Abstract
This study determined whether expression levels of a panel of biologically relevant microRNAs can be used as prognostic or predictive biomarkers in patients who participated in the International Adjuvant Lung Cancer Trial (IALT), the largest randomized study conducted to date of adjuvant chemotherapy in patients with radically resected non-small cell lung carcinoma (NSCLC). Expression of miR-21, miR-29b, miR-34a/b/c, miR-155 and let-7a was determined by quantitative real-time PCR in paraffin embedded formalin fixed tumor specimens from 639 IALT patients. Prognostic and predictive value of microRNA expression for survival were studied using a Cox model, which included every factor used in the stratified randomization, clinicopathological prognostic factors and other factors statistically related to microRNA expression. Investigation of the expression pattern of microRNAs in situ was performed. We also analyzed association of TP53 mutation status and miR-34a/b/c expression, EGFR and KRAS mutation status and miR-21 and Let-7a expression, respectively. Finally, association of p16 and miR-29b expression was assessed. Overall, no significant association was found between any of the tested microRNAs and survival, with the exception of miR-21 where a deleterious prognostic effect of lowered expression was suggested. Otherwise, no single or combinatorial microRNA expression profile predicted response to adjuvant cisplatin-based chemotherapy. Together, our results indicate that the miRNA expression patterns examined were neither predictive nor prognostic in a large patient cohort of radically resected NSCLC randomized to receive adjuvant cisplatin-based chemotherapy versus follow-up only.
Keywords: non–small cell lung cancer, adjuvant chemotherapy, randomized trial, biomarker, drug resistance, microRNA
Introduction
MicroRNAs are a class of small non-coding RNA species of 20–22 nucleotides that have been implicated in the control of many fundamental cellular and physiological processes such as cellular differentiation, proliferation, apoptosis and stem cell maintenance (1). MicroRNAs have been shown to play key roles in carcinogenesis and some microRNAs have been categorized as “oncomiRs” as opposed to “tumor suppressor miRs” (2). Expression patterns of microRNAs are often tissue specific and certain cancer types can be classified based on microRNA expression profiles (3;4). Importantly, expression of certain microRNAs has been associated with chemoresistance (5;6).
Early stage NSCLC patients who undergo complete surgical tumor resection still develop distant metastases in 50% to 70% of cases, resulting in an overall 5-year survival rate of only 40% (7). The International Adjuvant Lung Cancer Trial (IALT) demonstrated that adjuvant cisplatin-based chemotherapy improves the 5-year survival rate in this patient category by an absolute value of 4.1% (8). Two additional randomized studies have confirmed the prolonged 5-year survival rate in stage IB to IIIA NSCLC patients treated with adjuvant cisplatin-based chemotherapy. A third study, using carboplatin-paclitaxel did not confirm this for stage IB disease (9–11). Currently, cisplatin-based adjuvant chemotherapy is considered part of standard management of patients with completely resected stage II and III NSCLC. Nevertheless, survival benefit of adjuvant chemotherapy remains limited to a subgroup of treated patients and a recent update by the IALT investigators showed that the benefit of chemotherapy does not exist beyond 5 years of follow-up (12).
