Abstract
Earlier studies have shown that psoriasis in Japan and Thailand is associated with two different MHC haplotypes—those bearing HLA-Cw6 and those bearing HLA-Cw1 and HLA-B46. In an independent case-control sample from Thailand, we confirmed association of psoriasis with both haplotypes. No association was seen in Thai HLA-Cw1 haplotypes lacking HLA-B46, nor was HLA-Cw1 associated with psoriasis in a large Caucasian sample. To assess whether these risk haplotypes share a common origin, we sequenced genomic DNA from a Thai HLA-Cw1-B46 homozygote across the ~300 kb MHC risk interval, and compared it to sequence of a HLA-Cw6-B57 risk haplotype. Three small regions of homology were found, but these regions share equivalent sequence similarity with one or more clearly non-risk haplotypes, and they contain no polymorphism alleles unique to all risk haplotypes. Differences in psoriasis phenotype were also observed, including lower risk of disease, greater nail involvement, and later age at onset in HLA-Cw1-B46 carriers compared to HLA-Cw6 carriers. These findings suggest locus heterogeneity at PSORS1, the major psoriasis susceptibility locus in the MHC, with HLA-Cw6 imparting risk in both Caucasians and Asians, and an allele other than HLA-Cw1 on the HLA-Cw1-B46 haplotype acting as an additional risk variant in East Asians.
Keywords: Psoriasis, Human Leukocyte Antigens, human genetics, Major Histocompatibility Complex
Introduction
Human leukocyte antigen associations with psoriasis have been known for nearly 40 years (1). Earlier studies localized the disease determinant to the Class I end of the MHC (2, 3) and assigned the name PSORS1 (psoriasis susceptibility 1, OMIM #177900) to this locus (4). Association with HLA-Cw6 is particularly strong in many different world populations (5), and recent sequencing and haplotype analyses of Caucasian and Chinese Han psoriatics have indicated that HLA-Cw6 itself is likely to be the susceptibility determinant on these chromosomes, rather than any of ten nearby genes in the 300 kb PSORS1 candidate interval (6, 7). The prevalence of psoriasis differs markedly throughout the world (8). While it is unclear whether genetic or environmental factors are primarily responsible for this variation, it has been suggested that the rarity of psoriasis in Australian aborigines and several Amerindian populations is correlated with the absence of Human Leukocyte Antigen (HLA) haplotypes carrying HLA-Cw6 (8).
Several studies have documented a strong association between psoriasis and another HLA haplotype that is common in Japan and Thailand, but extremely rare in Caucasians (HLA-A*0207, -B*4601, -Cw*01) (9–13). This haplotype was associated with psoriasis when found in cis to any of three HLA Class II haplotypes (10), suggesting that the disease determinant resides on the Class I end of these haplotypes. Interestingly, all studies but one (9) found that the HLA-Cw1-B46 haplotype imparts considerably lower risk for psoriasis than does HLA-Cw6. There is also some evidence that this determinant produces a different clinical form of psoriasis, since HLA-Cw1-B46 is equally associated with early or late-onset disease (9, 10), whereas HLA-Cw6 is much more strongly associated with early-onset psoriasis in both Thais (10) and Caucasians (14).
A major goal of this study was to determine whether the HLA-Cw1-B46 psoriasis risk haplotype found in Japanese (12, 13, 15) and Thai (9, 10) populations and the HLA-Cw6-bearing psoriasis risk haplotypes found in both Caucasians and Asians represent allelic or locus heterogeneity at the PSORS1 locus, or if they share a disease locus inherited identical by descent (IBD) from a common ancestor. To this end, we cloned and sequenced the PSORS1 candidate interval of the HLA-Cw1-B46 haplotype for comparison with sequences for the HLA-Cw6-B57 and HLA-Cw6-B50 risk haplotypes and nine non-risk MHC haplotypes that were derived by us (6) and the Sanger Centre (16). In addition, we looked for differences in associated relative risk and phenotype of the HLA-Cw1-B46 and HLA-Cw6-bearing haplotypes in a previously-unreported sample of 206 Thai cases and 114 Thai controls. Finally, we compared the haplotype compositions and odds ratios for association of HLA-Cw1 in our Caucasian and Thai samples. Together, these analyses confirm that HLA-Cw1-B46 is a psoriasis risk haplotype in the Thai population, demonstrate that HLA-Cw1 is unlikely to be a direct determinant of risk for psoriasis in the Caucasian or Thai populations, and strongly suggest that the disease determinants carried on these two ancestral haplotypes are not derived from a common ancestor.
Materials and Methods
Subjects
Informed consent was obtained from all subjects under protocols adherent to the Declaration of Helsinki principles and approved by the Institutional Review Boards of the participating institutions. In the Caucasian sample, which consisted of 2,438 cases and 2,311 controls, most affected individuals were identified through the dermatology services of the University of Michigan Medical Center, the Ann Arbor Veterans Affairs Medical Center, and Henry Ford Hospital of Detroit. A few psoriatics were also provided by the National Psoriasis Foundation Tissue Bank. Individuals were defined as affected if chronic plaque or guttate psoriasis lesions covered more than 1% of the total body surface area or if at least two skin, scalp, nail or joint lesions were clinically diagnostic of psoriasis (17). Controls were recruited from the southeast Michigan area, and were required to be unrelated to each other or to any case, and to be free of a family history of psoriasis. For this study, only cases and controls of self-reported European Caucasian origin were analyzed. The Thai sample consisted of 206 psoriasis cases and 114 normal controls, all collected at the Institute of Dermatology in Bangkok, Thailand, using the same inclusion and exclusion criteria used for the Caucasian sample.
DNA preparation
Genomic DNA was prepared from heparinized whole blood using previously established methods (18). Blood samples collected in Bangkok were transported to Ann Arbor for DNA preparation within four days.
Genotyping
Eight SNPs in exons 2 and 3 of the HLA-C gene were genotyped—rs1131151, rs28732105, rs1050409, rs1131123, rs1131118, rs1050384, rs17839985 and rs41547419 at positions 89, 213, 218, 341, 361, 387, 459 and 540 of the coding sequence. These SNPs allow absolute discrimination of HLA-C to a triallelic level (Cw1/Cw6/neither), even in the absence of external phasing information, for all known alleles in release 2.10.00 of the IMGT-HLA Sequence Database (19); URL http://www.ebi.ac.uk/imgt/hla). Six SNPs in exons 2 and 3 of the HLA-B gene were also genotyped—rs713031, rs41562914, rs1131204, rs41553715, rs1071652, and rs2308466 at positions 142, 206, 277, 299, 362, and 560 of the coding sequence. These SNPs provide typing of HLA-B to a biallelic level (B46/other) in the absence of phasing information for all known alleles in release 2.16.00 of IMGT-HLA database. All SNPs were typed by single-base primer extension, as implemented in the SnapShot assay protocol (Applied Biosystems), per the manufacturer’s instructions. PCR amplification and SnapShot extension primer sequences are provided in Supplementary Table 1. Microsatellites were genotyped by PCR amplification using fluorescently labeled forward and unlabeled reverse primers followed by size determination by capillary electrophoresis on an Applied Biosystems 3100 Genetic Analyzer.
