Abstract
M059J and M059K cells were isolated from different portions of the same human malignant glioma. M059J cells are more radiosensitive than M059K cells due to the absence of DNA-PKcs and low-expression of ATM. The mechanism concerning the absence of DNA-PKcs in M059J is due to the frameshift mutation in PRKDC (DNA-PKcs gene); however, the reason for the low expression of ATM in M059J cells remains unclear. We showed here that the main reason for the lower ATM level in M059J cells was not related to the transcriptional regulation or protein degradation but was related to post-transcriptional regulation. Based on database information, we found that the 3’-untranslational region (UTR) of ATM contains a miR-100-binding site. By using an RNase protection assay and qRT-PCR, we identified that miR-100 is highly-expressed in M059J cells. We further demonstrated that miR-100 bound to the 3’-UTR of ATM. Knocking-down miR-100 promotes ATM expression in M059J cells. Up-regulating miR-100 in M059K cells and other cancer cells reduces ATM expression and sensitizes these cells to ionizing radiation. These results indicate that ATM is a target of miR-100, elucidating that the low-expression of ATM in M059J cells is mainly due to the high-expression of miR-100. These results also suggest that miR-100 could be a useful tool to target ATM and sensitize tumor cells to ionizing radiation.
Keywords: ATM, miR-100, M059J, ionizing radiation, DNA damage
1. Introduction
The M059J and M059K cell lines were isolated from different portions of the same human malignant glioma biopsy specimen, while M059J cells are much more sensitive than M059K cells to radiation [1]. It was reported that the DNA-PK catalytic subunit (DNA-PKcs) was absent and ATM was low-expressed in the M059J cell line [2, 3], which is responsible for the radiosensitive feature of M059J cells. Ionizing radiation (IR) induced DNA double strand breaks (DSBs) are a severe threat for cell survival. There are two major pathways in mammalian cells to repair DNA DSBs: non-homologous end-joining (NHEJ) and homologous recombination repair (HRR). DNA-PKcs is a major component of NHEJ [4–6]. ATM is one of the most important checkpoint proteins in mammalian cells [7], which mainly promotes the HRR pathway [8, 9] although it is also partially involved in NHEJ [10]. The absence of DNA-PK is due to the frameshift mutation in PRKDC (the gene for DNA-PKcs in M059J cells) [11]; however, the low-expression of ATM in M059J cells remains unclear.
microRNAs (miRNAs), a class of small non-coding RNAs (ncRNAs) with ~22 nucleotides, are important post-transcriptional regulators in affecting different biological functions [12–14]. miRNAs bind to partially complementary sequences of 3’-UTR of mRNAs, targeting them for degradation and/or inhibiting translation. The importance of ncRNA including miRNA in the regulation of biological functions in mammalian cell has been more and more realized since ~98% of human genome is the non-coding sequence. It has been reported that most mammalian mRNAs are conserved targets of miRNAs [15]. In this study, after excluding the possibility of transcriptional and translational modification of ATM in M059J cells, we explored the main reason for the low level of ATM in M059J cells, which is related to the over-expression of miR-100. These data also suggest that miR-100 could be a useful tool to target ATM for many purposes.
2. Materials and methods
2.1 Plasmids construction
To construct a plasmid expressing miR-100, we amplified a DNA fragment carrying pri-miR-100, using genomic DNA from a healthy blood donor as a template, as we did for miR-145 previously [16] but with different primers (Supplementary information Table S1). The amplified fragment was first cloned into a PCR cloning vector and subsequently into the lentiviral vector: pCDHCMV-MCS-EF1-copGFP (System Biosciences, Mountain View, CA, USA) at the EcoR1 and NotI sites. Expression of miR-100 was verified by TaqMan® real-time RT-PCR. The luciferase-UTR reporter plasmid that contains the ATM 3’-UTR carrying a putative or a mutant miR-100 binding site was constructed as follows: Oligonucleotides (Invitrogen, Carlsbad, CA, USA) used in luciferase assay constructions were shown as in Supplementary Table S1. Briefly, complimentary oligonucleotides for each selected region containing either a putative or mutated hsa-miR-100 binding site in the 3’-UTR of ATM were hybridized to form double-stranded DNA and inserted into a pMIR-ReporterTM firefly luciferase vector (Applied Biosystems, Foster City, CA, USA) at the SacI and HindIII sites. All constructs were confirmed by sequencing.