The IALT Biologic Program (IALT-Bio) was established, aiming to define biomarkers predictive for outcome of adjuvant chemotherapy as well as biomarkers prognostic for NSCLC overall survival. A predictive marker refers to a patient or tumor characteristic which is associated with therapy response. A prognostic marker refers to characteristic of a patient or tumor at the time of diagnosis that can be used to estimate the outcome. It was shown that IALT cases with low ERCC1 (excision repair cross - complementation group 1), as well as low p27kip1 expression, benefit from adjuvant chemotherapy (7;13). Importantly, ERCC1 expression remained predictive at 8-year follow-up (12). Expression levels of multidrug resistance proteins MRP1 and MRP2 had no predictive value but MRP2 was shown to be a strong prognostic factor (14). Based on our prior work as well as literature, we analyzed in the IALT cohort the expression of seven biologically relevant microRNAs: miR-21, miR-29b, miR-34a/b/c, miR-155, and let-7a. Using a microRNA microarray approach, we have previously identified microRNA expression profiles unique for NSCLC subtypes and we demonstrated that high miR-155 and low let-7a expression were associated with poor patient survival (15). Other reports also suggested a protective effect of let-7a expression in terms of survival and treatment outcome (16;17), and showed that reduced let-7, which is known to interact with KRAS, is a prognostic factor in lung cancer and can contribute to carcinogenesis (18–20). Furthermore, we have previously reported on low miR-21 expression as a biomarker for favorable outcome of adjuvant chemotherapy in colon cancer, as well as increased miR-21 expression in EGFR mutant tumors (21;22). Numerous other studies support a prognostic or treatment predictive role for miR-21 (23;24).
MicroRNA-34a has been proposed as a prognostic marker of relapse in surgically resected NSCLC and tumor suppressor p53, frequently inactivated in NSCLC, is known to activate the transcription of miR-34a as well as miR-34b and miR-34c, containing in their promotor a p53 binding site (25–29). Finally, reduced expression of miR-29b, known to target de novo DNA methyltransferase 3A and 3B (DNMT3A and DNMT3B), can lead to global hypermethylation and silencing of various tumor suppressor genes (30;31). Methylation of tumor suppressor p16 is a prognostic indicator in lung cancer and so far a potential association between p16 expression and miR-29b expression has not been investigated (7).
Our main hypothesis was that expression of the seven selected microRNAs in tumor specimens from IALT patients could predict treatment response and survival benefit from adjuvant chemotherapy. Furthermore, we hypothesized that associations of microRNAs and prognosis would differ in lung adenocarcinoma patients compared to squamous cell carcinoma patients. As a secondary analysis, we evaluated whether microRNA expression status was associated with molecular markers relevant in lung cancer (EGFR, K-Ras and TP53 mutation status as well as p16 expression), for which data was available the IALT cohort. Finally, we performed in situ investigation of microRNA expression in FFPE tissue sections.
Materials and methods
Patients and Study Design
Patients were enrolled in the IALT study, which randomized 1867 patients with completely resected non-small cell lung cancer, stages I through III, to receive adjuvant cisplatin-based chemotherapy or follow-up. See Appendix 1 for a list of The IALT-Bio Participating Centers. Formalin fixed paraffin-embedded (FFPE) tumor specimens were collected from patients at 28 centers in 14 countries that had recruited more than 10 patients (13). A total of 867 samples were reviewed centrally at the Centre Hospitalier Universitaire Albert Michallon, according to the histopathological classification system adopted by the World Health Organization (WHO) in 2004. The amount and quality of 824 of 867 blocks were judged adequate for serial sectioning and experimental procedures. Ultimately, 783 were judged NSCLC after central review. Approval for the study was obtained from local Institutional Review Boards according to the legal regulations in each participating country. For in situ hybridization experiments, additional tissue sections of NSCLC cases were obtained through the Department of Pathology at University of Maryland, Baltimore, MD. The use of these sections was granted approval by the Institutional Review Board of the National Cancer Institute, and was approved by the Institutional Review Board of the University of Maryland.
RNA isolation and qRT-PCR
Sections were all processed at the National Cancer Institute by the Lung Cancer Laboratory (Medical Oncology Branch) and the Laboratory of Human Carcinogenesis. In order to ensure consistency in experimental conditions, uniform procedures, reagents and equipment were used. Staff members involved in experimental procedures were jointly trained. All reagents, including all qRT-PCR reagents, were ordered in one batch, centrally stored and distributed among staff members of the two participating laboratories. Furthermore, all qRT-PCR microRNA Taqman assays (see also below) were performed on the same equipment. Samples with insufficient or necrotic tumor material were omitted from further processing (n=86). In total 697 tumor samples, as well as 79 adjacent normal tissue specimens were processed.