Cloning and sequencing
A Thai psoriatic who was homozygous for the HLA-Cw1-B46 haplotype throughout the 300 kb PSORS1 candidate region (31.129–31.429 Mb on build 36.3 of the human reference sequence for chromosome 6) was identified by genotyping HLA-C, HLA-B, and 10 microsatellite markers extending from MICA to telomeric of CDSN. A fosmid library was prepared from the genomic DNA of this individual and screened for the region of interest, as previously described (6). Thirteen overlapping fosmid clones that provided complete coverage of the risk interval were selected, with no attempt made to distinguish clones from the maternal or paternal chromosome. Inserts of each fosmid clone were subjected to shotgun sequencing, as described previously (6). High quality sequence coverage from at least two different plasmid subclones—and from both strands, whenever possible—was required for the entire fosmid insert, which resulted in a 22-fold average depth of coverage. The published sequence of the HLA-Cw7-B8 haplotype of the COX homozygous cell line (20) was used as both a reference for sequence alignment and for the numbering of the coordinate system used in the tables and figures of the present study, which starts at the first base of the 5′ primer (GCAACTTTTCTGTCAATCCA) used to amplify microsatellite marker D6S273 and extends in the telomeric direction. Overlapping fosmid clone sequences were assembled into a single contig; the resulting 337.1 kb of HLA-Cw1-B46 haplotype sequence (spanning 31.105–31.446 Mb on human reference) has been deposited in Genbank (accession number GQ472773).
Association analysis
Single marker association was evaluated using a chi-square contingency test of allelic counts; asymptotic p-values are reported. Haplotype inference and haplotype-based association tests were carried out with v 1.07 of PLINK (21); URL http://pngu.mgh.harvard.edu/purcell/plink/). For standard haplotype association, a logistic regression model with an allele dosage term was utilized, and p-values were determined with 1 million permutations of case-control status. For conditional haplotype-based association, a test of whether HLA-Cw1 has effects independent of HLA-B46 was constructed as a likelihood ratio test comparing an alternative model with separate effects for each of the three HLA-Cw1-B46 haplotypes to a null model which groups the Cw1+/B46– and Cw1–/B46– haplotypes together. An analagous conditional test for independent effect of HLA-B46 compares association of the Cw1+/B46+ and Cw1+/B46–haplotypes. Meta-analysis of disease associations in the two Thai studies used Cochran-Mantel-Haenszel test procedures. Power calculations were carried out with version 3.1 of G*Power (22); URL http://www.psycho.uni-duesseldorf.de/aap/projects/gpower/).
Sequence analysis
The HLA-Cw1-B46 sequence was aligned with the other eighteen MHC haplotype sequences by use of SeqMan (DNAStar, version 8.0.2); whenever necessary, sequence alignments were manually adjusted to yield the minimum possible number of polymorphisms. For each polymorphism, its location in the COX HLA-Cw7-B8 reference sequence and its alleles for all 19 haplotypes were recorded. MHC haplotype sequences were then compared with that of the HLA-Cw6-B57 haplotype by determining the percentage difference of polymorphic alleles over 2.5-kb intervals. Regions of similarity between pairs of MHC haplotype sequences were delineated by a two step approach (23). Rough bounds were first obtained using a moving window of 2.5 kb with a lag of 100 bp and a criterion of at least 80% identity of alleles for all included polymorphisms. Recursive entropic segmentation (24) with a stopping criterion based on the Bayesian information criterion (25) was then applied as a second-stage refinement. Version 0.97-600–1000 of the MINCOV program (26); URL http://www.stanford.edu/group/molepi/free_software.html) was used to search for minimal combinations of polymorphism alleles unique to risk haplotypes. MHC haplotype sequences within regions of homology were clustered using an average-distance agglomerative hierarchical method with a metric of percentage-difference of polymorphism alleles. Multiple instances of a single MHC haplotype (viz., two HLA-Cw6-B57, two Cw7-B7, two Cw8-B65, three Cw7-B8, two Cw3-B62, and two Cw12-B38 haplotypes) were consolidated into a single representative consensus sequence before comparison of sequence similarity and combinatorial analysis. All nineteen available MHC sequences were used for clustering.
Phenotype analysis
Five phenotypic aspects of psoriasis were measured at entry into the study: age at onset of disease, TBSA involvement of lesions, toenail involvement, fingernail involvement, and arthritis. All traits were compared for four different HLA-CB phenotypes (carriage of HLA-Cw1-B46 but no HLA-Cw6, carriage of HLA-Cw6 but no HLA-Cw1-B46, carriage of both HLA-Cw6 and HLA-Cw1-B46, carriage of neither HLA-Cw1-B46 nor HLA-Cw6), as well as all six possible HLA-CB genotypes involving these two alleles. Age at onset and TBSA were analyzed by one-way analysis of variance (ANOVA) after transforming the variables to approximate normality with the optimal Box-Cox power transformation (power of 0.8 for age at onset and 0.3 for TBSA); p-values were determined using 10,000 randomizations of the response variable observations. There were no significant departures from the assumption of homogeneity of variances for either of the tranformed variables, as assessed by a randomization version of Levene’s test. Sheffe’s modified S-method (27, 28) was used for an unplanned comparison of mean age at onset of HLA-Cw6 carriers vs. non-carriers; it controls the experimentwise error rate at the nominal level for all possible linear contrasts of the group means. Nail involvement and arthritis were analyzed by unordered two-way contingency tables, using Fisher’s exact test to determine the significance of association between phenotype variables and HLA-C status. Standardized Pearson residuals were examined to determine the relative contributions of different cells of the contingency table to the overall test result, and a 2 × 2 contingency table was used to analyze nail involvement for HLA-Cw1-B46 carriers vs. non-carriers.
Results
Association analysis
As shown in Table 1, in our Thai sample HLA-Cw6, HLA-Cw1 and HLA-B46 are all significantly associated with psoriasis (P = 3.2 × 10−6, 0.0011 and 0.0017, respectively). The associations with HLA-Cw1 and HLA-B46 are driven entirely by association with an underlying HLA-Cw1-B46 haplotype (P = 0.0016, Table 2). In this sample HLA-B46 is invariably linked with HLA-Cw1 (i.e., no HLA-Cw1−-B46+ haplotypes); however, HLA-Cw1 haplotypes lacking HLA-B46 do occur at a low frequency of ~3%, but they show a much smaller effect size (odds ratio (OR) = 1.34) and are not significantly associated with psoriasis (P = 0.63). Similarly, conditional haplotype-based association testing found no evidence that HLA-Cw1 is associated independently of HLA-B46 (p = 0.33), or vice versa (p = 0.52).
Table 1.
Single marker analysis of HLA-Cw1, HLA-Cw6, and HLA-B46 associations with psoriasis in Thais and Caucasians
| Allele | Thais | Caucasians | ||||||
|---|---|---|---|---|---|---|---|---|
| Frequency (proportion) in |
OR (95% CI)a | Pb | Frequency (proportion) in |
OR(95% CI)a | Pb | |||
| cases | controls | cases | controls | |||||
| HLA-Cw1 | 94 (.2338) | 26 (.1238) | 2.16 (1.35, 3.46) | 0.0011 | 161 (.0330) | 182 (.0394) | 0.83 (0.67, 1.03) | 0.097 |
| HLA-Cw6 | 71 (.1766) | 9 (.0429) | 4.79 (2.34, 9.80) | 3.2 × 10−6 | 1139 (.2336) | 420 (.0909) | 3.05 (2.70, 3.44) | 1.3 × 10−78 |
| HLA-B46 | 79 (.1955) | 22 (.0991) | 2.21 (1.33, 3.66) | 0.0017 | 0c (.0000) | 0c (.0000) | — | — |
Odds ratio and its 95% confidence interval for the allelic association test
P-value for allelic association test; multiallelic p-value for HLA-C is 3.4 × 10−8 in Thais and 3.2 × 10−77 in Caucasians
Frequency of HLA-B46 in Caucasians is based on typing all HLA-Cw1-positive individuals (160 cases and 174 controls successfully typed) and a large subsample of HLA-Cw1-negative individuals (505 cases and 667 controls successfully typed)
Table 2.
