Abstract
Significant strides in the understanding of the role of epigenetic regulation in asthma and allergy using both epidemiological approaches as well as experimental ones have been made. This review focuses on new research within the last two years. These include advances in determining how environmental agents implicated in airway disease can induce epigenetic changes, how epigenetic regulation can influence T helper cell (Th) differentiation and T regulatory (Treg) cell production, and new discoveries of epigenetic regulation associated with clinical outcomes.
Introduction
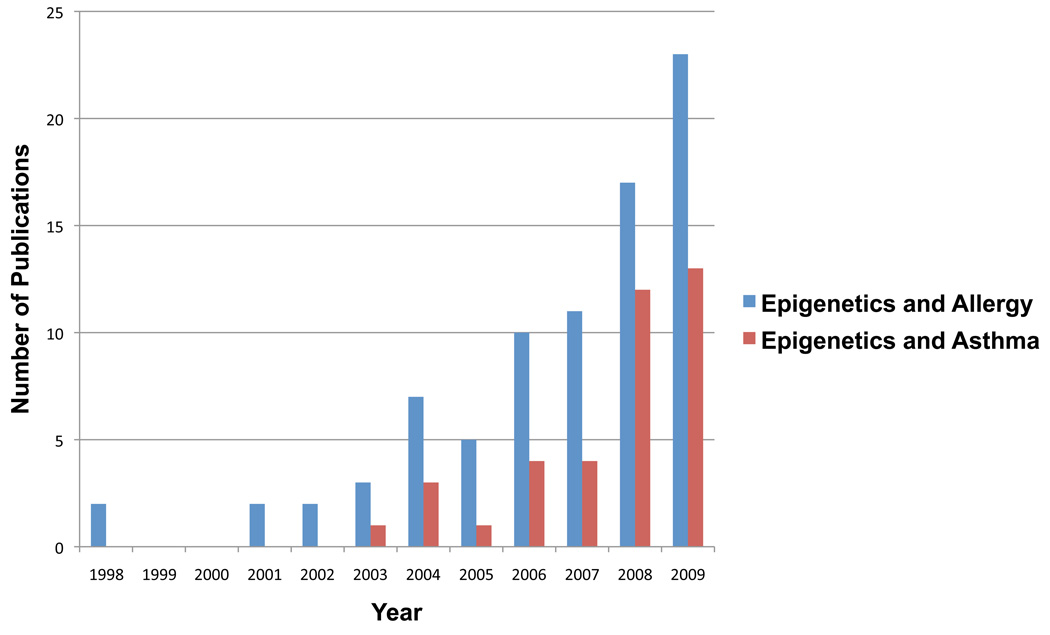
The field of epigenetics provides a new look at the old debate of nature versus nurture and reexamines the question “What health risks are heritable?” Publication rates on epigenetics and the complex diseases asthma and/or allergy have increased rapidly over the last several years (Figure 1). This new attention is a result of a growing recognition that epigenetic regulation has the potential to explain many mysteries in the pathogenesis of allergy, such as its susceptibility stemming from prenatal environmental exposures and its subsequent variable phenotype. Significant strides in the understanding of the role of epigenetic regulation in asthma and allergy using both epidemiological approaches as well as experimental ones have been made.
Figure 1.
Publications on Epigenetics and Allergy, Asthma through 2009. Number of publications was determined through PudMed search using search phrases “epigenetics and allergy” or “epigenetics and asthma”.
In this article we will review two areas where significant advances have been made: 1) epigenetic regulation in response to environmental exposures, and 2) epigenetic regulation associated with the development of asthma and allergy, both at the molecular and clinical level. This review will focus on new research within the last two years. These works include recent advances in determining how environmental agents implicated in airway disease can induce epigenetic changes, how epigenetic regulation can influence T helper cell (Th) differentiation and T regulatory (Treg) cell production, and new discoveries of epigenetic regulation associated with clinical outcomes.
Defining Epigenetic Regulation
The modern definition of epigenetics is the inheritance of changes occurring in gene expression that does not depend on changes to the DNA sequence.[1] However, the term epigenetics as coined by C.H. Waddington in the 1940’s is ‘the causal interactions between genes and their products, which bring the phenotype into being’.[2] Waddington’s definition initially referred to the role of epigenetics in embryonic development; however, the definition of epigenetics has evolved over time and is implicated in a wide variety of biological processes. The most common epigenetic mechanisms include DNA methylation, histone modifications, and noncoding RNAs. These molecular changes have the potential to modify the transcription of genes involved in the host response to environmental compounds, the ensuing proinflammatory response, or even the efficacy of pharmacological treatment.
Epigenetic modifications may be heritable across multiple generations such that prenatal parental or grandparental exposures impact gene expression in offspring without altering DNA sequences.[3] Early seminal work using mice models performed by Cooney and colleagues and Waterland and Jirtle demonstrated this inheritance for the kinked tail (AxinFu) allele and agouti-viable yellow (Avy) allele.[4–6] Alternatively, epigenetic modifications may occur postnatally leading to sustained effects on gene transcription.[7] In other work, Weaver and colleagues found that after exposure to histone deacetylase inhibitors such as trichostatin A or methionine in adulthood, rats underwent reversal of early life epigenetic programming in the hippocampus associated with glucocorticoid receptor expression and hypothalmic pituitary-adrenal and anxiety mediated behaviors.[8] Hence, epigenetic modifications also may be reversible during certain time periods according to a model that may fit the remitting course of childhood asthma.