2.2. PCR/RT-PCR and quantitative RT-PCR (qRT-PCR)
PCRs were performed to amplify pri-microRNA sequences or the ATM 3′-UTR sequence according to the standard three-step procedure. For RT-PCR, total RNA was isolated by using a Trizol reagent (Invitrogen, Carlsbad, CA, USA), and small RNA by using a miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). RNA (1 μg) was used to synthesize cDNA by using a TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed in triplicate with a TaqMan® Universal PCR Master Mix and a specific TaqMan® MicroRNA assay (Applied Biosystems) on an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). Samples were normalized to an RNU48 small RNA and relatively quantified using a 2−ΔΔCT method [17].
2.3. RNase protection assay
RNA probes for this experiment were constructed by PCR and in vitro transcription. Briefly, forward and reverse primers were designed to include a T7 promoter upstream to mature sequence (hsa-miR-100 and RNU48) with 10 over-lapping nucleotides (Supplementary information Table S1). Amplified PCR was purified using a QIAquick spin column (Qiagen) and proceeded with a Megashortscript™ kit (Ambion, Austin, TX, USA) for in vitro transcription reaction according to the manufacturer’s protocol. The RNA probes were hybridized to the total RNA from M059J or M059K cells with a mirVanaTM miRNA detection kit (Ambion) according to the manufacturer’s instruction. Gel was exposed directly to a phosphor screen overnight and the signals were detected by using a Typhoon™ 9210 (GE, Bio-Sciences, Piscataway, NJ, USA).
2.4. Cell lines and transfection/transduction
M059J and M059K cells were obtained from Dr. Allalunis-Turner’s laboratory [2]. U87MG and 293T cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The lung cancer cell lines, 95C and 95D were obtained from Dr. Lu’s laboratory [18]. 95C or 95D cells were directly co-transfected with the lentiviral vector-miR100 and the pCDHCMV-MCS-EF1 plasmid encoding a puromycin (Puro) marker (System Biosciences) at a ratio of 20:1 by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The Puro resistant colonies were selected and the miR-100 levels were measured by qRT-PCR. The glioma cell lines: U87MG or M059K cells were transduced by the packaged lentivirus. Briefly, approximately 2×106 293T cells were seeded in a 100 mm dish overnight. The lentiviral vector-miR-100 or lentiviral vector alone (2 μg) and pPACKH1 Packaging Plasmid Mix (10 μg) (System Biosciences) were transfected to 293T cells by using Lipofectamine™ 2000 according to the manufacturer’s instructions. The culture medium containing the packaged viruses was harvested at 48 hr after transfection and was spun at 4 ºC, 3000 rpm for 10 min. The supernatant was collected and polybrene was added to the final concentration 8 μg/ml. The mixture (5 ml) was added to the glioma cell culture in a 100 mm dish with 5 ml of medium. The transduced cells were harvested after 72–96 hr post-infection for further experiments. Cells transfection with 100 nM siRNA of PRKDC, ATM, Dicer (Santa Cruz Biotech Inc, Santa Cruz, CA, USA) or hsa-miR-100 inhibitor (Thermo Fisher Scientist Inc, Waltham, MA, USA) [19] was performed with the lipofectamineTM 2000 according to the manufacturer’s instructions. Cells were harvested at 36 hr after transfection for further experiments.
2.5. Antibodies and reagents
The DNA-PKcs antibody (MS-370-P1) was purchased from Thermo Fisher Scientific Inc (Waltham, MA, USA). The ATM antibody (2837S) and the mTor antibody (2927) were purchased from Cell Signaling (Danvers, MA, USA). The Ku70 antibody (SC-17789) was purchased from Santa Cruz Biotech Inc (Santa Cruz, CA, USA). Cycloheximide was purchased from Sigma-Aldrich Inc (St. Louis, MO, USA).