Association of HLA-Cw1-B46 haplotypes with psoriasis in Thais
| HLA Haplotype |
Frequency cases | Frequency controls | OR (95% CI)a | Pb | |
|---|---|---|---|---|---|
| Cw1 | B46 | ||||
| + | + | 0.1965 | 0.0991 | 2.25 (1.34, 3.80) | 0.0016 |
| + | − | 0.0373 | 0.0283 | 1.34 (0.51, 3.57) | 0.63 |
| − | + | 0.0000 | 0.0000 | — | — |
| − | − | 0.7662 | 0.8726 | 0.46 (0.28, 0.75) | 0.0012 |
Odds ratio and its 95% confidence interval in logistic regression dosage model for association
Global p-value = 0.0036; all p-values based on 1 million permutations
Although HLA-Cw6 appears to be more strongly associated with psoriasis than HLA-Cw1-B46 (OR = 4.79 vs. 2.16), the difference in odds ratios is not significant as their 95% confidence intervals (CI) overlap. Because of increased power, a meta-analysis that combines the allelic counts in our Thai sample with those of a previous Thai study (10) is able to demonstrate a significantly greater association of psoriasis with HLA-Cw6 (OR = 5.10, 95% CI = 3.23–8.06, P = 2.1 × 10−14) than with HLA-Cw1-B46 (OR = 2.16, 95% CI = 1.61–2.88, P = 1.4 × 10−7). The other Asian studies (9, 11–13) used serological typing, so their results could not be easily combined with the more recent allele-based studies, but it is noteworthy that three of four of these older studies also show a greater strength of association for HLA-Cw6.
We also looked for HLA-C associations in a Caucasian sample of 2,438 cases and 2,311 controls (Table 1). While HLA-Cw6 was very strongly associated with psoriasis in this sample (OR = 3.05, P = 1.3 × 10−78), HLA-Cw1 was clearly unassociated (OR = 0.83, P = 0.097). Despite the lower allele frequency of HLA-Cw1 in the Caucasian cohort (3.9% in Caucasian controls versus 12.4% in the Thai controls), our large sample had essentially 100% power to detect association if it indeed exists, assuming a multiplicative model, a significance level α = 0.05, and an OR of 2.16 (similar to that in Thais); our sample had 80% power to detect an association with an OR as low as 1.32. HLA-B46 did not occur in our Caucasian sample, neither among all 334 HLA-Cw1-positive individuals nor among a large typed subset of 1172 HLA-Cw1-negative individuals (95% CI for HLA-B46 frequency = 0.0000–0.0026 in 730 randomly selected controls and 0.000–0.0035 in 545 randomly selected cases).
Sequence comparisons
In order to determine the DNA sequence of the PSORS1 risk region on the HLA-Cw1-B46 haplotype, an affected Thai individual homozygous for this haplotype was identified by HLA typing and microsatellite genotyping, and a fosmid library was prepared and screened as described in Methods. A total of 13 overlapping fosmid clones were isolated that provided complete coverage of a 337 kb region extending from 15 kb telomeric of HLA-B to 90 kb centromeric of CDSN. The sequenced interval fully includes a 298 kb candidate region for PSORS1 shared by all known HLA-Cw6 risk haplotypes (6). We then compared this sequence to a collection of genomic DNA sequences generated by ourselves (6) and by the MHC Haplotype Project (16). Besides providing new examples of haplotypes we previously sequenced (HLA-Cw7-B8, Cw7-B7, Cw3-B62, Cw6-B57), inclusion of the MHC Haplotype Project sequences contributed sequences for four new haplotypes (HLA-Cw5-B18, Cw5-B44, Cw16-B44, and Cw3-B60) that all appear to be non-risk from our previous analysis (6). All together, 19 sequences were available, including those for eleven distinct MHC haplotypes that were complete enough for sequence comparison in the candidate interval.
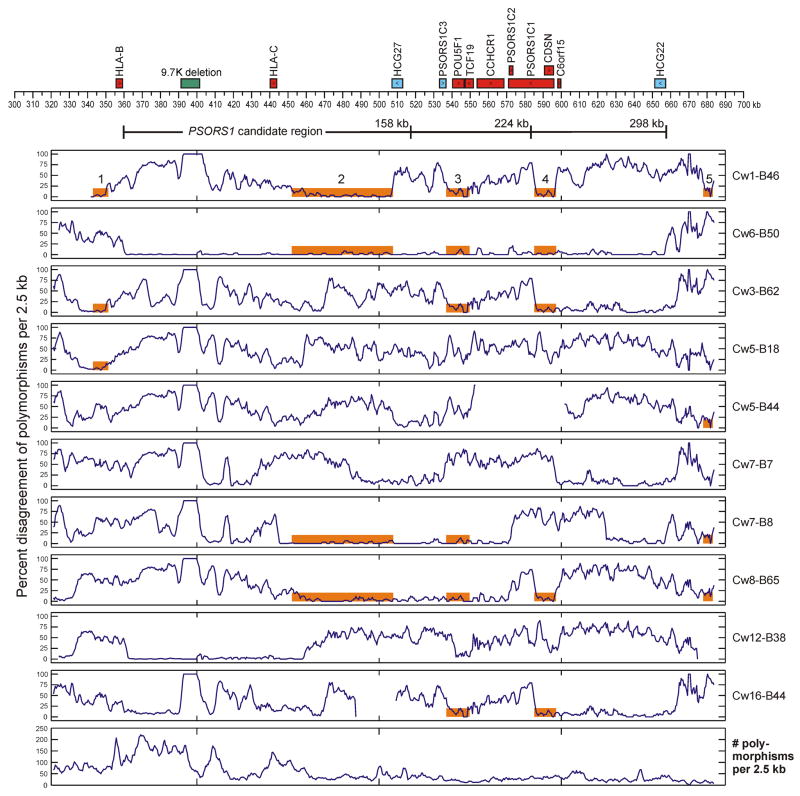
As shown in Figure 1, the HLA-Cw1-B46 and HLA-Cw6-B57 haplotypes exhibit substantial allelic divergence for more than three-quarters of the sequenced interval. Nevertheless, within the 331 kb region encompassing most sequences, four of 7,364 qualifying polymorphisms carry an allele common to all three psoriasis risk haplotypes (HLA-Cw6-B57, Cw6-B50, Cw1-B46) that is not found on any of the eight nonrisk haplotypes (Table 3). Furthermore, there are more than 59,000 two-way combinations and two billion three-way combinations of polymorphism alleles fulfilling this same criterion for a potential IBD disease locus. Progressively narrowing the region of comparison to each of three different PSORS1 candidate regions (298, 224, and 158 kb), which were delineated by previous work (6), only modestly reduces the number of potential disease loci (Table 3). However, if HLA-Cw6 and HLA-Cw1-B46 risk haplotypes are indeed descended from a common PSORS1-bearing ancestor, then the disease locus should occur within a region of sequence homology. Five such regions at least 5 kb in length, marked in orange in Figure 1, could be delineated. Two of these (regions 1 and 5) occur outside the 298 kb candidate interval that is the shortest region common to all known HLA-Cw6 risk haplotypes, and two of the remaining three (regions 3 and 4) are unpromising candidates for an identical-by-descent disease region as these short (12.8 and 12.0 kb, respectively) intervals bear no polymorphism alleles or combinations of alleles unique to the risk haplotypes. Furthermore, region 4 falls ouside of a 224-kb PSORS1 candidate interval firmly established by ancestral recombinant haplotype analysis, and region 3 falls outside of a probable though not definitively established 158-kb candidate interval.