Specimens consisted of 10µ m FFPE sections. We disposed only of one section per patient case. Glass slides containing the tissue section were cut using a diamond pen in two parts. One part was stained with hematoxylin and eosin (HE) and used for qRT- PCR. The other unstained part was available for in situ hybridization. HE-stained tissues were marked by a lung pathologist for tumor area and, if present, normal tissue area under a BX40 light microscope (Olympus, Tokyo, Japan). Each area was macro-dissected with sterile disposable scalpels (Cincinnati Surgical Company, Cincinnati, OH) for RNA isolation using the RecoverAll Total Nucleic Acid Isolation kit (Ambion, Austin, TX). Forty nanogram of RNA was required for expression analysis of miR-21, miR-29b, miR-34a, miR-34b, miR-34c, let-7a, and miR-155.
Quantitative RT-PCR of microRNAs was performed using TaqMan microRNA assays (Applied Biosystems, Foster City, CA) and the 7900 HT-Fast real-time PCR system (Applied Biosystems). We used small nuclear RNA U66 as endogenous normalization control, consistent with our prior report on miR-34a, miR-34b, and miR-34c expression (32). All assays were performed in triplicate by investigators who were blinded to clinical data of the sample cohort. MicroRNA expression was quantified as delta Ct values, where Ct = threshold cycle, delta Ct = (Ct target miRNA minus Ct RNU66). Delta CT was calculated using RQ manager software, version 1.2 (Applied Biosystems)..
Replicates with a Ct standard deviation greater than 1, or, in case of U66 only, with an average Ct greater than 35, were omitted from further analysis (n=58), resulting in a dataset of 639 cases. In case expression of a microRNA was available for both tumor and normal tissue, microRNA expression was additionally quantified as delta delta Ct values, delta delta Ct = (delta Ct target miRNA tumor tissue minus delta Ct target miRNA matched normal tissue), for a separate analysis.
In Situ Hybridization
In situ investigation of microRNA expression in lung cancer was performed on a selection of the IALT-Bio cases as well as on lung cancer tissue sections procured from the University of Maryland (see also above). Probes for human miR-21, miR-34a, miR-155, and let-7a were used (Exiqon, Woburn, MA). U6 and Scramble probes were used as positive and negative control, respectively. For in situ investigation of microRNA expression we used the method as previously reported (21). In situ hybridization conditions for each individual probe were optimized using serial tissue sections from University of Maryland lung cancer cases, since serial sections of IALT-Bio cases were not available. Methodology updates included the use of biotin-labeled microRNA probes (Exiqon), and a biotinyl tyramide-based system (GenPoint™, Catalyzed Signal Amplification System, DAKO, Glostrup, Denmark) and Vector NovaRed (Vector Laboratories, Burlingame, CA) as a substrate (brown/red). Tissues were counterstained with Mayer’s hematoxylin (blue). Images were taken on a BX40 light microscope using the Olympus DP70 digital camera and DP controller software (Olympus). Staining results were confirmed by an independent pathologist at the National Cancer Institute.
Statistical analysis
National Cancer Institute Investigators involved in experimental procedures remained blinded to any of the clinical data. All statistical analyses were performed at the Institute Gustave Roussy. Initial analysis of microRNA expression values revealed heterogeneity in the data distribution of the tumor specimens between the two laboratories. The data were therefore standardized by subtracting the subgroup mean and division by the subgroup standard deviation. Subgroups were well balanced with respect to treatment (adjuvant chemotherapy arm and control arm). To remove the potential to bias the results, the median standardized value was a priori chosen as cut-off to determine microRNA expression status. MicroRNA expression was defined as negative when the expression value was lower than the median and positive when the expression value was equal or greater than the median. To test for differences between microRNA negative and positive samples, comparisons had to take study center into account. Therefore logistic regression stratified by center was used both for univariate and multivariate analyses. The prognostic value of microRNA status and chemotherapy for survival were studied using a Cox model. As in the original IALT analysis, the Cox model included every factor used in the stratified randomization (center, tumor stage, and type of surgery (pneumonectomy, lobectomy, segmentectomy), plus clinical and histological prognostic factors (age (<55 yrs, 55–64 yrs, >64 yrs), sex, WHO performance status, nodal status, lymphoid infiltration (not intense, intense) and the revised histopathological type (adenocarcinoma, squamous cell carcinoma, other NSCLC)) (8). All other factors that were statistically related to microRNA expression in the multivariate logistic model (P <.05) were added to the survival Cox model. Trend tests were performed using the continuous standardized values instead of the dichotomized values (positive vs. negative). Analyses were also performed using distribution quartiles of the standardized values.