2.6. Luciferase assay
293T cells were transfected with the appropriate plasmids with or without 100 nM hsa-miR-100 mimics (Thermo Fisher Scientific, Pittsburgh, PA, USA) in 48-well plates. The cells were harvested 48 h after transfection, lysed and analyzed with a luciferase assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol and were measured on a luminescence microplate reader LUMIstar Galaxy (BMG labtechnologies, Cary, NC, USA). β-galactosidase or renilla luciferase was used for normalization.
2.7. Cell survival assay
Cell sensitivity to radiation was determined by the loss of colony-forming ability. Briefly, after the cells were irradiated by using an x-ray machine (X-RAD 320, N. Branford, CT, USA) at 320 kV, 10 mA, with the filtration of 2-mm aluminum. The dose rate was 2 Gy/min. After IR, the cells were collected and plated, aiming at a density of 20–100 colonies per dish. Two replicate dishes were prepared for each datum point, and cells were incubated for 2 weeks to allow colonies to develop. Colonies were stained with crystal violet (100% methanol solution) before counting.
2.8. Statistical analysis
Statistical analysis of data was done using the Student's t test. Differences with p < 0.05 are considered significant.
3. Results and Discussion
3.1. miR-100 is over-expressed in M059J cells
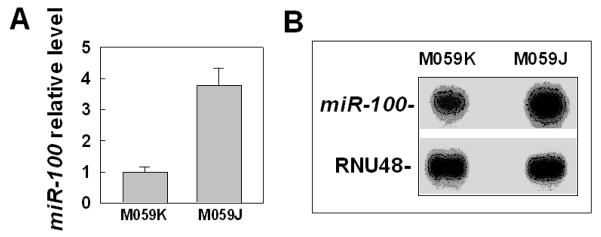
To look for the main reason for the low level of ATM in M059J cells, we first tested whether there was any difference in ATM during the transcriptional process between M059J and M059K cells by comparing the ATM mRNA levels in the two cell lines. The results showed that there was no apparent difference in ATM mRNA levels between M059J and M059K cells (Supplemantary information Figure S1). Further real-time RT-PCR data confirmed the results (data no shown), which is consistent to the previous report [20], indicating that the low level of ATM in M059J cells is not due to the low mRNA level. These results are different from previous report that the levels of mRNA and ATM protein were affected by DNA-PKcs [21], which might be due to the different cell lines that we detected. We next examined the post-translational degradation of ATM by testing the effects of cycloheximide (CHX) on ATM protein level changes at different times between M059J and M059K cells. The results showed there was no apparent difference in the ATM level change between M059J and M059K cells (Supporting information Figure S2), suggesting that the low level of ATM in M059J cells might not be due to the post-translational modification. These results led us to consider whether epigenetic modification plays any role for the low expression of ATM in M059J cells. The epigenetic modification mainly includes methylation and miRNA modification. We first tested the hypothesis that miRNAs might play a role for the low expression of ATM in M059J cells. For this purpose, we searched three databases (microCosm target, Targetscan and Pictar) for the miRNA candidates that could target the 3’-UTR of ATM. As a result, we found more than ten miRNAs that could be candidates. After comparing the expression levels of these miRNAs between M059J and M059K cells by using a real time PCR approach, we found that only miR-100 was over-expressed in M059J cells as compared with M059K cells (figure. 1A), suggesting that ATM might be the target of miR-100. The over-expression of miR-100 in M059J cells was further confirmed by an RNase protection assay (figure 1B). These results suggest that ATM might be the target of miR-100.
Fig. 1.
miR-100 is over-expressed in M059J cells. (A) Different expression of miR-100 between M059J and M059K cells was detected by using qRT-PCR. RNU48 was used as an internal loading control. (B) Different expression of miR-100 between M059J and M059K cells was detected by using an RNase protection assay. RNU48 was used as an internal loading control.