Figure 1.
Sequence comparison of ten MHC class I haplotypes with the HLA-Cw6-B57 risk haplotype. Known genes and their direction of transcription, as well as a 9.7-kb indel, are shown above the coordinate axis. Genes expressing non-coding RNA are colored cyan, and those expressing protein are colored red. Three delineations of the PSORS1 candidate region (Nair et al. 2006) are shown below the coordinate axis. The percent disagreement of polymorphism alleles, when compared with the HLA-Cw6-B57 haplotype, is plotted for each haplotype using a moving 2.5-kb window with a 500 bp lag. The bottom panel plots the number of polymorphisms that are variable among all sequenced haplotypes; only these polymorphisms were considered when computing percent disagreement. Regions of sequence homology at least 5 kb in length between the HLA-Cw1-B46 and HLA-Cw6-B57 haplotypes are mapped as five numbered orange bars in the top panel; these bars are also shown on all other haplotypes sharing the same regions of homology.
Table 3.
Polymorphism analysis of PSORS1 candidate regions and regions of sequence homology between HLA-Cw1-B46 and HLA-Cw6-B57.
| Regiona | Bounds (kb) | Length (kb) | No. of Haplo.b | No. of polymorphismsc |
Pct. disagreement Cw1-B46 vs. Cw6-B57d |
No. of combinations of polymorphisms common and unique to risk haplotypese |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| all | SNPs & indels | all | SNPs & indels | 1-way | 2-way | 3-way | ||||
| 1 | 343.0–351.3 | 8.3 | 11 | 218 | 214 | 4.1 | 3.7 | 0 | 0 | 0 |
| 2 | 452.1–507.6 | 55.5 | 10 | 915 | 881 | 4.5 | 4.0 | 0 | 31 | 13,272 |
| 3 | 536.8–597.1 | 12.8 | 11 | 92 | 91 | 12.0 | 11.0 | 0 | 0 | 0 |
| 4 | 585.1–597.1 | 12.0 | 10 | 159 | 158 | 5.0 | 5.1 | 0 | 0 | 0 |
| 5 | 677.9–683.1 | 5.2 | 11 | 34 | 33 | 11.8 | 12.1 | 0 | 0 | 0 |
| 158 kb | 359.9–517.1 | 157.6 | 10 | 4983 | 4912 | 46.9 | 46.7 | 4 | 13,817 | 3.8 × 107 |
| 224 kb | 359.9–583.4 | 223.6 | 9 | 5673 | 5571 | 46.1 | 45.7 | 4 | 17,421 | 5.4 × 107 |
| 298 kb | 359.9–657.6 | 297.7 | 9 | 6608 | 6470 | 46.5 | 46.0 | 4 | 38,109 | 1.8 × 108 |
| entire | 343.0–673.8 | 330.8 | 9 | 7364 | 7211 | 44.7 | 44.1 | 4 | 59,555 | 2.5 × 109 |
Region of MHC sequence as shown in Fig. 1; “entire” refers to the full interval for which most MHC haplotypes were sequenced.
Number of different haplotype sequences being compared that are fully sequenced for the region; this number varies among regions because three of the Sanger Centre sequences have gaps in coverage.
Number of polymorphisms that are variable for sequences being compared; separate tallies are given for all types of polymorphisms and for SNPs and indels only (i.e., excluding STRs and polyA/T variations, which tend to have higher mutation rates).
Percent disagreement of polymorphism alleles between the HLA-Cw1-B46 and HLA-Cw6-B57 risk haplotypes.
Number of one-way, two-way, and three-way allelic combinations of polymorphisms that are both common and unique to the three psoriasis risk haplotypes (Cw1-B46, Cw6-B57, and Cw6-B50) when compared to all nonrisk haplotypes.
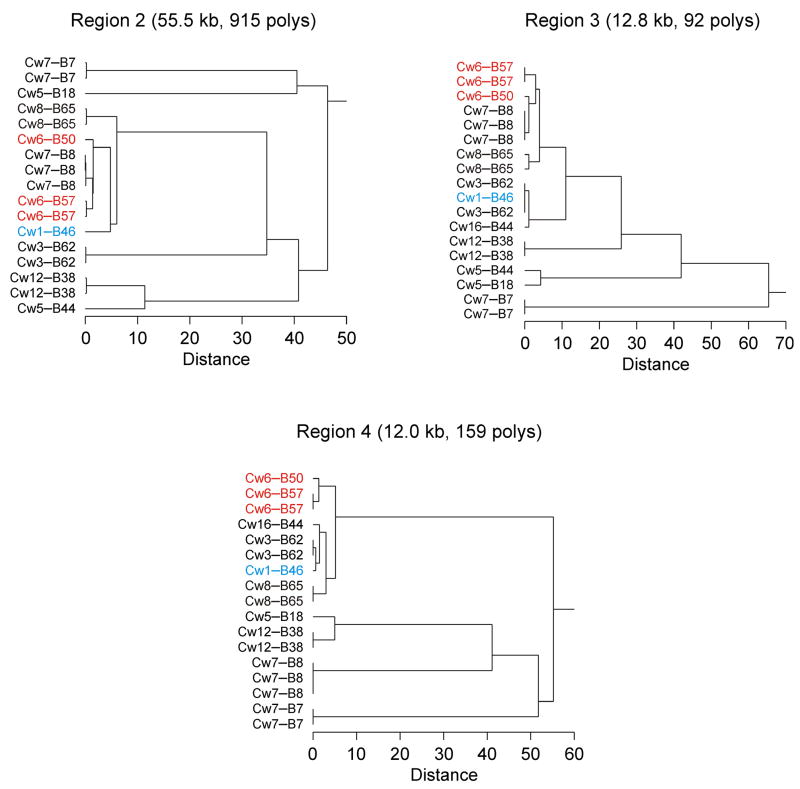
The final and largest region of homology, region 2, is a 55.5 kb interval between HLA-C and HCG27 with 95.5% allelic identity at all 915 variable polymorphisms and a 96.0% allelic identity among the 881 more stable SNPs and indels (i.e., excluding highly mutable poly-A/T and STR variations). Region 2 bears no single polymorphism with an allele restricted to risk haplotypes, but it does have 31 two-way and 13,272 three-way combinations of polymorphism alleles unique to risk. However, as can be seen in Figure 1, the three risk haplotypes share roughly equivalent levels of sequence similarity in region 2 with two clearly nonrisk haplotypes (HLA-Cw7-B8 and Cw8-B65). This visual comparison is confirmed more rigorously by the clustering dendograms of Figure 2. For region 2, the clustering distance separating the nonrisk HLA-Cw7-B8 haplotypes from any of the HLA-Cw6 risk haplotypes is substantially less than the distance between the HLA-Cw6 and HLA-Cw1-B46 risk haplotypes, and the former distance is actually slightly less than that between the two different HLA-Cw6 haplotypes, which are almost certainly IBD in this region. Furthermore, the nonrisk HLA-Cw8-B65 haplotype is only slightly more different from HLA-Cw1-B46 in region 2 than is the latter haplotype from the HLA-Cw6 haplotypes. Figure 2 shows a similar situation for regions 3 and 4. Extended regions of sequence similarity where only a few common haplotypes are observed (haplotype blocks) are commonplace within the MHC (29) and elsewhere in the human genome (30), which makes it difficult to test for identity by descent among these risk haplotypes. The lack of any known expressed genes or single polymorphism alleles unique to risk in region 2 argues strongly against an IBD disease locus in this interval, but a risk-specific haplotype of an unknown gene or of an intergenic regulatory element within the 55 kb defined by region 2 could conceivably be common to all three risk haplotypes, as long as it spans at least 2.3 kb (the minimum interval encompassed by any of the two-way or three-way allelic combinations unique to risk haplotypes).
