of osteoclastogenesis that occurs in myeloma is primarily mediated via RANKL. In myeloma, both the myeloma tumor cells and the bone marrow stroma of patients exhibit elevated levels of RANKL expression (40-45). The high levels of membrane-associated RANKL by myeloma cells have been correlated with the presence of multiple bone lesions in myeloma patients (44, 45). In addition, a soluble form of RANKL, released from tumor cells by protease activity, can diffuse away from the local tumor microenvironment to promote widespread osteoclast activation, thereby likely contributing to the systemic bone loss that is also a feature of myeloma (46). This idea is consistent with the present study where both local and systemic osteolysis is observed following upregulation of RANKL by the tumors formed from heparanase-high cells. Interestingly, we observed that in heparanase-high tumors growing in bone (SCID-hu model) there were much higher levels of RANKL than in heparanase-high tumors growing subcutaneously (Figure 3). This finding indicates that the increased expression of heparanase by myeloma cells may also affect bone marrow stromal cells and osteoblasts, resulting in increased RANKL expression from these cells. These possibilities remain to be explored.
The relationship between elevated heparanase expression and increased myeloma osteolysis is of significance because most patients express active heparanase enzyme within their bone marrow (21). Expression of heparanase within the myeloma bone marrow cells has been demonstrated by Affymetrix gene arrays where 92% of the 39 patients tested expressed heparanase mRNA in whole bone marrow samples (18). This study confirmed the presence of heparanase expression by myeloma tumor cells and demonstrated that heparanase was also expressed by monocytes and osteoclasts within the bone marrow-tumor microenvironment. As demonstrated in Fig. 5, heparanase released by cells can be taken up and utilized by neighboring cells. Thus, it is conceivable that bone marrow-derived heparanase in patients may be taken up by neighboring myeloma tumor cells where it stimulates RANKL expression. Interestingly, in a previous study, upregulated heparanase expression was also detected in osteoclasts in a murine model of bone fracture repair (47). Thus, during osteolysis that is characteristic of myeloma bone disease, heparanase produced directly by osteoclasts may contribute to events leading to bone resorption.
Our finding that heparanase stimulates RANKL expression, leading to both local and systemic osteolysis adds another important function of heparanase in regulating the myeloma microenvironment and promoting the aggressive disease phenotype. It will be important to determine if inhibitors of heparanase which have been shown to block myeloma growth (30) can also interfere with myeloma-induced osteolysis in vivo.
Supplementary Material
Acknowledgments
The authors thank Dr. Israel Vlodavsky (Technion, Haifa, Israel) for providing rHPSE and Dr. Renee Desmond (Department of Medicine and Comprehensive Cancer Center, UAB) for the statistical analysis of the data.
Grant Support NIH grant CA135075 and CA138340 (to RDS), a Multiple Myeloma Research Foundation (MMRF) Senior Research Award (to YY), the Carl L. Nelson Chair in Orthopaedic Creativity (to LJS) and a MMRF Research Fellow Award (to VCR).
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Jagannath S. Pathophysiological underpinnings of multiple myeloma progression. J Manag Care Pharm. 2008;14:7–11. doi: 10.18553/jmcp.2008.14.S7-A.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roodman GD. Pathogenesis of myeloma bone disease. Leukemia. 2009;23:435–41. doi: 10.1038/leu.2008.336. [DOI] [PubMed] [Google Scholar]
- 3.Edwards CM, Zhuang J, Mundy GR. The pathogenesis of the bone disease of multiple myeloma. Bone. 2008;42:1007–13. doi: 10.1016/j.bone.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanderson RD, Epstein J. Myeloma bone disease. J Bone Miner Res. 2009;24:1783–8. doi: 10.1359/jbmr.090901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends in Bioch Sci. 2009;34:511–9. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Dang YW, Luo DZ, Feng ZB, Tang XL. Expression of heparanase in hepatocellular carcinoma has prognostic significance: a tissue microarray study. Oncol Res. 2008;17:183–9. doi: 10.3727/096504008785114138. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann AC, Mori R, Vallbohmer D, et al. High expression of heparanase is significantly associated with dedifferentiation and lymph node metastasis in patients with pancreatic ductal adenocarcinomas and correlated to PDGFA and via HIF1a to HB-EGF and bFGF. J Gastrointest Surg. 2008;12:1674–81. doi: 10.1007/s11605-008-0628-2. [DOI] [PubMed] [Google Scholar]