DNA methylation
DNA methylation is an epigenetic mechanism that allows regulation of transcription via the addition of methyl groups to the fifth carbon of the nucleotide cytosine. It is the most frequent covalent modification of DNA.[9] DNA methylation often takes place in repetitive, high frequency CpG sites known as islands. These are regions with at least 500 base pairs and contain greater than 50% of cytosine and guanine nucleotides. Such regions are located at the 5’-terminal. CpG methylation inhibits gene transcription by either blocking the ability of transcription factors to bind to the recognition sites on the CpG nucleotides or by facilitating the binding of transcription inhibiting proteins.[10]
Histone Modification
Chromatin is structured within the cell nucleus into units called nucleosomes, composed of histone octamers around which DNA is coiled. Post-translation modifications of histones by means of acetylation, methylation, and phosphorylation are key elements in the chromatin packaging of DNA. As a result of this tight packaging, RNA polymerase II and transcription factors cannot reach their recognition sequence to turn on transcription. With acetylation, the DNA around the histone core unwinds, activators of transcription obtain access to DNA, and gene expression can then proceed.[11] Conversely, removal of acetyl groups as mediated via histone deacetylases (HDAC) induces gene silencing. HDACs operate as part of large multiprotein complexes so that when they are associated with other proteins they can form transcriptional repressor complexes.[12] Histone methylation is another epigenetic modification that is generally associated with transcriptional repression by adding a methyl group to amino acids in a histone protein; however, methylation of some histone residues may result in transcriptional activation, such as in the case of methylation of lysine 4 of histone 3 (H3K4). In contrast, methylation of lysine 9 or 27 of histone 3 (H3K9 and H3K27) is transcriptionally repressive.[13]
Histone modifications may occur in association with DNA methylation. Histone methylation differs from DNA methylation in that histone methylation directly alters chromatin structure to repress transcription where DNA methylation induces transcriptional silencing through recruitment of methyl-CpG binding proteins. These proteins selectively recognize and bind methylated DNA to repress transcriptional activity through recruitment of histone deacetylase-containing complexes.[14] Regulatory elements that aid in chromatin remodeling over long distances by binding of transcription factors and formation of chromatin loops also fall into the category of posttranslational histone modifications. Enhancers bind transcription factors and stimulate the formation of chromatin loops allowing promoter-transcription factor interaction, whereas silencers make chromatin structure inaccessible, thereby inducing opposing effects on gene transcription.[15, 16]
Epigenetic Regulation, Prenatal Exposures, and Asthma
So why consider epigenetic regulation in the pathogenesis of asthma? The variable natural history of asthma is determined by complex genetic and environmental influences and epigenetics may be a link to this intricate interplay. Environmental exposures during time periods when there is greater susceptibility to epigenetic regulation may be responsible for developmental plasticity. These time periods when epigenetic modifications may be more likely to occur include prenatal, early childhood and adolescence.[17] These time windows of susceptibility coincide with time periods when the asthma phenotype is known to be variable and changing.[1, 18]. During the prenatal period in particular, epigenetic reprogramming may lead to a generation of cells with a broad developmental potential.[19]
In the first case (i.e. prenatal time period), growing scientific evidence has supported a role for intrauterine environmental influences in the risk for later pediatric asthma.[20, 21] This premise has been described poignantly as the rationale behind the Barker hypothesis that postulates that organs undergo developmental programming in utero. This programming predetermines the subsequent physiologic and metabolic adaptations during adult life. Thus, through epigenetic regulation, in utero events have the potential to program persisting disease phenotypes and also determine the subsequent risk of disease.[17]
Another way to consider prenatal epigenetic regulation is as a mechanism underlying the heritable component of allergic disease that is not attributable to the DNA sequence alone.[22] Gene expression may be determined by its parental origin. This functional inequality of expression between two parental alleles of a gene is defined as genomic imprinting. The mechanism of imprinting is complex and not completely understood, however the “imprint mark” may be a parental-specific methylation of CpG-rich domains that silences gene transcription and is established during gametogenesis. These imprinted marks on a gene, however must be erased in the germline when transmitted through individuals of the opposite sex, but maintained during somatic cell division.[23, 24] Imprinting has been implicated in embryogenesis. For example, Barton and colleagues showed that eggs with only the maternal or paternal genome can only develop up to the blastocyst stage.[25] This finding suggests the developing embryo needs both the maternal and paternal genomes so that if the maternal copy of a gene has been imprinted and is inactive there will still be an active paternal copy of the gene.[26]
Gene expression in asthma and atopy also may be influenced by imprinting of parental genomes such that the genetic predisposition to allergic disease can be silenced by DNA methylation when transmitted from the father but expressed when transmitted by the mother. Some epidemiological literature supports this phenomenon. For example, Litonjua and colleagues reported the risk for childhood asthma increased with maternal, but not paternal history of asthma.[27] Furthermore, in a meta-analysis by Lim and colleagues, maternal asthma predisposes offspring to disease more so than paternal asthma. [28] Liu and colleagues found that maternal, but not paternal, total immunoglobulin (IgE) levels significantly correlated with elevated IgE levels in cord blood at age 6 months.[29] However, in more recent work, Ferreira and colleagues investigated the role of global DNA methylation patterns in this “maternal effect” of atopy, namely the greater predominance of asthma or allergy if the mother, as opposed to father, is atopic. In their essentially negative study, they found that the AluSp element (a CpG rich stretch of DNA) in β-chain of the IgE receptor gene was hypermethylated across all children regardless of atopic status of the parents or children.[30, 31] Hence, the findings that support imprinting as the explanation for the disproportionate inheritance of atopy from the mother are mixed, and more correlative than mechanistic.