3.2. ATM is the target of miR-100
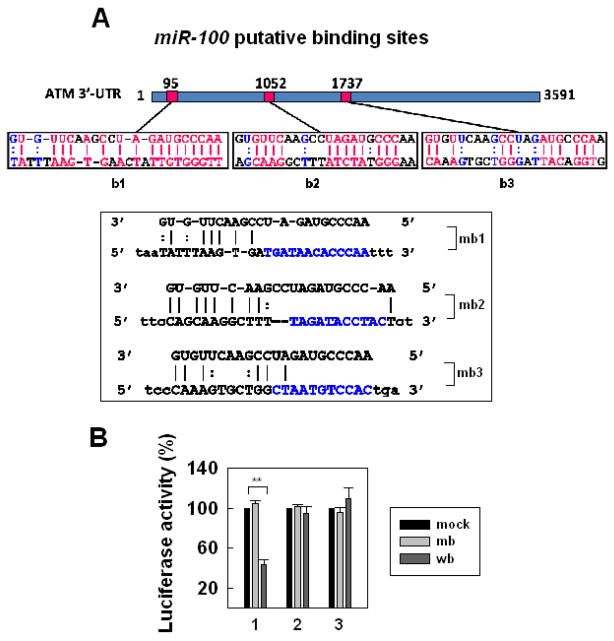
There are three putative miR-100 binding sites of the ATM 3’-UTR region (figure 2A). We made the constructs encoding the ATM 3’-UTR region carrying a putative miR-100 binding site and we labeled them as b1, b2 or b3; and the constructs containing a corresponding mutated site, we named as mb1, mb2 or-mb3 (figure 2A). To investigate whether ATM was the target of miR-100, we examined the effects of miR-100 on translation inhibition by using a luciferase assay with the vector encoding the putative (b1-b3) or mutant (mb1-mb3) miR-100 binding site of ATM 3’-UTR. The results showed that the translation activity was dramatically inhibited by the putative site of 3’-UTR of ATM, b1, otherwise, the translation activity was not affected at all by b2, b3 or mb1-mb3 that was mutated at the feed region (figure 2B). These results suggest that miR-100 inhibited ATM expression in M059J cells by targeting the specific b1 site of the 3’-UTR of ATM.
Fig. 2.
ATM is the target of miR-100. (A) Three putative regions of ATM 3’-UTR for miR-100 binding (b1, b2, b3) and corresponding mutant miR-100 binding sites (mb1, mb2, mb3) in which the seed sequences were mutated as indicated. (B) Effects of miR-100-binding sites on luciferase activity. 293T cells were co-transfected with the firefly luciferase reporter plasmid containing wild-type (b1, b2 or b3) or mutant ATM 3'-UTR (mb1, mb2 or mb3), with (b or mb) or without miR-100 (mock). Luciferase activity was assayed 48 h after transfection. The data represent mean ± SE of three independent experiments and normalized to their respective controls (mock) as 100%. **, P < 0.01, compared with cells treated with control RNA.