- 9.Cohen E, Doweck I, Naroditsky I, et al. Heparanase is overexpressed in lung cancer and correlates inversely with patient survival. Cancer. 2008;113:1004–11. doi: 10.1002/cncr.23680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shafat I, Pode D, Peretz T, Ilan N, Vlodavsky I, Nisman B. Clinical significance of urine heparanase in bladder cancer progression. Neoplasia. 2008;10:125–30. doi: 10.1593/neo.07875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerner I, Baraz L, Pikarsky E, et al. Function of heparanase in prostate tumorigenesis: potential for therapy. Clin Cancer Res. 2008;14:668–76. doi: 10.1158/1078-0432.CCR-07-1866. [DOI] [PubMed] [Google Scholar]
- 12.Cohen I, Pappo O, Elkin M, et al. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int J Cancer. 2006;118:1609–17. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 13.Mahtouk K, Hose D, Raynaud P, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–23. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T, Yamaguchi A, Goi T, et al. Heparanase expression in human colorectal cancer and its relationship to tumor angiogenesis, hematogenous metastasis, and prognosis. J Surg Oncol. 2004;87:174–81. doi: 10.1002/jso.20097. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Quiros RM, Maxhimer JB, et al. Inverse correlation between heparan sulfate composition and heparanase-1 gene expression in thyroid papillary carcinomas: a potential role in tumor metastasis. Clin Cancer Res. 2003;9:5968–79. [PubMed] [Google Scholar]
- 16.El-Assal ON, Yamanoi A, Ono T, Kohno H, Nagasue N. The clinicopathological significance of heparanase and basic fibroblast growth factor expressions in hepatocellular carcinoma. Clin Cancer Res. 2001;7:1299–305. [PubMed] [Google Scholar]
- 17.Koliopanos A, Friess H, Kleeff J, et al. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–9. [PubMed] [Google Scholar]
- 18.Mahtouk K, Hose D, Raynaud P, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–23. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Macleod V, Bendre M, et al. Heparanase promotes the spontaneous metastasis of myeloma cells to bone. Blood. 2005;105:1303–9. doi: 10.1182/blood-2004-06-2141. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Macleod V, Miao HQ, et al. Heparanase enhances syndecan-1 shedding: A novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–33. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 21.Kelly T, Miao HQ, Yang Y, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–56. [PubMed] [Google Scholar]
- 22.Purushothaman A, Uyama T, Kobayashi F, et al. Heparanase enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood: Epub ahead of print PMID: 20097882. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly T, Suva LJ, Huang Y, et al. Expression of heparanase by primary breast tumors promotes bone resorption in the absence of detectable bone metastases. Cancer Res. 2005;65:5778–84. doi: 10.1158/0008-5472.CAN-05-0749. [DOI] [PubMed] [Google Scholar]
- 24.Kelly T, Suva LJ, Nicks KM, Macleod V, Sanderson RD. Tumor-derived syndecan-1 mediates distal cross-talk with bone that enhances osteoclastogenesis. J Bone Miner Res. 25:1295–304. doi: 10.1002/jbmr.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Song B, Qin WJ, et al. Heparanase promotes bone destruction and invasiveness in prostate cancer. Cancer Lett. 2008;268:252–9. doi: 10.1016/j.canlet.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Kram V, Zcharia E, Yacoby-Zeevi O, et al. Heparanase is expressed in osteoblastic cells and stimulates bone formation and bone mass. J Cell Physiol. 2006;207:784–92. doi: 10.1002/jcp.20625. [DOI] [PubMed] [Google Scholar]
- 27.Borset M, Hjertner O, Yaccoby S, Epstein J, Sanderson RD. Syndecan-1 is targeted to the uropods of polarized myeloma cells where it promotes adhesion and sequesters heparin-binding proteins. Blood. 2000;96:2528–36. [PubMed] [Google Scholar]