Figure 2.
Sequence variation among MHC class I haplotypes within three regions of homology between HLA-Cw1-B46 and HLA-Cw6-B57. Hierarchical clustering dendrograms are shown. HLA-Cw6 haplotypes are shown in red, and the HLA-Cw1-B46 haplotype in cyan. The clustering distance metric is percent disagreement of polymorphism alleles within the region.
Phenotype comparisons
We next undertook a comparison of psoriasis phenotype of Thais carrying HLA-Cw1-B46 versus HLA-Cw6, under the hypothesis that if the two HLA risk haplotypes share a common causative variant, then the resulting disease phenotype should be similar in the same genetic population. As shown in Table 4, among the four HLA-CB phenotypes significant differences were observed for mean age at onset, toenail involvement, and fingernail involvement (P = 0.043, 0.0048, and 0.0070, respectively). No significant differences in total body surface area (TBSA) involvement or arthritis were observed (P = 0.098 and 0.84, respectively).
Table 4.
Variation of disease phenotype in Thai psoriatics as a function of HLA-CB phenotype
| HLA-CB phenotypea | nb | Age at onsetc (mean ± se) | TBSAc (mean ± se) | Pct. toenail involvement (mean ± se) | Pct. fingernail involvement (mean ± se) | Pct. arthritis (mean ± se) |
|---|---|---|---|---|---|---|
| Cw1-B46 only | 55 | 36.9 ± 1.6 | 26.2 ± 2.4 | 36.4 ± 6.5 | 36.4 ± 6.5 | 1.8 ± 1.8 |
| Cw6 only | 49 | 30.0 ± 2.3 | 34.2 ± 3.6 | 10.2 ± 4.3 | 14.3 ± 5.0 | 0.0 ± 0.0 |
| Cw6 + Cw1-B46 | 19 | 30.2 ± 3.2 | 34.2 ± 6.3 | 26.3 ± 10.1 | 31.6 ± 10.7 | 0.0 ± 0.0 |
| neither | 78 | 35.3 ± 1.8 | 24.7 ± 2.1 | 15.4 ± 4.1 | 14.1 ± 3.9 | 2.6 ± 1.8 |
| missing | 5 | 34.8 ± 8.3 | 28.2 ± 8.9 | 40.0 ± 21.9 | 40.0 ± 21.9 | 0.0 ± 0.0 |
| total | 206 | 34.0 ± 1.1 | 28.3 ± 1.5 | 21.4 ± 2.9 | 22.3 ± 2.9 | 1.5 ± 0.8 |
| Pd | 0.043 | 0.098 | 0.0048 | 0.0070 | 0.84 | |
HLA-CB phenotype is based on the carriage of HLA-CB haplotypes by the individual, where HLA-C is typed to a Cw1/Cw6/neither level and HLA-B to a B46/other level; i.e., “Cw1-B46 only” designates individuals with one or two copies of a HLA-Cw1-B46 haplotype but no copies of a HLA-Cw6 haplotype, “Cw6 only” means carriage of one or two copies of HLA-Cw6 but no copies of HLA-Cw1-B46, “Cw6 + Cw1-B46” means carriage of one HLA-Cw6 and one HLA-Cw1-B46 haplotype, “neither” means carriage of neither a HLA-Cw6 nor a HLA-Cw1-B46 haplotype, and “missing” means the HLA-CB haplotypes are unknown because of typing failures. In this sample, HLA-B46 haplotypes always carry HLA-Cw1 (see Table 3).
Number of individuals
Mean and standard error for raw variable values are shown, but before analysis data were transformed to approximate normality using the optimal Box-Cox power transformation (power of 0.8 for age at onset and 0.3 for TBSA).
P-values for age at onset and TBSA are for one-way ANOVA, based on 10,000 random permutations of the response variable observations; p-values for toenail and fingernail involvement and arthritis are for Fisher’s exact test on an unordered two-way contingency table. All tests excluded individuals with a missing HLA-CB phenotype.
Inspection of group means in Table 4 shows that age at onset is about six years earlier in the two groups of HLA-Cw6 carriers (30.0 and 30.2 years) than for either the HLA-Cw1-B46 only carriers (36.9 years) or carriers of neither risk haplotype (35.3 years). The contrast of the average of the mean transformed age at onset for the two groups of HLA-Cw6 carriers compared to the average of the mean transformed onset for the two groups of HLA-Cw6 non-carriers is significant (P = 0.037). Conversely, inspection of standardized Pearson residuals for the four HLA-CB phenotype groups indicates that greater nail involvement for the two groups of HLA-Cw1-B46 carriers (36.4% and 26.3% for toenail, 36.4% and 31.6% for fingernail) versus lesser involvement for either HLA-Cw6 only carriers (10.2% and 14.3% for toenail and fingernail) or carriers of neither risk haplotype (15.4% and 14.1% for toenail and fingernail) is largely responsible for the significant test findings of the 4 × 2 contingency table. Collapsing the contingency table to a 2 × 2 format based on HLA-Cw1-B46 carriage yields a strong positive association for both toenail involvement (OR = 3.30, P = 0.0010) and fingernail involvement (OR = 3.28, P = 0.00074). TBSA trends higher in HLA-Cw6 carriers (34.2% vs. 25.5%), and the marginal lack of significance for variation among groups may be due to inadequate power of our Thai sample, since increased TBSA has been shown to be associated with HLA-Cw6 in Caucasians (31). The findings for arthritis have little meaning given the low incidence of this trait (1.5%) among Thai affecteds in the sample.
Differences of disease phenotype among HLA-CB genotypes were similar to those seen among HLA-CB phenotypes, with mean age at onset lower in all groups carrying one or more copies of HLA-Cw6, and toenail and fingernail involvement higher in all groups carrying one or more copies of HLA-Cw1-B46 (data not shown). However, only the differences in toenail and fingernail involvement were significant (P = 0.018 and 0.024, respectively). The weaker significances for HLA-CB genotype compared to HLA-CB phenotype may be a simple outcome of subdividing a relatively small sample into six versus four categories with a concomitant reduction in power.