3.3. Over-expressed miR-100 is mainly responsible for the low-expression of ATM in M059J cells
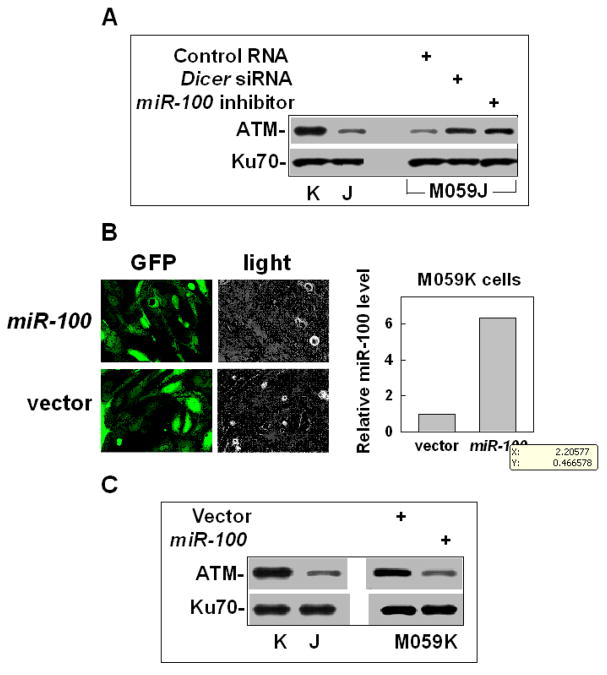
To investigate whether the over-expressed miR-100 in M059J cells is the main reason to inhibit ATM expression, we examined the effects of the miR-100 inhibitor or Dicer siRNA on the ATM expression in M059J and M059K cells. The results showed that when the expression of miR-100 or the miRNA forming process was inhibited in M059J, ATM was up-regulated (figure 3A), indicating that ATM is the target of miR-100. At the same time, we did not observe any apparent changes of ATM in M059K cells after the cells were treated with the miR-100 inhibitor or Dicer siRNA, which might be because the ATM level is normal in such cells and the cells might be less sensitive to any stimulator for further increasing the ATM level. To confirm the relationship between miR-100 and ATM, we made the construct encoding the pri-miR-100 in lentivirus vector (Supporting information Figure S3) and examined the effect of up-regulating miR-100 on the ATM expression in M059K cells. The results showed that when miR-100 was over-expressed in M059K cells (figure 3B), the level of ATM dramatically decreased (Fig. 3C). Similar results were observed from other glioma cell lines, U87MG cells and lung cancer cell lines, 95C and 95D cells (Supporting information Figure S4). These results confirm that the low expression of ATM in M059J cells is mainly due to the over-expression of miR-100. However, at this moment, we cannot exclude another possibility that methylation may also play a role in the low expression of ATM because the miR-100 inhibitor could not completely restore the ATM level of M059J cells shown in M059K cells (figure 3A), which needs future experiments to test. To address the question whether the levels of miR-100 and ATM was affected by DNA-PKcs, we detected the effects of the specific siRNA against PRKDC (encoding DNA-PKcs) on the levels of miR-100 and ATM in M059K cells. The results showed that neither the level of miR-100 nor the level of ATM protein changed after DNA-PKcs was efficiently knocked down in M059K cells (Supporting information Figure S5). These results exclude the possibility that the lower expression of ATM in M059J cells is a direct consequence of absent DNA-PKcs.
Fig. 3.
Over-expressed miR-100 is responsible for the low-expression in M059J cells. (A) The effects of the miR-100 inhibitor or the Dicer siRNA on ATM expression in M059J cells. Ku70 was used as an internal loading control. (B) Up-regulating miR-100 in M059K cells. The images reflect GFP signals, which represent the infection efficiencies of the lentivirus vectors. The miR-100 level was measured by qRT-PCR. (C) The effects of up-regulation of miR-100 on ATM expression in M059K cells.
At this moment, we still could not answer how miR-100 expression is regulated because there is no difference in the transcript sequence of miR-100 between M059J and M059K cells (data not shown), which needs more experiments to find the answer. We measured miR-100 levels in several brain tumor cell lines. The results show that the level of miR-100 varies in different cell lines (Supporting information Figure S6) although the levels of miR-100 were not affected by radiation (data not shown). The later results are consistent with that ATM activity is affected, but ATM expression level is not affected by the general stress including DNA damage response. The level of miR-100 in M059J is higher than in M059K but lower than in U87MG. The reason for the high level of miR-100 in U87MG cells not causing the lower level of ATM (compared with M059J) might be due to the heterogeneous features of cancer cell lines. Similar to MO59K cells, the inhibitor of miR-100 could not further increase the ATM level in U87MG cells (data not shown). This might be due to the same reason as mentioned above. The gene expression is regulated by many positive or negative factors including transcriptional factors, enhancers and inhibitors etc. These factors could be proteins or small non-coding RNA including miRNA. Most human genes are regulated by miRNA [15]. MiRNA genes make up ~1% of the human genomes [13]. Each miRNA has hundreds of mRNA targets, and individual mRNAs may be regulated by several miRNAs. The impact of this regulatory network on cellular physiology is conceivably enormous. Altered regulation of miRNAs is common in human cancers. Therefore, ATM expression is controlled by many factors. In this manuscript, we were interested in addressing why compared with M059K cells, the ATM level was so low in M059J cells since these two cell lines are derived from the same tumor specimen and their genotype backgrounds are supposed to be less heterogeneous. Next, we were interested in studying whether targeting ATM by miR-100 could sensitize the cells to ionizing radiation (IR)-induced killing because ATM plays an important role in promoting the HRR pathway [8, 9], and AT cells without the ATM function are very sensitive to IR-induced killing.