The predictive value of each microRNA was studied by testing the interaction between microRNA expression and the attributed treatment (chemotherapy or no chemotherapy) in the same Cox model. To study association between microRNAs and other markers, logistic regression of the marker on either positivity or standardized value of microRNA expression (denoted "trend"), stratified by center, was used. For analysis using normal and/ or normal-tumor matched expression levels, the Cox survival model had to be simplified due to a low number of available cases. Stratification by 2 regions of the world (Western Europe vs. other parts of the world) was used. Furthermore, a smaller number of adjustment variables were entered in the model: only stage (the only significant prognostic factor), plus histology and the variable(s) correlated with each microRNA. To study variation of the prognostic effect with histology, only squamous-cell carcinoma and adenocarcinoma were considered.
All analyses were performed with long term survival data (12). All reported P values were two-sided. P values below 0.01 were considered statistically significant in order to limit the risk of false positive results. All analyses were performed using SAS software, version 9.1 (SAS Institute Inc. Cary, NC).
Results
After quality control, 639 IALT-Bio samples remained that had measurements of at least one microRNA. Patient characteristics are listed in Supplemental Table 1. Patients characteristics varied in relation to the sample size per evaluated microRNA.
Association of microRNA expression and clinicopathological covariates
As microRNA expression is thought to vary according to histotypes and could be associated with tumor characteristics, we explored associations between individual microRNA expression patterns and the clinicopathological variables. The associations remaining significant in the multivariate analysis are summarized in Table 1. MiR-21 status was associated with histology (p=0.04) and lymphoid infiltration (p=0.04); miR-29b status was associated with age (p=0.03), histology (p=<0.0001) and lymphoid infiltration (p=0.005); miR-34a status was associated with histology (p=0.0002), lymphoid infiltration (p=0.03) and lymphatic invasion (p=0.04); miR-34b status was associated with disease stage (p=0.04) and histology (p=0.01), miR-34c status was associated with histology (p=0.0002); miR-155 status was associated with lymphoid infiltration (p=0.001), and no covariates were associated with let-7a status. Supplemental Table 2 summarizes the association of microRNA expression with covariates (univariate analysis or trend test). See also Supplemental Tables 3–9 for a complete overview of all univariate analyses per microRNA.
Table 1.