Discussion
Genome-wide linkage scans (17, 32) as well as more recent genome-wide association studies (33–36) have made it clear that the major genetic determinant of psoriasis resides within the MHC. We have identified HLA-Cw6 as the predominant PSORS1 disease allele in the Caucasian population (6), and this has been confirmed in the Han Chinese (7). However, considerable evidence indicates that HLA-Cw6 is not the only psoriasis susceptibility allele in the MHC. Psoriatic arthritis has also repeatedly been associated with HLA-B38 and HLA-B39 (splits of HLA-B16) (37–43) and with HLA-B27, especially when axial involvement is present (39, 40, 42, 44). Moreover, we have recently shown that additional, albeit less genetically robust, association signals are present in the MHC Class III region (35, 45). Together with the HLA-Cw1-B46 association that is the focus of this study, these findings suggest that genetic heterogeneity is likely to be present at PSORS1, with various effects on the phenotype.
Our interest in the HLA-Cw1-B46 haplotype stemmed from several prior demonstrations of disease association in Asian populations (9, 10, 12, 13, 15, 46). Taking advantage of a collection of Thai psoriasis patients and normal controls independent of those collected previously, we were able to robustly confirm the association of psoriasis with HLA-Cw6, HLA-Cw1, HLA-B46 and the HLA-Cw1-B46 haplotype in our Thai sample (Table 1). In order to assess whether these associations might be due to allelic heterogeneity at HLA-C, we tested for HLA-Cw1-specific associations with psoriasis in the Thai and Caucasian populations. In our Thai sample, haplotypes carrying HLA-Cw1 but lacking HLA-B46 showed no significant association with psoriasis (Table 2), but our sample lacks adequate power given the relatively low frequency of this haplotype. However, similar findings in a Japanese study (11) where HLA-Cw1 not on HLA-B46 haplotypes trended toward negative association with psoriasis (OR = 0.45, P = 0.068) and in a Thai study (9) where the strength of association of HLA-B46 with psoriasis (OR = 4.23, P = 1.4 × 10−6) was much greater than that for HLA-Cw1 (OR= 1.70, P = 0.083), increase the likelihood that HLA-Cw1 is not a direct determinant of psoriasis in East Asians.
In our Caucasian sample, which showed highly significant evidence for association with HLA-Cw6, we found no evidence for association with HLA-Cw1 despite >99% power to detect an association of the strength observed in the Thai population. No occurrences of the HLA-B46 allele were seen for a large genotyped subset of our Caucasian sample, which includes all HLA-Cw1 carriers, confirming the specificity of HLA-Cw1-B46 for Asian populations. While two small studies have reported association of HLA-Cw1 with psoriatic arthritis (47, 48), this may reflect the fact that HLA-Cw1 is in linkage disequilibrium with HLA-B27 in Caucasian populations. However, HLA-Cw1 was clearly unassociated with 493 psoriatic arthritis cases in our own much larger Caucasian sample (P = 0.38), with an effect size nearly identical to that seen for 1,549 purely cutaneous psoriasis cases in the same sample (OR = 0.84 vs. 0.85, respectively). One other study of 50 pediatric Kuwaiti psoriatics and 120 controls yielded a positive association with HLA-Cw1, but no association with HLA-Cw6 (49). Whether this divergent finding is the result of small sample size, different ethnicity (predominantly Arab), very early onset (<12 years), or the presence of arthritis (which was not reported) remains to be determined. Overall, it appears highly unlikely that HLA-Cw1 itself is a psoriasis risk determinant in either Thais or Caucasians. Hence another MHC locus, perhaps HLA-B46 itself, is driving the observed associations with the HLA-Cw1-B46 haplotype.
Consistent with data presented by others, we noted that HLA-Cw6 appears to be more strongly associated with psoriasis than is HLA-Cw1-B46 in the Thai population. While the 95% confidence intervals for the two odds ratios estimated from our sample overlapped (Table 1), the greater strength of the HLA-Cw6 association could be statistically established after combining our study with the only other relevant Thai study with allele-based HLA genotyping (10). In addition, three of four of the older serological studies (11–13) corroborate the greater risk of disease imparted by HLA-Cw6 compared to HLA-Cw1-B46 in Asian populations.
To our knowledge there is no evidence for an association between guttate psoriasis and the HLA-Cw1-B46 haplotype, in contrast to its strong association with HLA-Cw6 (50). It is also notable that the HLA-Cw1-B46 haplotype has been associated with other autoimmune diseases, including myasthenia gravis and Graves Disease (51), whereas HLA-Cw6 has not. Moreover, Romphruk et al reported that HLA-Cw1-B46 is equally associated with early and late-onset disease in Thai psoriatics, whereas HLA-Cw6 is more strongly associated with early onset disease (46). Taken together with the aforementioned difference in strength of association, these findings suggested that the psoriasis susceptibility determinants carried on these two haplotypes are different. We tested this hypothesis in two ways: by performing a sequence analysis of the two haplotypes, and by comparing the phenotypes of known carriers of each haplotype.
Detailed sequence comparison with eight nonrisk haplotypes (Figure 1 and Table 3) found no single variants unique to the HLA-Cw1-B46 and HLA-Cw6 risk haplotypes within potential IBD regions of homology in the PSORS1 candidate interval. Although two-way and three-way combinations of variants unique to these two risk haplotypes do exist, they are confined to a 55 kb region that contains no known genes and that has equivalent similarity with two nonrisk haplotypes (Figure 2). Nevertheless, based on sequence analysis alone, we cannot completely exclude the possibility that this region contains a variant that is identical by descent within a regulatory element or a novel expressed gene.
Phenotypic analysis provided additional support for the hypothesis of genetic heterogeneity, though its conclusions must be tempered by sample size considerations. We found that Thai psoriatics carrying HLA-Cw1-B46 have a later age at onset and greater nail involvement than do carriers of the HLA-Cw6 risk haplotype (Table 4). However, it is important to note that age at onset correlates with the presence or absence of HLA-Cw6 but not of HLA-Cw1-B46, and likelihood of nail involvement with the presence or absence of HLA-Cw1-B46 but not of HLA-Cw6. Together, the weight of evidence from these sequencing and phenotype comparisons strongly favors the hypothesis that the HLA-Cw1-B46 and HLA-Cw6 risk haplotypes do not derive from a common ancestral risk chromosome.
Given these findings in support of genetic heterogeneity, the evolutionary history of HLA-B46 is of interest. In 1992, Parham and colleagues dissected a complicated serological determinant known as Cw1×3 antigen (also called Cw11, CwB, Cx46, Cw1+3, C-Bangkok, and CSH1). In doing so, they demonstrated that the HLA-B46 allele is the result of an unusual gene conversion event in which a 31 bp segment of HLA-Cw1 encoding residues 66 to 76 of the α1 helix replaced the corresponding sequences of the HLA-B62 allele (52). Haplotypes containing HLA-B62 and HLA-Cw1 are not uncommon in Asian populations (53), supporting the notion of a gene conversion event rather than recombination. HLA-B62 has a worldwide distribution, whereas HLA-B46 is specific for Asian populations, demonstrating that HLA-B62 is the ancestral allele and that the gene conversion occurred in an individual of Asian descent. HLA-B46 is a common allele in Asian populations, suggesting that this event was followed by marked expansion in the population. Whether this expansion reflects positive selection for pathogen resistance, analogous to the postulated selection for HLA-Cw6 in resistance to Streptococcal pneumonia (54), is unknown.