3.4. Targeting ATM by miR-100 sensitizes the cells to IR-induced killing
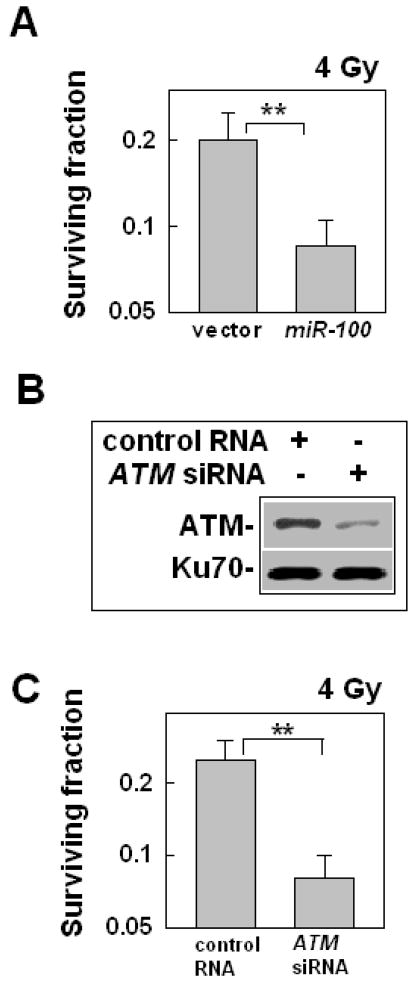
To determine the effect of miR-100 on cell sensitivity to IR, we used the clonogenic assay. The results showed that when miR-100 were up-expressed in M059K cells (figure 3B), the cells became more sensitive to IR than the cells transfected with the empty vector (figure 4A), suggesting that miR-100 could be used as a tool to sensitize cells to IR. mTOR is also a target of miR-100 [22], mTOR expression is lower in M059J cells than in M059K cells, and up-regulating miR-100 in M059K cells resulted in the down-regulation of mTOR in the cells (Supporting information Figure S7). To determine whether the low expression of mTOR by miR-100 in M059K also contributed to the effects of miR-100 on the sensitization of the cells to IR, we examined the effect of rapamycin, an mTor inhibitor, on cell radiosensitivity. The results showed that when mTOR in the cells was inhibited by rapamycin, the cells did not change their sensitivity to IR (data shown in another manuscript that we submitted). Based on these results, we could conclude that mTOR does not affect cell radiosensitivity and over-expression of miR-100 in the M059K cells induced-radiosensitivity is not due to the low-expression of mTOR. To confirm that the low-expression of ATM induced by the over-expression of miR-100 in M059K cells was the sole reason for the cell radiosensitization, we examined the effect of siRNA of ATM on the radiosensitivity of M059K cells because single miRNA could target multi-genes and miR-100 might target many other genes that also play a role in affecting the cell radiosensitivity. The results showed that when the ATM level in M059K cells was down-regulated by the siRNA (figure 4B), M059K cells became more sensitive to IR-induced killing (figure 4C), and the sensitization level is similar to that induced by miR-100 (figure 4A). These results confirm that up-regulating miR-100 in M059K cells induced radiosensitization is the consequence of the low-expression of ATM.
Fig. 4.
