Abstract
Purpose
To compare clinical, immunohistochemical and gene expression models of prognosis applicable to formalin-fixed, paraffin-embedded blocks in a large series of estrogen receptor positive breast cancers, from patients uniformly treated with adjuvant tamoxifen.
Methods
qRT-PCR assays for 50 genes identifying intrinsic breast cancer subtypes were completed on 786 specimens linked to clinical (median followup 11.7 years) and immunohistochemical (ER, PR, HER2, Ki67) data. Performance of predefined intrinsic subtype and Risk-Of-Relapse scores was assessed using multivariable Cox models and Kaplan-Meier analysis. Harrell’s C index was used to compare fixed models trained in independent data sets, including proliferation signatures.
Results
Despite clinical ER positivity, 10% of cases were assigned to non-Luminal subtypes. qRT-PCR signatures for proliferation genes gave more prognostic information than clinical assays for hormone receptors or Ki67. In Cox models incorporating standard prognostic variables, hazard ratios for breast cancer disease specific survival over the first 5 years of followup, relative to the most common Luminal A subtype, are 1.99 (95% CI: 1.09–3.64) for Luminal B, 3.65 (1.64–8.16) for HER2-enriched and 17.71 (1.71–183.33) for the basal like subtype. For node-negative disease, PAM50 qRT-PCR based risk assignment weighted for tumor size and proliferation identifies a group with >95% 10 yr survival without chemotherapy. In node positive disease, PAM50-based prognostic models were also superior.
Conclusion
The PAM50 gene expression test for intrinsic biological subtype can be applied to large series of formalin-fixed paraffin-embedded breast cancers, and gives more prognostic information than clinical factors and immunohistochemistry using standard cutpoints.
Introduction
A number of gene expression technologies and statistical models have reported methodologies to identify breast cancer patients with estrogen receptor positive, node negative (N0) disease that may be adequately managed with five years of tamoxifen monotherapy (1–5). However, these studies often included patients with tumors already associated with established low risk biomarkers, for example low grade histology, low Ki67 based proliferation index and favorable surgical stage. It therefore remains controversial whether genomic assays should be applied routinely, or whether surgical stage and a limited number of immunohistochemical markers will, in most cases, be adequate and less costly (6).
The clinical significance of continued efforts in this area is relevant for decisions regarding both chemotherapy and endocrine agents, as patients at low risk after five years of tamoxifen monotherapy could be spared the morbidity associated with extended aromatase inhibitor therapy (7). Studies that address this issue are few, because extremely long follow up and information on breast cancer specific mortality are required. Furthermore, since frozen tumor archives are unavailable from suitably large patient populations, gene expression technologies must be applicable to degraded RNA extracted from formalin fixed paraffin embedded tissues that are necessarily more than a decade old.
Our group has assembled and published several technological and statistical approaches to address prognosis in ER+ breast cancer. We therefore sought to compare clinicopathological, immunohistochemical and molecular methodologies in a single independent test set in order to identify the best approach. Importantly, we focused on fixed statistical models that were previously trained on independent data sets to avoid overoptimistic results. The models we report in this paper include the use of standard pathological factors, such as centrally-reviewed histological grade, as incorporated into Adjuvant! Online (8), models based on immunohistochemistry (IHC) for biomarkers of intrinsic subtypes (6), and a gene expression assay using fifty genes (PAM50). The latter represents a reduced gene set, amenable to assay by techniques such as quantitative real time reverse transcriptase PCR (qRT-PCR), that accurately identifies the major intrinsic biological subtypes of breast cancer and generates risk of relapse scores (9). The investigation utilized a large independent cohort of formalin-fixed, paraffin-embedded pathology specimens from patients with ER+ breast cancer, all M0 but otherwise representing a spectrum of T and N stages including a large fraction of node positive patients. All patients received adequate local treatment, five years of tamoxifen therapy but no adjuvant chemotherapy, and were followed for relapse free (RFS) and disease specific survival (DSS) for over a decade.
Methods
Patient and sample characteristics
The study cohort was accrued from female patients with invasive breast cancer, diagnosed in British Columbia between 1986 and 1992. Cancer tissue from these patients had been frozen and shipped to Vancouver Hospital for central ER and progesterone receptor (PR) testing by dextran-coated charcoal ligand binding assay. The PAM50 assay was conducted on the portion of this tissue that was formalin-fixed and paraffin-embedded for histologic correlation. Characteristics of this cohort have been previously described (6), and the same source blocks were used to assemble tissue microarrays for previously published studies on ER (10), HER2 (11), PR (12), Ki67, cytokeratin 5/6 and epidermal growth factor receptor (6, 13). Quantitative ER was determined using the Ariol automated digital imaging system (14), and the same method was applied for PR. For this study, we selected samples from patients with ER+ tumors by immunohistochemistry (IHC) who had received tamoxifen as their only adjuvant systemic therapy. Provincial guidelines from that time period recommended tamoxifen for women >50 years of age, whose ER status was positive or unknown, and who were either node positive or had lymphovascular invasion. Cohort identification and sample selection for this study are summarized as per REMARK criteria (15) in Supplementary Table 1.