Intrauterine exposures causing epigenetic regulation also may affect asthma risk. Recent epidemiological support for this premise was suggested by Li and Colleagues. Using retrospective questionnaires, they demonstrated that prenatal exposure to maternal tobacco smoking was associated with lower pulmonary function and increased asthmatic symptoms in childhood. Moreover combined maternal and grandmaternal smoking during pregnancy was associated with an even greater risk of childhood asthma, suggesting a multigenerational heritability for asthma.[32]
Prenatal Folate and Risk of Atopy and Asthma
One feature of the intrauterine environment that has been implicated in later childhood risk for atopy is prenatal diet and folate intake specifically. Folate is a relatively large source of methyl donors and prenatal supplementation may alter DNA methylation and consequently gene expression. In mice, the importance of a prenatal diet high in methyl donors was demonstrated by Hollingsworth and colleagues who found that this diet (combined with soy, also known to alter DNA methylation) given to mice during gestation and weaning was associated with greater airway hyperactivity, airway eosinophilic inflammation, chemokines, and IgE production in the offspring. The phenotype also was associated with greater levels of DNA methylation and RNA and protein suppression of runt-related transcription factor 3 (Runx3), a gene associated with silencing of CD4 during T cell lineage decisions.[33, 34] Consistent with Hollingsworth, Whitrow and colleagues found that women who received supplemental folic acid during late pregnancy as opposed to early supplementation, as measured by retrospective consumption questionnaires and interviews, had children with a greater risk of physician-diagnosed asthma.[35] However, Håberg and colleagues reported that use of folic acid supplements in pregnancy during the first trimester, also measured by retrospective questionnaires, was associated with a slight increase in the risk of early respiratory infections and wheeze.[36] Their differing findings in regards to timing of folate exposure during pregnancy may be explained by differences in reporting of folate intake as a bi-categorical (yes/no) value and also shorter follow-up (18 months versus 5.5 years). Nonetheless, they suggest that a critical window within pregnancy during which folate exposure can epigenetically influence the subsequent risk for asthma may be important to investigate prospectively and should be confirmed in more human studies. In other work, Steegers-Theunissen and colleagues found that periconceptional folic acid use of the mother was related to increased methylation of the insulin-like growth factor 2 (IGF2), a critical and widely expressed embryonic growth factor of the child. Furthermore, maternal S-adenosylmethionine (an endogenous source of methyl groups formed from demethylated methionine) concentrations were correlated positively with IGF2 methylation of the child’s peripheral blood DNA.[37] These findings suggest that prenatal dietary folate may act as an epigenetic regulator of gene expression and ultimately may be associated with altered disease risk.
Prenatal Environmental Exposures and Asthma Risk
A second intrauterine environmental exposure widely implicated with later pediatric asthma is air pollution. Prenatal environmental tobacco smoke (ETS) exposure has been associated with decreased pulmonary function and a greater risk for developing asthma in both children and adults.[38–41] Wu and colleagues showed that the mice exposed to sidestream smoke during earlier periods of gestation had significantly elevated methacholine responses for lung resistance, decreased lung compliance, increased substance P inervation in tracheal smooth muscle, and elevated levels of nerve growth factor in BAL.[42] In humans, our group showed that at age 2 years, more difficulty breathing and probable asthma was reported among children jointly exposed to prenatal polycyclic aromatic hydrocarbons (PAHs) and postnatal ETS.[43] As described below, data on how these intrauterine exposures may confer a greater risk for allergy and the role of epigenetic regulation are just beginning to emerge.
Epigenetic Regulation of the Effects of Environmental Exposures Implicated in Allergy, Asthma: The Latest Evidence
There is a growing body of literature implicating epigenetic mechanisms in the host response to several environmental exposures implicated in the development of atopy or asthma. For example, considerable work has been done recently on the induction of epigenetic changes following exposure to air pollution. For one, our group found that concomitant exposure of mice to diesel exhaust particles and allergen Aspergillus fumigatus (A. fumigatus) augmented immunoglobulin (Ig) E production potentially by inducing DNA hypermethylation at select CpG sites in the interferon (IFN)- γ promoter and DNA hypomethylation in the interleukin (IL)-4 promoter. The extent of altered methylation correlated with IgE production.[44] While the bulk of noncoding DNA may not be conserved between humans and mice, sequence comparisons reveal islands of highly conserved noncoding sequences (CNS). This has been seen at the site located approximately 5 kb 5′ of the transcription start site for the IFN-γ gene and also at the −53 CpG and −190 CpG of the IFN- γ promoter in CD4 cells.[45, 46] However, Janson and colleagues found interspecies differences in the methylation status of the IFN- γ promoter in naive CD4 T lymphocytes. In humans the IFN- γ promoter displays a high level of methylation in naive CD4 T lymphocytes, but becomes demethylated with Th1 differentiation, whereas in mice the IFN- γ promoter region of naive CD4 T lymphocytes is demethylated.[47]
In other work, Cau and colleagues documented altered chromatin modification following exposure to diesel exhaust particles. In their experiments, exposure of human bronchial epithelial cell lines to diesel exhaust particles increased acetylation of histone H4 associated with the cyclooxygenase (COX)-2 promoter. This led to the posttranslational degradation of histone deacetylase 1 (HDAC1) and recruitment of histone acetyltransferase (HAT) p300 to the COX2- promoter, ultimately activating COX-2 gene expression.[48] In a recent human cohort study by Baccarelli and colleagues, exposure of adults to diesel and fine particulate matter (PM)2.5 was associated with DNA demethylation in repetitive elements such as LINE-1, an indicator of global methylation.[49, 50] Similarly, Tarantini and colleagues studied the short- (after 2 consecutive days off from work in a steel production plant) and long-term (after 3 consecutive days of work in a steel production plant) effects of PM10 exposure on both global and asthma candidate gene (iNOS) DNA methylation in humans. They reported that short-term PM10 exposure may be associated with DNA demethylation of the iNOS promoter, and long-term PM10 exposure was associated with global DNA demethylation estimated in Alu repeated elements (another indicator of global methylation).[51] Our group showed that higher transplacental exposure to PAH was associated with altered DNA methylation of Acyl-CoA synthetase long-chain family member 3 (ACSL3), possibly a novel asthma candidate gene. [52] Combined, these studies implicate epigenetic regulation, including both DNA methylation and histone modification, in the individual response to exposure to air pollution, and diesel, PM and PAH specifically. Finally, a recent study by Breton and colleagues reported that prenatal tobacco smoke was associated with lower DNA methylation for AluYb8 repetitive elements in children. Interestingly, prenatal exposure was associated with lower LINE-1 methylation in the GSTM1 null children but higher methylation in the GSTM1-present children. This work suggests a novel interaction between exposure and genotype influencing susceptibility to DNA methylation.[53]