The effects of up-regulation of miR-100 on the cell radiosensitivity. (A) The effects of up-regulation of miR-100 on M059K cell radiosensitivity. At 72 h after infection with the lentivirus encoding pri-miR-100, the cells were exposed to 4 Gy. The clonogenic assay was performed as described in METHODS. Data shown are the mean ± SE from three independent experiments; **, P < 0.01, compared with the cells infected with the lentiviral vector without miR-100. (B) The effects of the ATM siRNA on protein expression. The cells were treated with the ATM siRNA or a control RNA (100 nM) for 48 h. The cells were collected for Western blot assay. (C) The effects of the ATM siRNA on the M059K cell radiosensitivity. The cells were treated with the RNAs as described above and then the cells were collected for clonogenic assay. The data represent mean ± SE of three independent experiments, **: P < 0.01.
In summary, our data, to the best of our knowledge, demonstrate for the first time that ATM is the target of miR-100, and indicate that over-expression of miR-100 is mainly responsible for the low expression of ATM in M059J cells. These data also demonstrate that miR-100 targeting ATM could sensitize the cells to IR-induced killing. In addition, based on these results, we could identify miRNAs that target DNA repair genes to sensitize tumor cells to radiotherapy or chemotherapy and thus improve cancer treatment [23].
Supplementary Material
Acknowledgments
We thank Drs Allalunis-Turner and Lu for providing cell lines, members of the Wang laboratory for helpful discussion and Doreen Theune for editing this manuscript. This work was supported by NIH grant GM80771 and Emory University School of Medicine start-up funds designated for Y.W.
Footnotes
All the authors declare that they have no competing interests.
Authors’ contribution
WLN and DY performed the experiments and analyzed the data. XZ and YM performed partial experiments. YW designed this study and wrote the manuscript. All the authors approved the final submission.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allalunis-Turner MJ, Barron GM, Day RS, Dobler KD, Mirzayans R. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat Res. 1993;134:349–354. [PubMed] [Google Scholar]
- 2.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, III, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 3.Gately DP, Hittle JC, Chan GKT, Yen TJ. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol Biol Cell. 1998;9:2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeggo PA, Taccioli GE, Jackson SP. Menage á trois: double strand break repair, V(D)J recombination and DNA-PK. BioEssays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 5.Chan DW, Chen BPC, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annual Review of Immunology. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 8.Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S, Povirk LF, Valerie K. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J Biol Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 10.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A Double-Strand Break Repair Defect in ATM-Deficient Cells Contributes to Radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 11.Anderson C, Dunn J, Freimuth P, Galloway A, Allalunis-Turner M. Frameshift Mutation in PRKDC, the Gene for DNA-PKcs, in the DNA Repair-Defective, Human, Glioma-Derived Cell Line M059J. Radiat Res. 2001;156:2–9. doi: 10.1667/0033-7587(2001)156[0002:fmiptg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natil Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y. Spontaneous metastasis of clonal cell subpopulations of human lung giant cell carcinoma after subcutaneous inoculation in nude mice. Chinese Journal of Oncology. 1989;11:1–7. [PubMed] [Google Scholar]
- 19.Dalby B, Catesa S, Harrisa A, Ohkia EC, Tilkinsb ML, Priceb PJ, Ciccarone VC. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33:95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Chan DW, Gately DP, Urban S, Galloway AM, Lees-Miller SP, Yen T, Allalunis-Turner J. Lack of correlation between ATM protein expression and tumour cell radiosensitivity. Int J Radiat Biol. 1998;74:217–224. doi: 10.1080/095530098141591. [DOI] [PubMed] [Google Scholar]
- 21.Peng Y, Woods RG, Beamish H, Ye R, Lees-Miller SP, Lavin MF, Bedford JS. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 2005;65:1670–1677. doi: 10.1158/0008-5472.CAN-04-3451. [DOI] [PubMed] [Google Scholar]
- 22.Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters expression of cellular microRNA species that affect its replication. J Virol. 2008;82:6524–6535. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan D, Ng1 W, Zhang X, Wang P, Zhang Z, Mo Y, Mao H, Hao C, Van Meir E, Olson J, Curran W, Wang Y. Targeting DNA-PK and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE. 2010:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.