RNA preparation, qRT-PCR, and assignment of biological subtype and Risk-of-elapse (ROR) score
H&E sections from each block were reviewed by a pathologist (TON). Areas containing representative invasive breast carcinoma were selected and circled on the source block. Using a 1.0 mm punch needle, at least two tumor cores were extracted from the circled area. Details of RNA preparation from paraffin cores, the qRT-PCR assay for the PAM50 panel and reference genes, and how these results allow assignment into Luminal A, Luminal B, HER2-enriched and Basal-like subtypes, and the independently-trained ROR-S (Risk Of Relapse based on Subtype), ROR-T (-Tumor size weighted model), ROR-P (-Proliferation weighted model) and ROR-PT (Proliferation and Tumor size weighted) risk score assignments are presented in Supplementary Methods. For clarity, the term ROR-T is now used for the same model described in our earlier publication as ROR-C (“clinical”) (9).
Relation of clinicopathological factors, intrinsic subtypes and risk of relapse (ROR) scores to clinical outcome
Statistical analyses were conducted using SPSS v16.0 and R v2.8.0. Univariable analyses of tumor subtype against breast cancer relapse-free survival (RFS) and disease-specific survival (DSS) were performed by Kaplan-Meier analysis with log rank test. Multivariable analyses were performed against the standard clinical parameters of tumor size, nodal status, histological grade, patient age and HER2 status. HER2 scores were centrally-determined based on assay of adjacent cores from the same source blocks, assembled into tissue microarrays and subjected to IHC and fluorescent in situ hybridization (FISH) analysis using clinical-equivalent protocols (11). Cox regression models (16) were built to estimate the adjusted hazard ratios of the qRT-PCR-assigned breast cancer subtypes, as well as ROR scores categorized by published cut-points and as a continuous variable. IHC-based subtypes were assigned as previously defined (6). The online decision making tool Adjuvant! Online (www.adjuvantonline.com), previously validated on the British Columbia population cohort (8), was used to generate breast cancer relapse-free and disease-specific survival estimates for each patient in this cohort. Only cases with information for all the covariates were included in the analyses. Smoothed plots of weighted Schoenfeld residuals were used to assess proportional hazard assumptions (17) and time stratifications were employed where hazards were not proportional over the entire follow up period.
The C-index (18) is defined as the probability that risk assignments to members of a random pair are accurately ranked according to their prognosis. The number of concordant pairs (order of failure and risk assignment agree), discordant pairs (order of failure and risk assignment disagree), and uninformative pairs are tabulated to calculate the measure. C-index values of 0.5 indicate random prediction and higher values indicate increasing prediction accuracy. Variability in the C-index for each predictor and p-values from comparisons were estimated from 1000 bootstrap samples of the risk assignments. Calculation was performed using the rcorr.cens function implemented in the Hmisc (19) library for R statistical software version 2.8.1 (http://www.R-project.org)
Results
Intrinsic subtyping of ER positive, tamoxifen-treated breast cancer using the PAM50 assay
RNA was extracted from pathologist-guided tissue cores from 991 formalin-fixed, paraffin-embedded breast cancer specimens. 811 samples yielded sufficient RNA for analysis (at least 1.2 ug total RNA at a concentration ≥ 25 ng/uL). Template was technically sufficient in 786 cases, based on all internal housekeeper gene controls being expressed in the sample above background. Clinical characteristics for the patients included in the PAM50 analysis are presented in Table 1 (Supplementary Tables S2 and S3 provide details stratified by node status). Based on the nearest PAM50 centroid algorithm, intrinsic breast cancer subtypes were assigned using gene expression as follows: 372 samples (47.3%) were Luminal A, 329 (41.9%) Luminal B, 64 (8.1%) HER2-enriched, 5 (0.6%) Basal-like, and 16 (2.0%) Normal-like. Thus, while all cases in this study were positive for ER by centrally-assessed IHC analysis on a tissue microarray (10), and 98.8% were also positive by dextran-charcoal coated biochemical assay (Table 1), the gene expression panel nevertheless assigned 9% of cases into non-luminal subtypes, mostly HER2-enriched. This phenomenon has been previously observed when interrogating published datasets for expression of the PAM50 genes (9). For the sixteen cases assigned as Normal-like, histology was reviewed from adjacent tissue cores, and in 14 of 16 cases invasive cancer cells were absent or rare. Normal-like cases were therefore excluded from outcome analyses, as a breast cancer subtype could not be confidently assigned due to insufficient tumor content.
Table 1.
Clinical characteristics of the whole cohort.