Epigenetic Regulation and T Cell Changes Related to the Allergic Immune Response: The Latest Evidence
Th1 v Th2
The role of Th2 polarization in allergic disease has been well-studied since Mosmann and colleagues seminal reports in mice in 1986.[54] Despite this abundance of research, it still remains unclear what molecular processes are important for the persistence of T cell polarization, especially in vivo. Previous studies have examined the role of intrauterine cytokine environments in transplacental transmission of T cell polarization. For example, using a mouse model, Hamada and colleagues reported that postnatal ovalbumin challenge induced airway hyperresponsiveness in the offspring of mothers sensitized to ovalbumin prenatally, whereas no effect was seen in the offspring of nonsensitized mothers. Furthermore, postnatal challenge to unrelated allergens induced a similar response in the offspring of ovalbumin-allergic mothers, suggesting that maternal inheritance of an asthma-like phenotype may be allergen independent. Importantly, treatment of asthmatic mothers with neutralizing anti-IL-4 antibody prior to conception abrogated this response. [55] This finding associates IL-4 and potentially other intrauterine cytokines as a mediator of maternal inheritance of susceptibility to airway hyper-responsiveness, at least in mice. The role of epigenetic regulation in the intrauterine cytokine environment has yet to be demonstrated.
Recent advances have been made in the understanding of the regulation of levels of methylation of CpG sites within the IFN-γ promoter. Jones and colleagues documented that during Th2 polarization, the IFN-γ promoter undergoes de novo methylation, most notably in the CpG located at the −53 position proximal to the transcription start site (TSS); which is highly conserved across species. DNA methylation at this site prevents transcription factor (c-Jun, CREB) binding required for IFN-γ gene expression during Th1 polarization.[46] White and colleagues also demonstrated that under Th1 polarizing conditions, CpG sites −295, −186 and −54 upstream to the (TSS) of the IFN-γ promoter in neonatal CD4+ T cells were demethylated, whereas under Th2 polarizing conditions the majority of these sites remained methylated, again providing a epigenetic mechanism for possible suppression of the IFN-γ locus in Th2 polarized cells.[56] Recent advances also have focused on chromatin remodeling of the IFN-γ promoter. Zhang and colleagues found that nucleosomes at the IFN-γ gene promoter undergo repositioning to develop a more open topography which creates new access to transcription factor binding in Th1 cells; this occurs in a STAT4 dependent, T-bet independent manner.[57] The activation of STAT4 allows for histone acetylation to accumulate at the conserved regions of the IFN- γ promoter, facilitating STAT4 transcription factor binding, thereby committing naïve T cells towards a Th1 lineage and not that of Th2. These results suggest IFN-γ transcription is determined in part by the balance of the activity of histone acetylases and deacetylases recruited to the IFN- γ promoter. However, the IFN- γ promoter also can acquire repressive epigenetic modifications, such as histone methylation, particularly in developing Th2 cells. For example, Chang and colleagues found that methylation of lysine 9 of histone 3 (H3K9) was sustained in Th1 cells, but rapidly extinguished in developing Th2 cells, whereas lysine 27 of histone 3 (H3K27) became methylated during Th2 differentiation. Both events occurred via STAT6-and GATA-3-dependent mechanisms.[58]
Previous work also has focused on regulatory elements of the IFN- γ locus. For example, Lee and colleagues reported that Th1 cells display DNAse I hypersensitivity and histone modifications at conserved noncoding sequence (CNS)1, whereas Th2 cells do not. This finding, indicates that this region is more permissive to transcription in Th1 differentiated cells compared to Th2 cells.[45] Hatton and colleagues found that CNS22 is particularly susceptible to permissive histone modifications and selective T-bet binding, and subsequently functions to increase IFN- γ expression in Th1 cells. Deletion of CNS22 resulted in decreased IFN- γ expression in Th1 effector cells, cytotoxic T lymphocytes and natural killer cells.[59] These findings implicate CNS22 as an epigenetic regulator of IFN- γ expression. Furthermore, Shnyreva and colleagues reported histone acetylation in the IFN- γ promoter, intronic regions, CNS1 and CNS2 increased as naïve T cells differentiated into IFN-γ-producing effector CD8+ and Th1 T cells, but not into Th2 T cells.[60] In other work, Schoenborn and colleagues performed a large scale epigenetic profiling using DNA methylation analysis and DNase hypersensitivity site mapping of the IFN-γ gene. They found that regulatory elements such as CNS−54 and +46 functioned mainly as boundary elements where DNA sequences prevent promoter-enhancer interactions. In contrast, more proximal CNSs such as CNS–34, CNS–22, CNS–6 functioned to increase transcription of the IFN-γ gene.[15]
DNA methylation also plays an important role in the control of Th2 cytokine expression and stabilization during T helper cell development. Most notably, when naïve T cells are activated under Th2 polarizing conditions, demethylation occurs at the IL-4 gene promoter.[16, 61] More recently investigations have focused on the epigenetic regulation of IL-13 expression. Webster and colleagues reported that in naïve CD4+ T cells, DNA hypomethylation was limited to the distal promoter of IL-13, however with Th2 differentiation the proximal IL-13 promoter preferentially was hypomethylated. Furthermore, histone H4 acetylation levels and hypersensitivity sites at the IL-13 proximal promoter were considerably higher in Th2 cells.[62] These results suggest that differential IL-13 expression may depend on the epigenetic control and accessibility of the proximal promoter in Th2 cells.