- 28.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283:32628–36. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Yaccoby S, Liu W, et al. Soluble syndecan-1 promotes growth of myeloma tumors in vivo. Blood. 2002;100:610–7. doi: 10.1182/blood.v100.2.610. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, MacLeod V, Dai Y, et al. The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood. 2007;110:2041–8. doi: 10.1182/blood-2007-04-082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hildebrand T, Laib A, Muller R, Dequeker J, Ruegsegger P. Direct three-dimensional morphometric analysis of human cancellous bone: microstructural data from spine, femur, iliac crest, and calcaneus. J Bone Miner Res. 1999;14:1167–74. doi: 10.1359/jbmr.1999.14.7.1167. [DOI] [PubMed] [Google Scholar]
- 32.Abdallah BM, Stilgren LS, Nissen N, Kassem M, Jorgensen HR, Abrahamsen B. Increased RANKL/OPG mRNA ratio in iliac bone biopsies from women with hip fractures. Calcif Tissue Int. 2005;76:90–7. doi: 10.1007/s00223-004-0074-4. [DOI] [PubMed] [Google Scholar]
- 33.Lee K, Qian DZ, Rey S, Wei H, Liu JO, Semenza GL. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc Natl Acad Sci U S A. 2009;106:2353–8. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Heider U, Zavrski I, Jakob C, et al. Expression of receptor activator of NF-kappaB ligand (RANKL) mRNA in human multiple myeloma cells. J Cancer Res Clin Oncol. 2004;130:469–74. doi: 10.1007/s00432-004-0578-3. [DOI] [PubMed] [Google Scholar]
- 36.Edwards CM, Zhuang J, Mundy GR. The pathogenesis of the bone disease of multiple myeloma. Bone. 2008;42:1007–13. doi: 10.1016/j.bone.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Sanderson RD. Heparanase regulates levels of syndecan-1 in the nucleus. PLoS ONE. 2009;4:e4947. doi: 10.1371/journal.pone.0004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakob C, Sterz J, Liebisch P, et al. Incorporation of the bone marker carboxy-terminal telopeptide of type-1 collagen improves prognostic information of the International Staging System in newly diagnosed symptomatic multiple myeloma. Leukemia. 2008;22:1767–72. doi: 10.1038/leu.2008.159. [DOI] [PubMed] [Google Scholar]
- 39.Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 Patients With Newly Diagnosed Multiple Myeloma. Mayo Clinic Proceedings. 2003;78:21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 40.Lai FPL, Cole-Sinclair M, Cheng W-J, et al. Myeloma cells can directly contribute to the pool of RANKL in bone bypassing the classic stromal and osteoblast pathway of osteoclast stimulation. British Journal of Haematology. 2004;126:192–201. doi: 10.1111/j.1365-2141.2004.05018.x. [DOI] [PubMed] [Google Scholar]
- 41.Yaccoby S, Pearse RN, Johnson CL, Barlogie B, Choi Y, Epstein J. Myeloma interacts with the bone marrow microenvironment to induce osteoclastogenesis and is dependent on osteoclast activity. Br J Haematol. 2002;116:278–90. doi: 10.1046/j.1365-2141.2002.03257.x. [DOI] [PubMed] [Google Scholar]
- 42.Pearse RN, Sordillo EM, Yaccoby S, et al. Multiple myeloma disrupts the TRANCE/ osteoprotegerin cytokine axis to trigger bone destruction and promote tumor progression. Proc Natl Acad Sci U S A. 2001;98:11581–6. doi: 10.1073/pnas.201394498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barille-Nion S, Bataille R. New insights in myeloma-induced osteolysis. Leuk Lymphoma. 2003;44:1463–7. doi: 10.3109/10428190309178765. [DOI] [PubMed] [Google Scholar]
- 44.Heider U, Langelotz C, Jakob C, et al. Expression of Receptor Activator of Nuclear Factor {kappa}B Ligand on Bone Marrow Plasma Cells Correlates with Osteolytic Bone Disease in Patients with Multiple Myeloma. Clin Cancer Res. 2003;9:1436–40. [PubMed] [Google Scholar]
- 45.Farrugia AN, Atkins GJ, To LB, et al. Receptor Activator of Nuclear Factor-{kappa}B Ligand Expression by Human Myeloma Cells Mediates Osteoclast Formation in Vitro and Correlates with Bone Destruction in Vivo. Cancer Res. 2003;63:5438–45. [PubMed] [Google Scholar]
- 46.Wilson TJ, Nannuru KC, Futakuchi M, Sadanandam A, Singh RK. Cathepsin G enhances mammary tumor-induced osteolysis by generating soluble receptor activator of nuclear factor-kappaB ligand. Cancer Res. 2008;68:5803–11. doi: 10.1158/0008-5472.CAN-07-5889. [DOI] [PubMed] [Google Scholar]
- 47.Saijo M, Kitazawa R, Nakajima M, Kurosaka M, Maeda S, Kitazawa S. Heparanase mRNA expression during fracture repair in mice. Histochem Cell Biol. 2003;120:493–503. doi: 10.1007/s00418-003-0589-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