| Clinical parameter | Total | PAM50 subtype (all n=786) | |||||
|---|---|---|---|---|---|---|---|
| Luminal A | Luminal B | Her2 | Basal | Normal | |||
| Sample Size | N | 786 | 372 | 329 | 64 | 5 | 16 |
| follow-up times in recurrence-free patients (yr) | median (min. – max.) | 9.7 (0.12–18) | 12 (0.25–18) | 7.6 (0.12–18) | 7.3 (0.47–18) | 2.3 (0.6–4.1) | 13 (3.2–18) |
| followup-times in disease specific surviving patients (yr) | median (min. – max.) | 12 (0.55–18) | 13 (0.57–18) | 10 (0.64–18) | 8.8 (0.55–18) | 5 (1.6–16) | 14 (3.2–18) |
| age (years) | median | 67 | 67 | 68 | 66.5 | 65 | 68.5 |
| pre-menopausal | yes | 20 | 10 | 7 | 2 | 1 | 0 |
| no | 752 | 358 | 314 | 62 | 4 | 14 | |
| unknown/pregnant | 14 | 4 | 8 | 0 | 0 | 2 | |
| surgery | Complete mastectomy | 468 | 210 | 203 | 39 | 5 | 11 |
| partial mastectomy | 306 | 159 | 119 | 23 | 0 | 5 | |
| other | 12 | 3 | 7 | 2 | 0 | 0 | |
| axillary node | yes | 745 | 349 | 313 | 62 | 5 | 16 |
| dissection | no | 41 | 23 | 16 | 2 | 0 | 0 |
| radiation therapy | yes | 419 | 207 | 164 | 40 | 1 | 7 |
| no | 367 | 165 | 165 | 24 | 4 | 9 | |
| tumour size (cm) | median | 2.1 | 2.0 | 2.5 | 2.5 | 3.5 | 2.3 |
| T stage (clinical) | T0/IS | 1 | 0 | 0 | 0 | 0 | 1 |
| T1 | 331 | 180 | 118 | 27 | 3 | 3 | |
| T2 | 380 | 165 | 179 | 28 | 2 | 6 | |
| T3 | 18 | 9 | 5 | 3 | 0 | 1 | |
| T4 | 34 | 10 | 17 | 3 | 0 | 4 | |
| TX | 22 | 8 | 10 | 3 | 0 | 1 | |
| # positive nodes | 0 | 222 | 95 | 97 | 19 | 1 | 10 |
| 1–3 | 360 | 182 | 148 | 26 | 1 | 3 | |
| 4–9 | 125 | 55 | 53 | 12 | 2 | 3 | |
| 10+ | 26 | 10 | 14 | 2 | 0 | 0 | |
| unknown | 53 | 30 | 17 | 5 | 1 | 0 | |
| grade | grade 1: well differentiated | 34 | 25 | 5 | 1 | 1 | 2 |
| grade 2:moderately differentiated | 338 | 186 | 129 | 14 | 0 | 9 | |
| grade 3: poorly differentiated | 370 | 135 | 179 | 48 | 3 | 5 | |
| unknown | 44 | 26 | 16 | 1 | 1 | 0 | |
| histologic subtype | ductal NOS | 708 | 329 | 302 | 60 | 4 | 13 |
| lobular | 61 | 32 | 21 | 4 | 1 | 3 | |
| mucinous | 7 | 4 | 3 | 0 | 0 | 0 | |
| tubular | 7 | 6 | 1 | 0 | 0 | 0 | |
| medullary | 2 | 1 | 1 | 0 | 0 | 0 | |
| apocrine | 1 | 0 | 1 | 0 | 0 | 0 | |
| lymphovascular | yes | 485 | 210 | 220 | 44 | 2 | 9 |
| invasion | no | 262 | 139 | 94 | 19 | 3 | 7 |
| unknown | 39 | 23 | 15 | 1 | 0 | 0 | |
| clinical estrogen | missing | 9 | 5 | 3 | 0 | 0 | 1 |
| receptor status (DCC ligand binding assay) | negative (0–9 fmol/mg) | 9 | 3 | 2 | 4 | 0 | 0 |
| positive (>10 fmol/mg) | 768 | 364 | 324 | 60 | 5 | 15 | |
| median (fmol/mg) | 254.5 | 255.5 | 327.0 | 74.0 | 32.0 | 54.0 | |
| clinical progesterone | missing | 161 | 84 | 53 | 15 | 2 | 7 |
| receptor status (DCC ligand binding assay) | negative (0–9 fmol/mg) | 72 | 15 | 39 | 18 | 0 | 0 |
| positive (>10 fmol/mg) | 553 | 273 | 237 | 31 | 3 | 9 | |
| median (fmol/mg) | 129 | 202 | 84.5 | 17 | 153 | 239 | |
| immunohistochemical | negative | 696 | 348 | 294 | 34 | 4 | 16 |
| HER2 with FISH | positive | 75 | 15 | 30 | 29 | 1 | 0 |
| correction on 2+ cases | unknown | 15 | 9 | 5 | 1 | 0 | 0 |
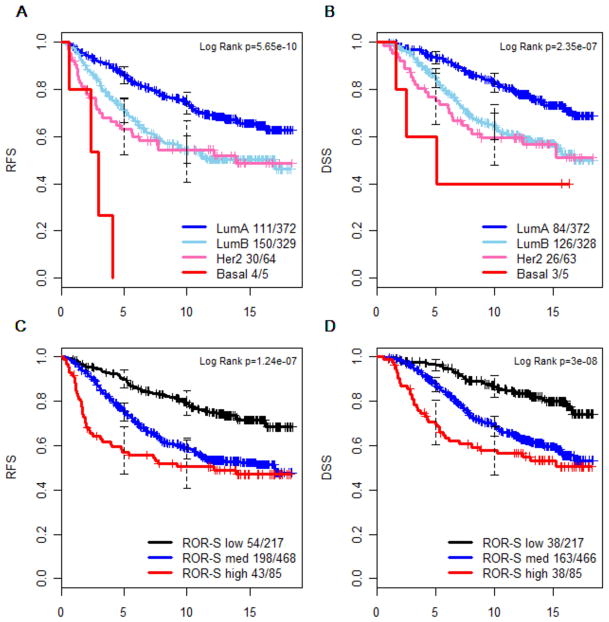
The intrinsic biological subtypes were strongly prognostic by Kaplan-Meier analysis (Figures 1A and 1B). In the British Columbia population at the time these samples were originally acquired, many patients with a clinically low-risk profile received no adjuvant systemic therapy (8). In contrast, those receiving adjuvant tamoxifen (the subjects of this study) had tumors that were mostly node positive, high grade and/or exhibited lymphovascular invasion, and therefore constitute a higher risk group with overall 10 year RFS of 62% and DSS of 72%. Those assigned by the PAM50 assay to Luminal A status had a significantly better outcome (10 year relapse free survival 74%, disease-specific survival = 83%) than Luminal B, HER2-enriched or basal like tumors (Figure 1A for RFS and Figure 1B for DSS). The ROR risk-of-relapse algorithms (9) were originally trained on microarray data from node negative patients who received no adjuvant systemic therapy, and have not previously been applied to a population homogeneously-treated with adjuvant tamoxifen, nor to a series containing large numbers of node positive cases, nor to the endpoint of disease specific survival. In this data set ROR-S (a model based solely on gene expression) nevertheless showed performance consistent with our previous report (Figures 1C and 1D). Multivariable Cox models were constructed to test the independent value of PAM50 subtyping against standard clinical and pathological factors including age, histological grade, lymphovascular invasion, HER2 expression, nodal status, and tumor size. To meet proportional hazard assumptions, multivariable models were assessed with the time axis split at 5 years (20), as HER2-enriched and basal-like tumors (Figure 1A and 1B) and ROR-S high category tumors (Figure 1C and 1D) had a much higher event rate in the first five years than subsequently. The intrinsic biological subtype and ROR-S remained significant in the multivariable models for DSS (Table 2) and RFS (Supplementary Table S4), particularly in the first five years, as did pathological staging variables (tumor size and node status). However histological grade, lymphovascular invasion and clinical HER2 status, significant in univariable analysis in this cohort, no longer contributed significant independent prognostic information when the multivariable analysis included the PAM50 assignments.