In other work, Kim and colleagues characterized demethylation events for the Th2 cytokine locus control region rad50 hypersensitive site 7 (RHS7), an area where IL-4, IL-5, and IL-13 expression is regulated.[63] Under Th2 polarizing conditions, RHS7 becomes demethylated in a STAT6 dependent manner in CD4+ T cells. This demethylation appears to require a signaling contribution from both the IL-4 receptor and CD28, whereas GATA3 mediated signaling also involving demethylation appears to be insufficient for this remodeling.[64] Additionally, Chong and colleagues found that loss of the proximal enhancer of the CD4 gene (E4p) in immature CD4+CD8+ thymocytes resulted in unstable expression of CD4 in mature T cells. However, deletion of E4p in cells already committed to the helper T cell lineage resulted in stable CD4 expression. Transcriptionally permissive histone modifications of histone 3, such as acetylation and trimethylation, required initial E4p enhancer activity. These findings suggest that E4p is an epigenetic regulator of CD4 expression[65] Dispirito and colleagues found that the level of diacetylation of histone 3 increased as naive CD8+ T cells developed into T memory cells, thereby implicating epigenetic regulation in CD8 differentiation. [66] In other recent work, Hughes and colleagues reported that paradoxically DNA methylation may be associated with transcriptional permissiveness, particularly in areas of increased promoter CpG island density. Using DNA methylation and gene expression analysis, they found that 27% of methylated genes (55 genes) in primary human CD4+ T cells are expressed. Of the methylated, but transcriptionally expressed genes, methylation peaks were located approximately 9 nucleosome away from the transcription start site (TSS) and had higher maximum CpG island densities within promoter sequences of target genes. In contrast methylated and transcriptionally repressed genes were located only 6 nucleosomes away from the TSS and had a relatively lower maximum CpG island density.[67] This finding may be a consequence of poor recruitment of methyl binding proteins and ultimately histone deacetylases secondary to the longer distance from the TSS, however this has yet to be demonstrated. This alteration of repressive DNA methylation patterns at key CpGs may play a role in T cell polarization In summary, recent advances in epigenetic regulation have provided great insight in T helper cell lineage commitment; although our knowledge is still relatively limited in the area of in vivo changes.
T reg
There is growing evidence supporting the role for T regulatory (Treg) cells and the immunosuppressive cytokines they produce, as mechanisms by which allergen specific immunotherapy and healthy immune responses to allergens are mediated.[68] T reg cells are capable of suppressing effector cells and therefore likely play a role in allergen tolerance by harnessing allergen induced inflammation in early processes such as sensitization.[69] Specifically, T reg cells contribute to the control of allergen specific immune responses by suppressing of mast cells, basophils, eosinophils, and antigen-presenting cells that support the generation of effector Th2 and Th1 cells and by suppressing allergen-specific IgE, IgG4, IgA production, or both.[68]
Epigenetic mechanisms controlling Treg development are just beginning to be explored. Studies previously have focused on FoxP3, a central control element essential for Treg development and function. Epigenetic changes are a prerequisite for FoxP3 expression and T reg differentiation. Recently, Floess and colleagues documented that T regs induced by TGF-beta display incomplete demethylation despite high FoxP3 expression. Upon restimulation in the absence of TGF-beta, these Tregs lose both FoxP3 expression and suppressive activity. These results suggest that expression of FoxP3 must be stabilized by epigenetic modification to allow the development of a permanent suppressor cell lineage.[70] Polansky and colleagues also found that inhibition of DNA methylation by azacytidine, even in the absence of exogenous TGF beta, promoted de novo induction of FoxP3 expression. In contrast, in vitro methylation of Treg specific demethylated region (TSDR) diminished FoxP3 transcriptional activity. This work demonstrates that epigenetic regulation in this region is critical for the establishment of a stable Treg lineage.[71] Janson and colleagues examined the methylation profile of FoxP3 promoter in the commitment towards the T reg lineage. They found that human CD4+CD25hi T regs displayed a demethylated FoxP3 promoter in contrast to CD4+CD25lo T cells which were partially methylated. Moreover stimulated CD4+CD25lo T cells transiently expressed FoxP3, but remained partially methylated.[72] This characterization of the unique FoxP3 promoter methylation in Tregs suggests that a demethylated pattern is a prerequisite for stable FoxP3 expression and suppressive phenotype. Much more research is needed to illuminate the epigenetic markers needed to establish a lineage commitment towards that of Tregs and its role in atopic disease.