Summary of significant associations between covariates and dichotomized microRNA expression values (multivariate logistic model)
| Covariate | miR | Categories | % in miR negative group |
% in miR positive group |
multivariate p-value |
|---|---|---|---|---|---|
| Age | miR-29b | < 55 years | 29% | 31% | 0.03 |
| 55–64 years | 40% | 47% | |||
| >64 years | 31% | 22% | |||
| Stage | miR-34b | I | 33% | 38% | 0.04 |
| II | 20% | 26% | |||
| III | 47% | 36% | |||
| Histology | miR-21 | Adenocarcinoma | 28% | 41% | 0.04 |
| Squamous cell | 60% | 48% | |||
| Other NSCLC | 12% | 11% | |||
| miR-29b | Adenocarcinoma | 22% | 46% | <0.0001 | |
| Squamous cell | 68% | 41% | |||
| Other NSCLC | 11% | 13% | |||
| miR-34a | Adenocarcinoma | 24% | 45% | 0.0002 | |
| Squamous cell | 64% | 44% | |||
| Other NSCLC | 13% | 11% | |||
| miR-34b | Adenocarcinoma | 35% | 34% | 0.01 | |
| Squamous cell | 50% | 57% | |||
| Other NSCLC | 15% | 9% | |||
| miR-34c | Adenocarcinoma | 38% | 31% | 0.0002 | |
| Squamous cell | 47% | 61% | |||
| Other NSCLC | 16% | 8% | |||
|
Lymphoid infiltration |
miR-21 | Intense | 9% | 15% | 0.04 |
| Not intense | 91% | 85% | |||
| miR-29b | Intense | 8% | 15% | 0.005 | |
| Not intense | 92% | 85% | |||
| miR-34a | Intense | 9% | 14% | 0.03 | |
| Not intense | 91% | 86% | |||
| miR-155 | Intense | 7% | 17% | 0.001 | |
| Not intense | 93% | 83% | |||
| Lymphatic invasion | miR-34a | Yes | 67% | 75% | 0.04 |
| No | 33% | 25% | |||
| KRAS | Let-7a | Yes | 87% | 85% | 0.10* 0.03# |
| No | 13% | 15% |
univariate analysis dichotomized values;
trend test on let-7a
Prognostic analysis
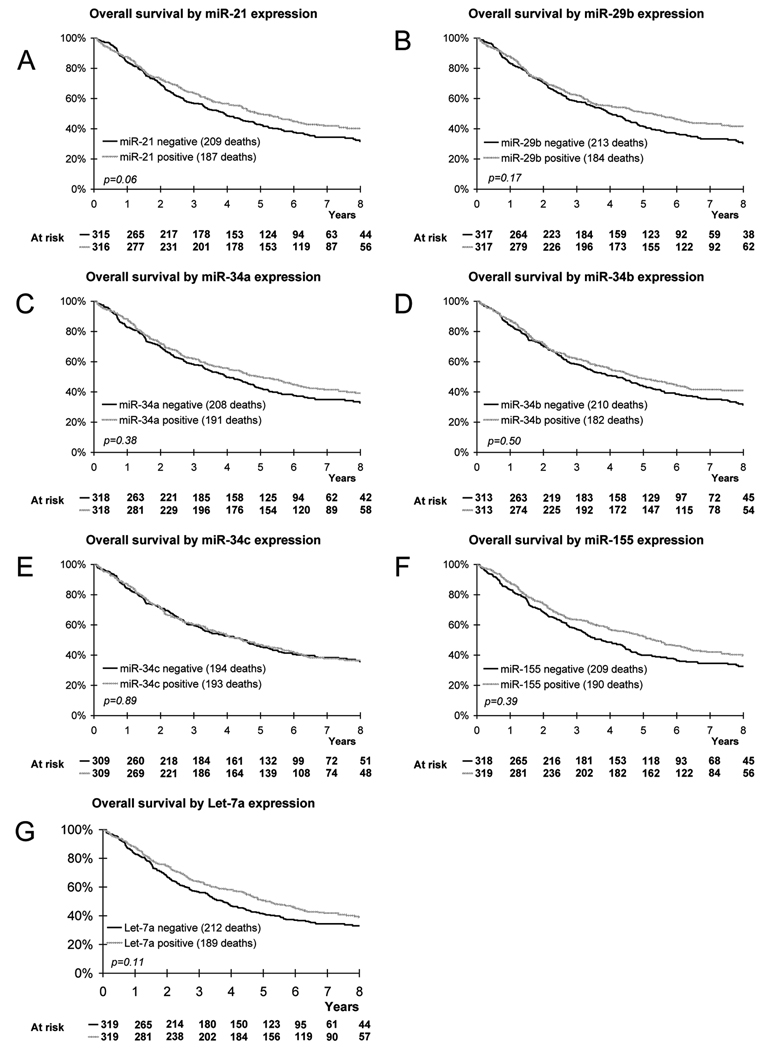
Next, we investigated the prognostic value of microRNA expression. The expression status of none of the microRNAs was significantly prognostic for outcome (See Figure 1, as well as Table 2 for a summary of results). However, there is a borderline prognostic effect of miR-21 expression on overall survival with a worse survival for miR-21 negative cases (p=0.06, trend: p=0.01). Although there is no prognostic effect of miR-34b as well as miR-34c positivity on survival, there is nevertheless a heterogeneous effect between quartiles (p=0.005 and p=0.004, respectively) which is not associated with a specific trend. On the population evaluable for both let-7a and KRAS (n=582), let-7a was a good prognostic indicator (HR=0.79 [0.64;0.99]; p=0.04). However, in a total of 638 samples evaluable for let-7a, this prognostic association was no longer significant (HR=0.84 [0.68;1.04]; p=0.11).