Figure 1.
Kaplan-Meier survival analysis of intrinsic subtype (panels A and B) and Risk of Relapse score (ROR-S, panels C and D), as determined by PAM50 gene expression measurement by qRT-PCR performed on paraffin blocks from women with invasive breast carcinoma, treated with adjuvant tamoxifen. The number of events and total number of patients in each group is shown beside each curve’s description. RFS = relapse-free survival (panels A and C). DSS = breast cancer disease-specific survival (panels B and D; excludes two cases with unknown cause of death).
TABLE 2.
Cox model multivariable analysis of breast cancer disease-specific survival among ER positive, tamoxifen-treated women, incorporating standard clinicopathological factors and (A) intrinsic subtype or (B) Risk of Relapse (ROR-S), as determined by PAM50 qRT-PCR measurements. p-values for multilevel categorical variables are derived from likelihood ratio tests between models with and without each these variables.
| A. Intrinsic subtype | ||||
|---|---|---|---|---|
| Clinical endpoint | multivariable disease-specific survival (0 – 5 years followup) | multivariable disease-specific survival (5 years to end of followup) | ||
| hazard ratio (95% CI) | p-value | hazard ratio (95% CI) | p-value | |
| Age | 1.02 (0.99–1.05) | 0.2665 | 1.00 (0.98–1.02) | 0.9786 |
| Grade (1–2 vs 3) | 1.51 (0.87–2.60) | 0.1405 | 1.05 (0.71–1.56) | 0.8109 |
| LV | 1.02 (0.58–1.81) | 0.9421 | 1.16 (0.77–1.75) | 0.4852 |
| HER2 (IHC) | 1.50 (0.77–2.91) | 0.2314 | 0.82 (0.40–1.69) | 0.5968 |
| Node status (0 as reference group) | < 0.0001 | 0.0012 | ||
| 1–3 | 2.07 (0.95–4.54) | 1.54 (0.96–2.47) | ||
| 4+ | 5.80 (2.64–12.71) | 2.78 (1.60–4.82) | ||
| Tumor size (T1 as reference group) | 0.049 | 0.0002 | ||
| T2 | 1.22 (0.71–2.09) | 1.62 (1.08–2.42) | ||
| T3 | 3.92 (1.50–10.22) | 5.11 (1.78–14.62) | ||
| T4 | 1.31 (0.38–4.50) | 4.02 (1.85–8.74) | ||
| Subtype (Luminal A as reference group) | 0.0018 | 0.0381 | ||
| Luminal B | 1.99 (1.09–3.64) | 1.70 (1.13–2.55) | ||
| Her2-enriched | 3.65 (1.64–8.16) | 1.52 (0.72–3.18) | ||
| Basal-like | 17.71 (1.71–183.33) | NA | ||
| B. Risk of relapse (ROR-S) | ||||
| Clinical endpoint | multivariable disease-specific survival (0 – 5 years followup) | multivariable disease-specific survival (5 years to end of followup) | ||
| hazard ratio (95% CI) | p-value | hazard ratio (95% CI) | p-value | |
| Age | 1.02 (0.99–1.05) | 0.2676 | 1.00 (0.98–1.02) | 0.9089 |
| Grade (1–2 vs 3) | 1.36 (0.79–2.36) | 0.2674 | 1.01 (0.68–1.51) | 0.9588 |
| LV | 0.95 (0.54–1.66) | 0.852 | 1.18 (0.78–1.79) | 0.4299 |
| HER2 (IHC) | 1.46 (0.77–2.77) | 0.2467 | 0.87 (0.43–1.77) | 0.6964 |
| Node status (0 as reference group) | < 0.0001 | 0.0014 | ||
| 1–3 | 2.14 (1.00–4.60) | 1.55 (0.97–2.48) | ||
| 4+ | 6.03 (2.79–13.05) | 2.78 (1.59–4.86) | ||
| Tumor size (T1 as reference group) | 0.0647 | 0.0003 | ||
| T2 | 1.19 (0.70–2.05) | 1.64 (1.10–2.45) | ||
| T3 | 3.34 (1.32–8.43) | 3.69 (1.30–10.46) | ||
| T4 | 0.90 (0.25–3.19) | 4.44 (2.01–9.78) | ||
| ROR-S (low as reference group) | <0.0001 | 0.0388 | ||
| Med | 2.04 (0.89–4.66) | 1.86 (1.15–3.00) | ||
| High | 6.48 (2.56–16.40) | 1.57 (0.71–3.46) | ||
Comparisons between gene expression and clinical assays for hormone receptors and proliferation
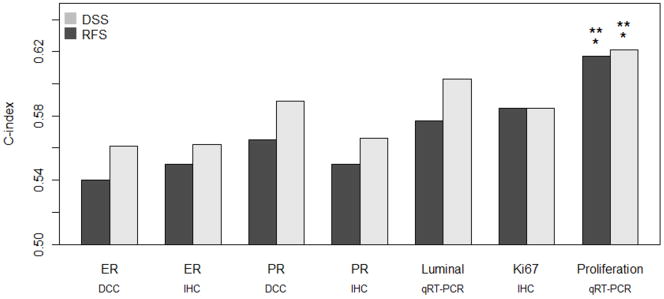
In a case that is ER positive by immunohistochemistry, additional information about hormone receptor expression can be obtained in several ways, including dextran-coated charcoal ligand binding assay, quantitative immunohistochemistry for ER, or equivalent measures of progesterone receptor (PR). Most published assays for breast cancer prognosis in ER+ disease include tumor growth rate as one of the parameters in the statistical model, and this dataset was previously assessed in detail for immunohistochemical Ki67 index (6). The PAM50 qRT-PCR data allows detailed quantitative assessment of the functionality of the estrogen response pathway (8 gene luminal signature) as well as a proliferation signature based on the mean expression of eleven genes linked to cell cycle progression (trained on published data, as per Supplementary Methods). The availability of all these measurements (10) provides an