Epigenetic Regulation and Clinical Allergic Disease
To date, only a few papers have examined the clinical consequences of epigenetic modifications on asthma and allergic disease. For example, our group found that the 59-CpG island methylation status of acyl-CoA synthetase long chain (ACSL)3 is not only associated with prenatal PAH exposure, but also is associated significantly with a parental report of asthma symptoms in children prior to age 5 years. ACSL3 belongs to the ACSL family of genes that encodes key enzymes in fatty acid metabolism.[52] It has been speculated that hypermethylation of this gene in T helper cells or lung tissues may diminish fatty acid utilization and beta-oxidation-energy production. This in turn may influence membrane phospholipid composition, with associated anti-inflammatory effects in asthma. However it is unknown how these changes directly airway inflammation and future studies will need to validate whether ACSL3 is a novel asthma gene. Nonetheless, this is the first study to examine the effects of prenatal exposure to ambient air pollutants on DNA methylation patterns in genes potentially associated with asthma phenotype of the offspring. Another cohort study by Kwon and colleagues examined the relationship between CpG methylation and IL-4 gene expression before and after allergen stimulation in human CD4+ lymphocytes from sensitized hosts. They found that after combined Dermatophagoides pteronyssinus/Dermatophagoides farinae stimulation, hypomethylation in the IL-4 promoter increased among asthmatics. The concentration of IL-4 was strongly correlated with the extent of hypomethylation.[73] In a prospective cohort study by White and colleagues, ex vivo IFN-γ promoter methylation was reduced in CD8+ T cells, but not CD4+ T cells from atopic children.[56]
Finally, Su and colleagues examined epigenetic regulatory functions in the maintenance of Th1 and Th2 immunity in children by ex vivo inhibition of endogenous HDAC activity. They found that increasing cellular acetylation via administration of trichostatin, histone deacetylase inhibitor, to peripheral blood mononuclear cells, and corresponding decreasing HDAC activity, shifted immune responses toward Th2 phenotype with increased production of IL-13 and IL-5 and increased expression of GATA-3.[74] These results suggest that endogeneous HDAC was important in preventing pre-established cytokine responses from deviating toward excessive Th2 polarization. Furthermore, other work by Su and colleagues demonstrated that ex vivo HAT activity was increased and HDAC activity was reduced among children with allergic asthma relative to atopic nonasthmatic controls. The intensity of cellular histone acetylation increased progressively with increasing bronchial hyperresponsiveness, thereby potentially implicating epigenetic regulatory control over varying atopic asthmatic phenotypes.[75]
Epigenetic modifications, including DNA methyation, are dynamic. For example, Tarantini and colleagues found that short term exposure to PM10 is associated with decreased methylation of the inducible nitric oxide synthase gene (iNOS) promoter over three days.[51] Furthermore, Baccarrelli and colleagues reported that exposure of adults to diesel and PM2.5 over 7 days (using moving averages) was associated with DNA demethylation in repetitive elements such as LINE-1, an indicator of global methylation.[50] In contrast, Bjornsson and colleagues reported that 8–10% of individuals on average had an absolute change in global DNA methylation of more than 20%. These results were found in 2 distinct populations over a duration of 11–16 years. [76] While the majority of individuals did not have significant changes in this study, there was familial clustering of longitudinal DNA methylation changes, thereby implicating a genetic component to methylation stability. Additionally, Christensen and colleagues studied methylation patterns in 1,413 CpG loci associated with 773 genes in 10 human tissue types. They found that loci in CpG islands were more likely to gain methylation within the 10 years of study follow-up compared to loci outside of CpG islands that were more likely to demethylate. These methylation changes were found consistently across tissue type. [77] While these findings may not best demonstrate the dynamic nature of these changes, it does demonstrate some plasticity of epigenetic modifications and furthermore implicates age-related losses of epigenetic patterns. Histone modification also are believed to be dynamic and associated with even shorter term changes to gene transcription compared to DNA methylation.[78] However future studies are needed to validate this notion.
Conclusion
The accumulated evidence has undoubtedly solidified the case for implicating epigenetics as a mediator of complex gene by environment interactions relevant to the development of asthma and allergic diseases. Advances have been made linking air pollution and ETS exposure with atopy via epigenetic mechanisms. Furthermore, considerable strides have been made implicating epigenetic mechanisms in T cell differentiation, especially at the IFN-γ locus. However, much more research is still needed, particularly to delineate the role of epigenetics in the lineage commitment of T reg cells and also to define the clinical consequences of such epigenetic alterations. Despite the acceptance of epigenetic regulation in the pathogenesis of complex diseases and the rapid rise in publications, the extent of environmental epigenetics in the pathogenesis of asthma and allergies is just being realized. Longitudinal cohort studies are needed to examine the time course and time period of susceptibility to epigenetic regulation following environmental exposures and their contribution to allergic disease. Ultimately, an individual’s epigenome early in life may be a helpful biomarker in determining later risk of asthma and atopy and initiating early intervention. Despite these challenges, the future holds exciting promise.