The eleven amino acids transferred from HLA-Cw1 to HLA-B62 by gene conversion differ from the corresponding residues of HLA-Cw6 only at amino acid residue 73 (threonine in HLA-Cw1 vs alanine in HLA-Cw6). Because we have shown here that HLA-Cw1 is not disease-associated, on the (unproven) hypothesis that this specific segment of HLA-C confers disease susceptibility, we could infer that alanine residue 73 is unlikely to be of critical importance, as previously suggested (55). However, there are many other possible explanations for the observed HLA-B46 association. HLA-B and HLA-C are very similar to each other, reflecting a relatively recent gene duplication (56). As MHC Class I genes, both HLA-B and HLA-C are involved in the presentation of peptides to CD8+ T-cells, whose emigration into the epidermis appears to be necessary for development of the epidermal hyperplastic response (57). One possibility would be that the two alleles could be presenting different antigens. Alternatively, another nearby gene in the MHC Class III region could be the causative agent on the HLA-Cw1-B46 haplotype. Several of these genes are strong functional candidates. For instance, MICA and MICB are nonclassical MHC genes that participate in the regulation of CD8+ T-cells and NK cells (58), and the tumor necrosis factor and lymphotoxin genes encode proteins whose blockade is highly therapeutically effective (59). While our earlier studies of recombinant ancestral haplotypes argue strongly against a primary role for MHC Class III genes as the drivers of the HLA-Cw6 association signal (6, 60), no comparable mapping studies exist as yet for the HLA-Cw1-B46 disease association in Asians. Thus, at this stage it is premature to speculate that the genetic heterogeneity suggested by our data must involve HLA-B46 itself, although it is certainly possible.
In conclusion, we have presented several lines of evidence for a distinct PSORS1 locus in the Thai population. Future genetic studies of this allele in Asian populations should focus on increasing sample size and high-density genotyping of HLA-B, its flanking sequences, and the MHC Class III region.
Supplementary Material
Acknowledgments
We thank all the psoriasis patients and normal controls who volunteered to participate in this study. This work was supported by R01 awards (AR042742 and AR050511) from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases, National Institutes of Health, by the Ann Arbor Veterans Affairs Hospital, by the Dudley and Dawn Holmes Fund, by the Babcock Memorial Trust, by the National Psoriasis Foundation, and by an award (M01 RR00042) from the National Center for Research Resources, National Institutes of Health, to the University of Michigan General Clinical Research Center.
Abbreviations
- ANOVA
analysis of variance
- CI
confidence interval
- HLA
Human Leukocyte Antigen
- MHC
Major Histocompatibility Complex
- OMIM
Online Mendelian Inheritance in Man
- OR
odds ratio
- PSORS1
Psoriasis Susceptibility 1
- TBSA
total body surface area
References
- 1.Russell TJ, Schultes LM, Kuban DJ. Histocompatibility (HL-A) antigens associated with psoriasis. N Engl J Med. 1972;287:738–40. doi: 10.1056/NEJM197210122871503. [DOI] [PubMed] [Google Scholar]
- 2.Jenisch S, Henseler T, Nair RP, et al. Linkage analysis of human leukocyte antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/-C region. Am J Hum Genet. 1998;63:191–9. doi: 10.1086/301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt-Egenolf M, Eiermann TH, Boehncke WH, Ständer M, Sterry W. Familial juvenile onset psoriasis is associated with the human leukocyte antigen (HLA) class I side of the extended haplotype Cw6- B57-DRB1*0701-DQA1*0201-DQB1*0303: a population- and family-based study. J Invest Dermatol. 1996;106:711–4. doi: 10.1111/1523-1747.ep12345600. [DOI] [PubMed] [Google Scholar]
- 4.Leder RO, Mansbridge JN, Hallmayer J, Hodge SE. Familial psoriasis and HLA-B: unambiguous support for linkage in 97 published families. Hum Hered. 1998;48:198–211. doi: 10.1159/000022802. [DOI] [PubMed] [Google Scholar]
- 5.Elder JT, Nair RP, Guo SW, Henseler T, Christophers E, Voorhees JJ. The genetics of psoriasis. Arch Dermatol. 1994;130:216–24. [PubMed] [Google Scholar]
- 6.Nair RP, Stuart PE, Nistor I, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–51. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X, Yang S, Huang W, et al. Fine mapping of the psoriasis susceptibility locus PSORS1 supports HLA-C as the susceptibility gene in the Han Chinese population. PLoS Genet. 2008;4:e1000038. doi: 10.1371/journal.pgen.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri SP, Farber EM. The prevalence of psoriasis in the world. J Eur Acad Dermatol Venereol. 2001;15:16–7. doi: 10.1046/j.1468-3083.2001.00192.x. [DOI] [PubMed] [Google Scholar]
- 9.Vejbaesya S, Eiermann TH, Suthipinititharm P, et al. Serological and molecular analysis of HLA class I and II alleles in Thai patients with psoriasis vulgaris. Tissue Antigens. 1998;52:389–92. doi: 10.1111/j.1399-0039.1998.tb03061.x. [DOI] [PubMed] [Google Scholar]
- 10.Choonhakarn C, Romphruk A, Puapairoj C, Jirarattanapochai K, Leelayuwat C. Haplotype associations of the major histocompatibility complex with psoriasis in Northeastern Thais. Int J Dermatol. 2002;41:330–4. doi: 10.1046/j.1365-4362.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa H, Asahina A, Akazaki S, et al. Association of Cw11 in Japanese patients with psoriasis vulgaris. Tissue Antigens. 1990;36:241–2. doi: 10.1111/j.1399-0039.1990.tb01835.x. [DOI] [PubMed] [Google Scholar]
- 12.Koizumi H, Fukaya T, Tsukinaga I, Ohkawara A, Wakisaka A, Aizawa M. HLA antigens in psoriasis vulgaris. Acta Dermatol Kyoto. 1988;83:483–8. [Google Scholar]
- 13.Ozawa A, Miyahara M, Sugai J, et al. HLA class I and II alleles and susceptibility to generalized pustular psoriasis: significant associations with HLA-Cw1 and HLA-DQB1*0303. J Dermatol. 1998;25:573–81. doi: 10.1111/j.1346-8138.1998.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 14.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13:450–6. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa H, Akazaki S, Asahina A, et al. Study of HLA class I, class II and complement genes (C2, C4A, C4B and BF) in Japanese psoriatics and analysis of a newly-found high-risk haplotype by pulsed field gel electrophoresis. Arch Dermatol Res. 1991;283:281–4. doi: 10.1007/BF00376613. [DOI] [PubMed] [Google Scholar]
- 16.Horton R, Gibson R, Coggill P, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair RP, Henseler T, Jenisch S, et al. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet. 1997;6:1349–56. doi: 10.1093/hmg/6.8.1349. [DOI] [PubMed] [Google Scholar]
- 18.Nair R, Guo S, Jenisch S, et al. Scanning chromosome 17 for psoriasis susceptibility: lack of evidence for a distal 17q locus. Hum Hered. 1995;45:219–30. doi: 10.1159/000154293. [DOI] [PubMed] [Google Scholar]