opportunity to determine which approach most accurately captures the prognostic effect of estrogen pathway biomarkers and tumor growth rate in a direct comparison (Figure 2). Given a randomly selected pair of subjects, the concordance index (C-index) is the probability that the patient assigned the more extreme risk score actually has a worse prognosis. A value of 0.5 indicates discrimination that is no better than chance prediction, and a value of 1 indicates perfect discrimination of samples. Using the C-index to compare prognostic capacity in this uniformly tamoxifen-treated cohort, the combination of luminal genes measured by the PAM50 yields more prognostic information than other methods of hormone receptor analysis, but the differences are not significant. Although Ki67 index by immunohistochemistry appears to outperform quantitative ER, the proliferation signature provides the most robust approach for the prediction of both RFS and DSS (Figure 2, Supplementary Table S5). Multivariable analysis indicated that the Ki67 immunohistochemical assay did not contribute significant independent information to prognostic models for either node negative or node positive breast cancer patients, when information on the proliferation signature is included (Supplementary Table S6).
Figure 2.
C-index estimates of relapse free (RFS) and disease-specific (DSS) survival for different measures of hormone receptors and proliferation. DCC = dextran-coated ligand binding assay. IHC = % positive nuclei by immunohistochemistry. The Luminal and Proliferation measures are the means of normalized qRT-PCR values across 8 and 11 signature genes respectively, as described in Supplementary Methods. P-values were estimated from 1000 bootstrap samples. Single asterisk (*) designates significant improvement (p < 0.05) in C-index relative to clinical quantitative ER by dextran-charcoal coated ligand binding assay (DCC). Double asterisk (**) designates significant improvement (p < 0.05) in C-index relative to visual quantitative Ki67 index.
Comparison of fixed models of prognosis in node negative breast cancer
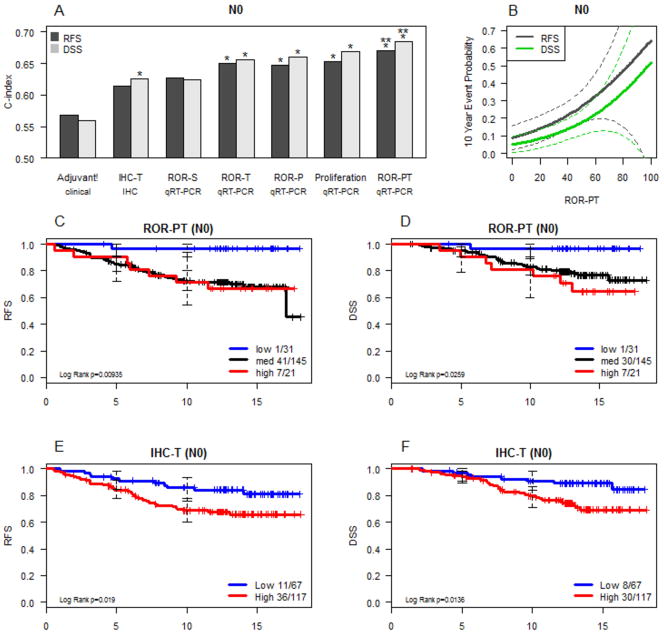
For formal model comparisons, data was generated on four fixed approaches, without any element of training within the test set: a) clinical model based on Adjuvant! Online, b) IHC-based (incorporating data on Ki67 and HER2), c) the ROR-S approach based on PAM50 gene expression alone, and d) the proliferation signature alone and as incorporated into the ROR-P risk model using a beta coefficient weighting for proliferation (described in Supplementary Methods). Adjuvant! Online incorporates full tumor size staging information; to account for the influence of tumor size the biomarker models were also weighted by a beta coefficient (T) that incorporated the prognostic information associated with T1 status versus higher T stage (the level of detail available in the independent training sets). This approach created IHC-T, ROR-T and ROR-PT models. In addition, the strong independent influence of N stage was accounted for by conducting the analysis separately in the N0 and N+ populations. C-index assessments showed superiority of the biomarker models over the purely clinical Adjuvant! Online model in the node negative population, with the ROR-PT approach providing the most prognostic information (Figure 3A). In multivariable analysis, the addition of ROR-P to a model of ROR-S results in a significant increase in explained prognostic variation (RFS p=0.0032; DFS p=0.0015); ROR-PT is also significant after conditioning on ROR-S (RFS p= 0.0023; DFS p=0.0015) but not ROR-P (RFS p=0.12; DFS p=0.13). A continuous score based on the ROR-PT was generated to translate the data into an individual RFS and DSS risk assessment tool (Figure 3B). Kaplan Meier analysis illustrates the ability of the ROR-PT model to identify patients who have an extremely high chance (>95%) of remaining disease free (Figure 3C) and alive beyond 10 years (Figure 3D). In contrast, our previously published IHC model (6), could not identify a group with sufficiently favorable outcomes that five years of tamoxifen might be considered adequate treatment (i.e. <90% 10 yr RFS; Figures 3E and 3F).