Acknowledgments
Supported by the National Institute of Environmental Health Sciences R01 ES013163 and P50ES015905
References
- 1.Esteller M. Epigenetics in evolution and disease. Lancet. 2008:590–596. [Google Scholar]
- 2.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 3.Tang W, Ho S. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–182. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolinoy D, Weidman J, Jirtle R. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Waterland R, Jirtle R. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooney C, Dave A, Wolff G. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz D. Epigenetics and environmental lung disease. Proc Am Thorac Soc. 2010;7:123–125. doi: 10.1513/pats.200908-084RM. [DOI] [PubMed] [Google Scholar]
- 8.Weaver I, Meaney M, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos K, Mazzola T, Carvalho H. The prima donna of epigenetics: the regulation of gene expression by DNA methylation. Braz J Med Biol Res. 2005;38:1531–1541. doi: 10.1590/s0100-879x2005001000010. [DOI] [PubMed] [Google Scholar]
- 10.Isidoro-García M, Dávila-González I, Pascual de Pedro M, Sanz-Lozano C, Lorente-Toledano F. Interactions between genes and the environment. Epigenetics in allergy. Allergol Immunopathol (Madr) 2007;35:254–258. doi: 10.1157/13112992. [DOI] [PubMed] [Google Scholar]
- 11.Miller R, Herbstman J. Epigenetic Mechanisms in AsthmaEpigenetic and Human Health: Linking Hereditary, Environmental and Nutrional Aspects. Wiley Publishers; 2008. [Google Scholar]
- 12.Johnson C, Turner B. Histone deacetylases: complex transducers of nuclear signals. Semin Cell Dev Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 14.Ballestar E, Esteller M. The epigenetic breakdown of cancer cells: from DNA methylation to histone modifications. Prog Mol Subcell Biol. 2005;38:169–181. doi: 10.1007/3-540-27310-7_7. [DOI] [PubMed] [Google Scholar]
- 15.Schoenborn J, Dorschner M, Sekimata M, Santer D, Shnyreva M, Fitzpatrick D, Stamatoyannopoulos J, Stamatoyonnapoulos J, Wilson C. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janson P, Winerdal M, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochim Biophys Acta. 2009;1790:906–919. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Miller R, Ho S. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177:567–573. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandhane P, Greene J, Cowan J, Taylor D, Sears M. Sex differences in factors associated with childhood- and adolescent-onset wheeze. Am J Respir Crit Care Med. 2005;172:45–54. doi: 10.1164/rccm.200412-1738OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 20.Herz U, Joachim R, Ahrens B, Scheffold A, Radbruch A, Renz H. Prenatal sensitization in a mouse model. Am J Respir Crit Care Med. 2000;162:S62–S65. doi: 10.1164/ajrccm.162.supplement_2.ras-1. [DOI] [PubMed] [Google Scholar]
- 21.Prescott S, Clifton V. Asthma and pregnancy: emerging evidence of epigenetic interactions in utero. Curr Opin Allergy Clin Immunol. 2009;9:417–426. doi: 10.1097/ACI.0b013e328330634f. [DOI] [PubMed] [Google Scholar]
- 22.Reamon-Buettner S, Borlak J. A new paradigm in toxicology and teratology: altering gene activity in the absence of DNA sequence variation. Reprod Toxicol. 2007;24:20–30. doi: 10.1016/j.reprotox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Falls J, Pulford D, Wylie A, Jirtle R. Genomic imprinting: implications for human disease. Am J Pathol. 1999;154:635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt M, Cookson W. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy. 1998;28 Suppl 1:56–61. doi: 10.1046/j.1365-2222.1998.0280s1056.x. discussion 65–6. [DOI] [PubMed] [Google Scholar]
- 25.Barton S, Surani M, Norris M. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 26.Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu Rev Genet. 1988;22:127–146. doi: 10.1146/annurev.ge.22.120188.001015. [DOI] [PubMed] [Google Scholar]
- 27.Litonjua A, Carey V, Burge H, Weiss S, Gold D. Parental history and the risk for childhood asthma. Does mother confer more risk than father? Am J Respir Crit Care Med. 1998;158:176–181. doi: 10.1164/ajrccm.158.1.9710014. [DOI] [PubMed] [Google Scholar]
- 28.Lim R, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta-analysis. PLoS One. 2010;5:e10134. doi: 10.1371/journal.pone.0010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Wang C, Chuang H, Ou C, Hsu T, Yang K. Prenatal prediction of infant atopy by maternal but not paternal total IgE levels. J Allergy Clin Immunol. 2003;112:899–904. doi: 10.1016/j.jaci.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 30.Cookson W, Young R, Sandford A, Moffatt M, Shirakawa T, Sharp P, Faux J, Julier C, Nakumuura Y, Nakumura Y. Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet. 1992;340:381–384. doi: 10.1016/0140-6736(92)91468-n. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira M, Oates N, van Vliet J, Zhao Z, Ehrich M, Martin N, Montgomery G, Whitelaw E, Duffy D. Characterization of the methylation patterns of MS4A2 in atopic cases and controls. Allergy. 2009 doi: 10.1111/j.1398-9995.2009.02135.x. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Langholz B, Salam M, Gilliland F. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest. 2005;127:1232–1241. doi: 10.1378/chest.127.4.1232. [DOI] [PubMed] [Google Scholar]
- 33.Hollingsworth J, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, Bailey N, Potts E, Whitehead G, Brass D, Schwartz D. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ehlers M, Laule-Kilian K, Petter M, Aldrian C, Grueter B, Würch A, Yoshida N, Watanabe T, Satake M, Steimle V. Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4-/CD8+ thymocytes. J Immunol. 2003;171:3594–3604. doi: 10.4049/jimmunol.171.7.3594. [DOI] [PubMed] [Google Scholar]
- 35.Whitrow M, Moore V, Rumbold A, Davies M. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol. 2009;170:1486–1493. doi: 10.1093/aje/kwp315. [DOI] [PubMed] [Google Scholar]
- 36.Håberg S, London S, Stigum H, Nafstad P, Nystad W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch Dis Child. 2009;94:180–184. doi: 10.1136/adc.2008.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steegers-Theunissen R, Obermann-Borst S, Kremer D, Lindemans J, Siebel C, Steegers E, Slagboom P, Heijmans B. Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS One. 2009;4:e7845. doi: 10.1371/journal.pone.0007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mannino D, Moorman J, Kingsley B, Rose D, Repace J. Health effects related to environmental tobacco smoke exposure in children in the United States: data from the Third National Health and Nutrition Examination Survey. Arch Pediatr Adolesc Med. 2001;155:36–41. doi: 10.1001/archpedi.155.1.36. [DOI] [PubMed] [Google Scholar]