- 19.Robinson J, Waller MJ, Parham P, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–4. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart CA, Horton R, Allcock RJ, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–87. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Bernaola-Galvan P, Haghighi F, Grosse I. Applications of recursive segmentation to the analysis of DNA sequences. Comput Chem. 2002;26:491–510. doi: 10.1016/s0097-8485(02)00010-4. [DOI] [PubMed] [Google Scholar]
- 24.Bernaoloa-Galvan P, Roman-Roldan R, Oliver JI. Compositional segmentation and long-range fractal correlations in DNA sequences. Phys Rev E. 1996;53:5181–9. doi: 10.1103/physreve.53.5181. [DOI] [PubMed] [Google Scholar]
- 25.Li W. New stopping criteria for segmenting DNA sequences. Phys Rev Lett. 2001;86:5815–8. doi: 10.1103/PhysRevLett.86.5815. [DOI] [PubMed] [Google Scholar]
- 26.Salamon H, Tarhio J, Ronningen K, Thomson G. On distinguishing unique combinations in biological sequences. J Comput Biol. 1996;3:407–23. doi: 10.1089/cmb.1996.3.407. [DOI] [PubMed] [Google Scholar]
- 27.Sheffe H. Multiple testing versus multiple estimation. Improper confidence sets. Estimation of directions and ratios. Annals of Mathematical Statistics. 1970;41:1–29. [Google Scholar]
- 28.Klockars AJ, Hancock GR. Scheffe’s more powerful F-protected post hoc procedure. Educ Behav Stat. 2000;25:13–9. [Google Scholar]
- 29.Dawkins R, Leelayuwat C, Gaudieri S, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 30.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudjonsson JE, Karason A, Antonsdottir AA, et al. HLA-Cw6-positive and HLA-Cw6-negative patients with Psoriasis vulgaris have distinct clinical features. J Invest Dermatol. 2002;118:362–5. doi: 10.1046/j.0022-202x.2001.01656.x. [DOI] [PubMed] [Google Scholar]
- 32.Trembath RC, Clough RL, Rosbotham JL, et al. Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet. 1997;6:813–20. doi: 10.1093/hmg/6.5.813. [DOI] [PubMed] [Google Scholar]
- 33.Capon F, Di Meglio P, Szaub J, et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum Genet. 2007;122:201–6. doi: 10.1007/s00439-007-0397-0. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang XJ, Huang W, Yang S, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–10. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 37.Murray C, Mann DL, Gerber LN, et al. Histocompatibility alloantigens in psoriasis and psoriatic arthritis. Evidence for the influence of multiple genes in the major histocompatibility complex. J Clin Invest. 1980;66:670–5. doi: 10.1172/JCI109903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinoza LR, Vasey FB, Gaylord SW, et al. Histocompatibility typing in the seronegative spondyloarthropathies: a survey. Semin Arthritis Rheum. 1982;11:375–81. doi: 10.1016/0049-0172(82)90058-0. [DOI] [PubMed] [Google Scholar]
- 39.Beaulieu AD, Roy R, Mathon G, et al. Psoriatic arthritis: risk factors for patients with psoriasis - a study based on histocompatibility antigen frequencies. J Rheumatol. 1983;10:633–6. [PubMed] [Google Scholar]
- 40.Gladman DD, Anhorn KA, Schachter RK, Mervart H. HLA antigens in psoriatic arthritis. J Rheumatol. 1986;13:586–92. [PubMed] [Google Scholar]
- 41.Crivellato E, Zacchi T. HLA-B39 and the axial type of psoriatic arthritis. Acta Derm Venereol. 1987;67:249–50. [PubMed] [Google Scholar]
- 42.McHugh NJ, Laurent MR, Treadwell BL, Tweed JM, Dagger J. Psoriatic arthritis: clinical subgroups and histocompatibility antigens. Ann Rheum Dis. 1987;46:184–8. doi: 10.1136/ard.46.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez S, Martinez-Borra J, Lopez-Vazquez A, Garcia-Fernandez S, Torre-Alonso JC, Lopez-Larrea C. MICA rather than MICB, TNFA, or HLA-DRB1 is associated with susceptibility to psoriatic arthritis. J Rheumatol. 2002;29:973–8. [PubMed] [Google Scholar]
- 44.Armstrong RD, Panayi GS, Welsh KI. Histocompatibility antigens in psoriasis, psoriatic arthropathy, and ankylosing spondylitis. Ann Rheum Dis. 1983;42:142–6. doi: 10.1136/ard.42.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng B-J, Soltani-Arabsahi R, Bowcock AM, et al. Multiple loci within the Major Histocompatibility Complex confer risk of psoriasis. Am J Hum Genet. 2009 doi: 10.1371/journal.pgen.1000606. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romphruk AV, Oka A, Romphruk A, et al. Corneodesmosin gene: no evidence for PSORS 1 gene in North-eastern Thai psoriasis patients. Tissue Antigens. 2003;62:217–24. doi: 10.1034/j.1399-0039.2003.00056.x. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Larrea C, Torre Alonso JC, Rodriguez Perez A, Coto E. HLA antigens in psoriatic arthritis subtypes of a Spanish population. Ann Rheum Dis. 1990;49:318–9. doi: 10.1136/ard.49.5.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerber LH, Murray CL, Perlman SG, et al. Human lymphocyte antigens characterizing psoriatic arthritis and its subtypes. J Rheumatol. 1982;9:703–7. [PubMed] [Google Scholar]
- 49.Nanda A, Al-Fouzan AS, El-Kashlan M, Al-Sweih N, Al-Muzairai I. Salient features and HLA markers of childhood psoriasis in Kuwait. Clin Exp Dermatol. 2000;25:147–51. doi: 10.1046/j.1365-2230.2000.00598.x. [DOI] [PubMed] [Google Scholar]
- 50.Mallon E, Bunce M, Savoie H, et al. HLA-C and guttate psoriasis. Br J Dermatol. 2000;143:1177–82. doi: 10.1046/j.1365-2133.2000.03885.x. [DOI] [PubMed] [Google Scholar]
- 51.Barber LD, Percival L, Valiante NM, et al. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. 1996;184:735–40. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zemmour J, Gumperz JE, Hildebrand WH, et al. The molecular basis for reactivity of anti-Cw1 and anti-Cw3 alloantisera with HLA-B46 haplotypes. Tissue Antigens. 1992;39:249–57. doi: 10.1111/j.1399-0039.1992.tb01943.x. [DOI] [PubMed] [Google Scholar]
- 53.Baur MP, Neuebauer M, Albert ED. Reference tables of two-locus haplotype frequencies and delta values in Caucasians, Orientals, and Negroids. Berlin: Springer-Verlag; 1984. [Google Scholar]
- 54.McFadden JP, Baker BS, Powles AV, Fry L. Psoriasis and streptococci: the natural selection of psoriasis revisited. Br J Dermatol. 2009;160:929–37. doi: 10.1111/j.1365-2133.2009.09102.x. [DOI] [PubMed] [Google Scholar]
- 55.Asahina A, Akazaki S, Nakagawa H, et al. Specific nucleotide sequence of HLA-C is strongly associated with psoriasis vulgaris. J Invest Dermatol. 1991;97:254–8. doi: 10.1111/1523-1747.ep12480361. [DOI] [PubMed] [Google Scholar]
- 56.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 57.Conrad C, Boyman O, Tonel G, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13:836–42. doi: 10.1038/nm1605. [DOI] [PubMed] [Google Scholar]
- 58.Ogasawara K, Lanier LL. NKG2D in NK and T cell-mediated immunity. J Clin Immunol. 2005;25:534–40. doi: 10.1007/s10875-005-8786-4. [DOI] [PubMed] [Google Scholar]
- 59.Krueger G, Callis K. Potential of tumor necrosis factor inhibitors in psoriasis and psoriatic arthritis. Arch Dermatol. 2004;140:218–25. doi: 10.1001/archderm.140.2.218. [DOI] [PubMed] [Google Scholar]
- 60.Nair RP, Stuart P, Henseler T, et al. Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet. 2000;66:1833–44. doi: 10.1086/302932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.