Figure 3.
Comparison of prognostic classifiers in node negative subjects. The C-index is used to compare accuracy of the prognostic classifiers (panel A and Supplemental Table S5). * designates significant improvement (p < 0.05) in C-index relative to the clinical model (Adjuvant!) and ** to the IHC-T model. Taking the best-performing model, ROR-PT values are related to actual 10 year event probabilities using a Cox proportional hazard model (panel B, dotted lines are 95% CI). Kaplan-Meier survival analysis of the size and proliferation weighted Risk of Relapse (ROR-PT) assignments are presented in panels C and D, and comparable information provided by a model of IHC subtype and tumor size is shown in panels E and F. RFS = relapse-free survival. DSS = breast cancer disease-specific survival (excludes two cases with unknown cause of death)
Comparison of fixed models of prognosis in node positive breast cancer
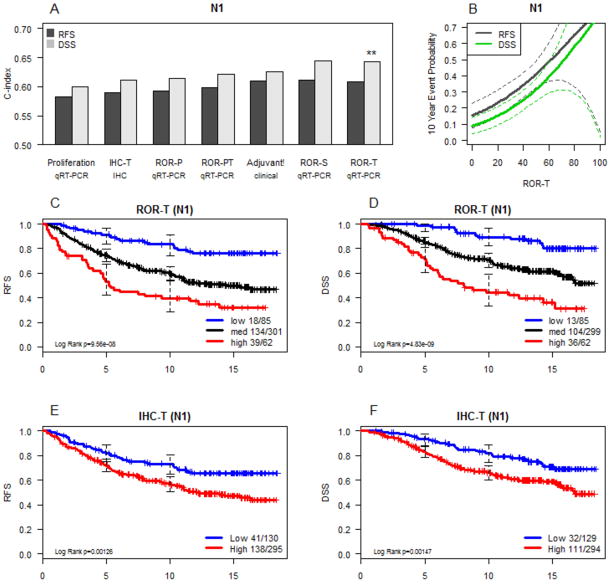
For node positive disease, C index analysis (Figure 4A) supports the conclusion that the ROR-T score produces the best prognostic model; in contrast to N0 disease, the proliferation signature added relatively little information and proliferation weighting (ROR-PT) did not yield a superior model. Adjuvant Online! performed almost as well, but had the advantage of incorporating the actual number of involved lymph nodes. This information was not available in the independent training sets used to build the ROR models, and so could not be used in the current analysis (which can however serve to train future models incorporating number of involved lymph nodes). The continuous score model for node positive disease (Figure 4B) produces a very broad range of prognosis, similar to N0 disease, although few patients have a prognosis in the range where tamoxifen monotherapy for five years would be considered sufficient treatment. While there were large and highly significant differences in survival in ROR-defined risk groups, Kaplan Meier analysis (Figures 4C–4D) illustrates that even patients in the lowest risk ROR group are still subject to relapses and late deaths from breast cancer, particularly after the fifth year of follow up. The immunohistochemistry-based risk model incorporating Ki67 and HER2 also produces a statistically significant prognostic impact for RFS (Figure 4E) and DSS (Figure 4F), although these differences are narrower than those achieved by the gene expression-based model.
Figure 4.
Comparison of prognostic classifiers in node positive subjects. A) C-index comparison of the accuracy of prognostic classifiers as described in Figure 3. B) Cox proportional hazard model relating the best performing model (ROR-T) to actual 10 year event probabilities. C) Kaplan-Meier survival analysis of the size weighted Risk of Relapse (ROR-T) assignments for RFS and (in panel D) DFS. E) and F) comparable information as provided by a model of IHC subtype and tumor size.
DISCUSSION
Previous studies have established that intrinsic biological signatures are present and have prognostic significance in breast cancer cohorts from multiple different institutions, profiled with several gene expression microarray platforms (21–24). In order to identify these subtypes on standard formalin-fixed, paraffin-embedded pathology specimens, we developed a qRT-PCR test based on a panel of 50 genes (9). The analysis reported here applied this test to a series of paraffin blocks with > 15 years detailed followup.