- 39.Mannino D, Homa D, Redd S. Involuntary smoking and asthma severity in children: data from the Third National Health and Nutrition Examination Survey. Chest. 2002;122:409–415. doi: 10.1378/chest.122.2.409. [DOI] [PubMed] [Google Scholar]
- 40.Eisner M. Environmental tobacco smoke exposure and pulmonary function among adults in NHANES III: impact on the general population and adults with current asthma. Environ Health Perspect. 2002;110:765–770. doi: 10.1289/ehp.02110765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alati R, Al Mamun A, O'Callaghan M, Najman J, Williams G. In utero and postnatal maternal smoking and asthma in adolescence. Epidemiology. 2006;17:138–144. doi: 10.1097/01.ede.0000198148.02347.33. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z, Hunter D, Kish V, Benders K, Batchelor T, Dey R. Prenatal and early, but not late, postnatal exposure of mice to sidestream tobacco smoke increases airway hyperresponsiveness later in life. Environ Health Perspect. 2009;117:1434–1440. doi: 10.1289/ehp.0800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller R, Garfinkel R, Horton M, Camann D, Perera F, Whyatt R, Kinney P. Polycyclic aromatic hydrocarbons, environmental tobacco smoke, and respiratory symptoms in an inner-city birth cohort. Chest. 2004;126:1071–1078. doi: 10.1378/chest.126.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, Chen L, Miller R. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102:76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee D, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 46.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janson P, Marits P, Thörn M, Ohlsson R, Winqvist O. CpG methylation of the IFNG gene as a mechanism to induce immunosuppression [correction of immunosupression] in tumor-infiltrating lymphocytes. J Immunol. 2008;181:2878–2886. doi: 10.4049/jimmunol.181.4.2878. [DOI] [PubMed] [Google Scholar]
- 48.Cao D, Bromberg P, Samet J. COX-2 expression induced by diesel particles involves chromatin modification and degradation of HDAC1. Am J Respir Cell Mol Biol. 2007;37:232–239. doi: 10.1165/rcmb.2006-0449OC. [DOI] [PubMed] [Google Scholar]
- 49.Nawrot T, Adcock I. The detrimental health effects of traffic-related air pollution: a role for DNA methylation? Am J Respir Crit Care Med. 2009;179:523–524. doi: 10.1164/rccm.200812-1900ED. [DOI] [PubMed] [Google Scholar]
- 50.Baccarelli A, Wright R, Bollati V, Tarantini L, Litonjua A, Suh H, Zanobetti A, Sparrow D, Vokonas P, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi P, Baccarelli A. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perera F, Tang W, Herbstman J, Tang D, Levin L, Miller R, Ho S. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breton C, Byun H, Wenten M, Pan F, Yang A, Gilliland F. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mosmann T, Cherwinski H, Bond M, Giedlin M, Coffman R. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 55.Hamada K, Suzaki Y, Goldman A, Ning Y, Goldsmith C, Palecanda A, Coull B, Hubeau C, Kobzik L. Allergen-independent maternal transmission of asthma susceptibility. J Immunol. 2003;170:1683–1689. doi: 10.4049/jimmunol.170.4.1683. [DOI] [PubMed] [Google Scholar]
- 56.White G, Hollams E, Yerkovich S, Bosco A, Holt B, Bassami M, Kusel M, Sly P, Holt P. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17:557–564. doi: 10.1111/j.1399-3038.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–1505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang S, Aune T. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 59.Hatton R, Harrington L, Luther R, Wakefield T, Janowski K, Oliver J, Lallone R, Murphy K, Weaver C. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Shnyreva M, Weaver W, Blanchette M, Taylor S, Tompa M, Fitzpatrick D, Wilson C. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee D, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 62.Webster R, Rodriguez Y, Klimecki W, Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J Biol Chem. 2007;282:700–709. doi: 10.1074/jbc.M609501200. [DOI] [PubMed] [Google Scholar]
- 63.Lee G, Spilianakis C, Flavell R. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 64.Kim S, Fields P, Flavell R. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A. 2007;104:17052–17057. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong M, Simpson N, Ciofani M, Chen G, Collins A, Littman D. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dispirito J, Shen H. Histone acetylation at the single-cell level: a marker of memory CD8(+) T cell differentiation and functionality. J Immunol. 2010;184:4631–4636. doi: 10.4049/jimmunol.0903830. [DOI] [PubMed] [Google Scholar]
- 67.Hughes T, Webb R, Fei Y, Wren J, Sawalha A. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010 doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akdis M, Blaser K, Akdis C. T regulatory cells in allergy: novel concepts in the pathogenesis, prevention, and treatment of allergic diseases. J Allergy Clin Immunol. 2005;116:961–968. doi: 10.1016/j.jaci.2005.09.004. quiz 69. [DOI] [PubMed] [Google Scholar]
- 69.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 70.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang H, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Polansky J, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 72.Janson P, Winerdal M, Marits P, Thörn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3:e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon N, Kim J, Lee J, Oh M, Choi D. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol. 2008;28:139–146. doi: 10.1007/s10875-007-9148-1. [DOI] [PubMed] [Google Scholar]
- 74.Su R, Becker A, Kozyrskyj A, Hayglass K. Epigenetic regulation of established human type 1 versus type 2 cytokine responses. J Allergy Clin Immunol. 2008;121:57–63. doi: 10.1016/j.jaci.2007.09.004. e3. [DOI] [PubMed] [Google Scholar]
- 75.Su R, Becker A, Kozyrskyj A, Hayglass K. Altered epigenetic regulation and increasing severity of bronchial hyperresponsiveness in atopic asthmatic children. J Allergy Clin Immunol. 2009;124:1116–1118. doi: 10.1016/j.jaci.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 76.Bjornsson H, Sigurdsson M, Fallin M, Irizarry R, Aspelund T, Cui H, Yu W, Rongione M, Ekström T, Harris T, Launer L, Eiriksdottir G, Leppert M, Sapienza C, Gudnason V, Feinberg A. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Christensen B, Houseman E, Marsit C, Zheng S, Wrensch M, Wiemels J, Nelson H, Karagas M, Padbury J, Bueno R, Sugarbaker D, Yeh R, Wiencke J, Kelsey K. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hazzalin C, Mahadevan L. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]