Whereas previously assessed cohorts consisted mainly of low risk women receiving no adjuvant systemic therapy, or were heterogeneously-treated, the cases in the current study are all women with estrogen receptor positive breast cancer who received endocrine therapy as their sole adjuvant treatment, a group of particular clinical importance and contemporary relevance. In this analysis we sought to compare different technologies for predicting long term outcomes for such patients. In this study cohort, patients were diagnosed with node positive or higher risk N0 disease. Only 8% of the N0 population had grade 1 disease and 55% exhibited lymphovascular invasion (Table S2). Under the current standard of care in most countries the majority of these patients would now be treated with adjuvant chemotherapy (25) and extended endocrine therapy. Using a series of fixed models trained in independent data sets, we compared a standard approach using clinico-pathological information (Adjuvant! Online), to our published Luminal B discriminator based on Ki67 and HER2 immunohistochemistry additionally weighted for T stage (IHC-T), and to PAM50 gene expression based ROR models weighted for T stage (ROR-T and ROR-PT). In node-negative patients, the ROR-PT approach was the most accurate and was able to identify patients in whom 5 years of tamoxifen may be adequate treatment based on the very low late relapse rate in the 5 to 10 year window (Figure 3C). In node positive disease, the PAM50 approach represents an advance in prognostication, but late relapses and deaths were seen even in the lowest risk group identified using the best ROR model. Unlike in N0 disease, proliferation signature weighting did not improve the C index in node positive disease.
On this cohort, detailed centrally-determined immunohistochemical analyses have previously been performed and published (6, 10–13, 26). C-index, Kaplan-Meier and Cox model analyses show that immunohistochemical approaches do work and provide significant prognostic information. However, the PAM50-based models are superior in terms of adding significant additional information and in their capacity to identify a particularly low-risk group of women.
We view these PAM50 models, derived from archival formalin fixed RNA, as a potential replacement for grade, hormone receptor, Ki67 and HER2 based prognostic models, but not as a replacement for pathological stage (as tumor size and nodal status remain independent predictors in multivariable models that include PAM50 based prognostic information). One weakness of our approach is that our current accounting for pathological stage is over-simplified due to the limited stage distributions and clinical information in our training sets. We analyzed the data as either node negative or node positive, and accounted for T stage by categorizing the samples as either T1 or greater. A future aim is to integrate the PAM50 data into the Adjuvant! Online approach (27) to more completely account for the prognostic influence of pathological stage. To achieve this we would need to construct a training set that adequately includes all the 5 categories of T size and four categories of N stage used in Adjuvant! Online, in order to gauge the prognostic weight of these pathological stage categories in the setting of PAM50 information. Additionally, incorporation of all immunohistochemical data as continuous variables in a combined model may improve its prognostic value. The current series contains sufficiently detailed clinical and immunohistochemical information to contribute to such detailed comparisons, as a training set requiring further validation.
An additional caveat to our study is that the population was strongly biased towards higher risk breast cancers and so likely underestimates of the number of patients in the broader, node negative population for whom adjuvant tamoxifen would represent adequate treatment. The current generation of adjuvant aromatase inhibitor trials would be an appropriate setting to address the value of our approach further. We accept the possibility that a better model using Ki67 at a different cut point could be developed. However since we were focused on comparing fixed models, we used our published approach. Further work on the Ki67 model and cut point optimization will require independent data sets.
In comparison with other signatures such as the recurrence score and genomic grade index (1, 28, 29), the PAM50 has the potential advantage of discriminating high risk patients into Luminal B, HER2-Enriched and Basal-like subtypes, who are likely to respond differently to the main systemic therapy options (endocrine, anti-HER2, and anthracycline vs. non-anthracycline vs. taxane chemotherapy regimens). The assay requires neither frozen tissue (30) nor manual microdissection of cut sections(1), and can be readily applied to standard paraffin blocks including archival tissues from clinical trials. Currently available assays such as Mammaprint (31) and OncotypeDX (32) were optimized to recognize particularly low risk patients from among a node negative early stage population who did not receive chemotherapy. Because intrinsic subtyping is designed to identify discriminative biological features of breast cancer, rather than being derived around clinical outcome in a specific population, this approach is particularly likely to extrapolate well onto other patient cohorts (33). The current study demonstrates the ability of PAM50 to recognize a very low risk prognostic group among women receiving tamoxifen and no chemotherapy, similar to the Oncotype Dx assay(34, 35). A direct comparison of different expression profile approaches may become possible in the future through a reanalysis of cohorts with the PAM50 that have already been analyzed by OncotypeDX, since both assays can be applied to the same source material.
Our inability to identify a group of patients with node positive disease in whom five years of tamoxifen is adequate is reminiscent of the recent findings from the Southwest Oncology Group, who also found that a molecular signature for good outcome in N0 disease failed in node positive disease in this regard (35). It would be relevant to study a series of patients treated with extended adjuvant aromatase inhibitor therapy, who will have even lower residual risk, as some of the patients in the low risk N+ group may simply require longer treatment with modern endocrine therapy rather than chemotherapy. The development of new approaches for defining prognosis in N+ disease is also warranted. We have already established the preoperative endocrine prognostic index (PEPI), which demonstrated that the “on endocrine treatment” Ki67 value is more effective than baseline Ki67 for the identification of patients with clinical stage 2 and 3 disease who have excellent long term outcomes after neoadjuvant endocrine therapy (36). A comparison between Ki67 and the PAM50 based proliferation signature in the neoadjuvant endocrine therapy setting is therefore one logical next step. The applicability of this test to formalin-fixed paraffin-embedded tissues will make possible its use on large clinical trial archives that address this issue (37). The results of our study highlight the feasibility of measuring multi-gene expression panels on such series as a means for demonstrating clinical utility, using a method readily applicable to prospective clinical samples that provides more prognostic information than clinical or standard immunohistochemical approaches.
Description of how this work might be applied to future practice of cancer medicine.
Molecular intrinsic subtyping reveals the major biological categories of breast cancer. Herein we demonstrate adaptation of a 50 gene intrinsic subtyping signature for testing standard paraffin blocks. Using a large, homogeneously treated cohort of breast cancer patients, we directly compare gene expression results to high quality clinical and central immunohistochemical data. We show the PAM50 approach to be superior as a prognostic test, specifically able to identify an ultra-low risk group who may not need chemotherapy. Based on these results, intrinsic subtyping tests are now being applied to randomized clinical trials series in Canada and the USA to assess predictive capacity (already underway for response to endocrine therapy, anthracyclines and taxanes, with further studies under consideration). Should such studies prove a predictive value for intrinsic subtyping, this test could be clinically implemented in a similar form, as it has been designed for application on standard laboratory specimens.
Supplementary Material
Acknowledgments
FUNDING
Torsten Nielsen is a Senior Scholar of the Michael Smith Foundation for Health Research. Grant support was provided by a National Cancer Institute (NCI) Strategic Partnering to Evaluate Cancer Signatures Grant No. U01 CA114722-01, the Canadian Cancer Society, the Huntsman Cancer Institute/Foundation (P.S.B.), the ARUP Institute for Clinical and Experimental Pathology (P.S.B.), an NCI Breast SPORE Grant No. P50-CA58223-09A1 (C.M.P.), a St Louis Affiliate of the Susan G. Komen Foundation CRAFT grant (M.J.E.), the Breast Cancer Research Foundation (C.M.P. and M.J.E.), and an unrestricted educational grant from sanofi-aventis Canada. Additional support provided by the TRAC facility and Informatics at the Huntsman Cancer Center, supported in part by the NCI Cancer Center Support Grant No. P30 CA42014-19, and the tissue procurement facility at the Alvin J. Siteman Cancer Center at Washington University School of Medicine, which is funded in part by the NCI Cancer Center Support Grant No. P30 CA91842.
The researchers would like to thank current and former members of the British Columbia Cancer Agency’s Breast Cancer Outcomes Unit, including S. Chia, K. Gelmon, H. Kennecke, I. Olivotto, and C. Speers for maintaining the clinical database.
References
- 1.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 2.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 3.Goetz MP, Suman VJ, Ingle JN, et al. A two-gene expression ratio of homeobox 13 and interleukin-17B receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin Cancer Res. 2006;12:2080–7. doi: 10.1158/1078-0432.CCR-05-1263. [DOI] [PubMed] [Google Scholar]
- 4.Ross JS. Multigene classifiers, prognostic factors, and predictors of breast cancer clinical outcome. Adv Anat Pathol. 2009;16:204–15. doi: 10.1097/PAP.0b013e3181a9d4bf. [DOI] [PubMed] [Google Scholar]
- 5.Tutt A, Wang A, Rowland C, et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer. 2008;8:339. doi: 10.1186/1471-2407-8-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goss PE, Ingle JN, Martino S, et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349:1793–802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- 8.Olivotto IA, Bajdik CD, Ravdin PM, et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23:2716–25. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 9.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheang MC, Treaba DO, Speers CH, et al. Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol. 2006;24:5637–44. doi: 10.1200/JCO.2005.05.4155. [DOI] [PubMed] [Google Scholar]
- 11.Chia S, Norris B, Speers C, et al. Human epidermal growth factor receptor 2 overexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J Clin Oncol. 2008;26:5697–704. doi: 10.1200/JCO.2007.15.8659. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Chia SK, Mehl E, et al. Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Treat. 2009;119:53–61. doi: 10.1007/s10549-009-0318-0. [DOI] [PubMed] [Google Scholar]
- 13.Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–76. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 14.Turbin DA, Leung S, Cheang MC, et al. Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases. Breast Cancer Res Treat. 2008;110:417–26. doi: 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- 15.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Urol. 2005;2:416–22. [PubMed] [Google Scholar]
- 16.Cox D, Oakes D. Monographs on Statistics and Probability. London, England: Chapman and Hall; 1984. Analysis of Survival Data. [Google Scholar]
- 17.Grambsch P, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–26. [Google Scholar]
- 18.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Harrell FE., Jr . R package version 2.3-0. 2009. Design Package. [Google Scholar]
- 20.Schemper M. Cox Analysis of Survival Data with Non-Proportional Hazard Functions. The Statistician. 1992;41:455–65. [Google Scholar]
- 21.Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8:R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 23.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapp AV, Jeffrey SS, Langerod A, et al. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol. 2007;18:1133–44. doi: 10.1093/annonc/mdm271. [DOI] [PubMed] [Google Scholar]
- 26.Jensen KC, Turbin DA, Leung S, et al. New cutpoints to identify increased HER2 copy number: analysis of a large, population-based cohort with long-term follow-up. Breast Cancer Res Treat. 2008;112:453–9. doi: 10.1007/s10549-007-9887-y. [DOI] [PubMed] [Google Scholar]
- 27.Ravdin PM, Siminoff LA, Davis GJ, et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19:980–91. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 28.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 29.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 30.Glas AM, Floore A, Delahaye LJ, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 32.Paik S. Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist. 2007;12:631–5. doi: 10.1634/theoncologist.12-6-631. [DOI] [PubMed] [Google Scholar]
- 33.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 34.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 35.Albain KS, Barlow WE, Shak S, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2009 doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis MJ, Tao Y, Luo J, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100:1380–8. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–52. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.