Fig. 1.
Kaplan-Meier estimates of overall survival according to miR-21 expression (A), miR-29b expression (B), miR-34a expression (C), miR-34b expression (D), miR-34c expression (E), miR-155 expression (F), Let-7a expression (G).
Table 2.
Prognostic analyses
| Variable | Comparison | No deaths/ No pts |
8-y OS rate [95% CI] |
Median OS (mo) |
HR for death [95% CI] |
p- value (trend) |
|---|---|---|---|---|---|---|
| miR-21 | negative | 209 / 315 | 32% [26%;38%] | 47 | REF=1 |
0.06 (0.01) |
| positive | 187 / 316 | 40% [35%;46%] | 59 | 0.81 [0.65;1.01] | ||
| miR-29b | negative | 213 / 317 | 30% [25%;36%] | 48 | REF=1 |
0.17 (0.11) |
| positive | 184 / 317 | 42% [36%;47%] | 63 | 0.85 [0.67;1.07] | ||
| miR-34a | negative | 208 / 318 | 33% [27%;39%] | 48 | REF=1 |
0.38 (0.07) |
| positive | 191 / 318 | 39% [34%;45%] | 60 | 0.90 [0.72;1.14] | ||
| miR-34b | negative | 210 / 313 | 31% [26%;37%] | 50 | REF=1 |
0.50 (0.42) |
| positive | 182 / 313 | 41% [36%;47%] | 58 | 0.93 [0.75;1.15] | ||
| miR-34c | negative | 194 / 309 | 36% [30%;42%] | 54 | REF=1 |
0.89 (0.29) |
| positive | 193 / 309 | 37% [31%;42%] | 53 | 1.02 [0.82;1.26] | ||
| miR-155 | negative | 209 / 318 | 40% [35%;45%] | 45 | REF=1 |
0.39 (0.09) |
| positive | 190 / 319 | 40% [34%;45%] | 64 | 0.91 [0.72;1.13] | ||
| Let-7a | negative | 212 / 319 | 33% [28%;39%] | 45 | REF=1 |
0.11 (0.25) |
| positive | 189 / 319 | 39% [33%;45%] | 61 | 0.84 [0.68;1.04] |
Abbreviations: HR: hazard ratio; CI: confidence interval; OS: overall survival; no: number; y: year; pts: patients
Predictive analysis
As microRNA expression status has been associated with treatment response, we next investigated the predictive value of the seven microRNAs (21;33). Expression status of none of the microRNAs had a predictive effect on survival. See Table 3 for a summary of results as well as Supplemental Figure 1 for Kaplan-Meier estimates of overall survival according to treatment in microRNA positive patients compared with microRNA negative patients.
Table 3.
Predictive analyses*
| Variable | Parameter | Chemotherapy group |
Control group | HR for death Chemotherapy vs. Control |
|---|---|---|---|---|
| miR-21 | miR-21 neg (no deaths/no pts) 8-y OS rate; median OS |
111 / 163 29% [22%;37%]; 50 |
98 / 152 35% [28%;44%]; 42 |
0.90 [0.67;1.19] p=0.45 |
| mir21 pos (no deaths/no pts) 8-y OS rate; median OS |
96 / 161 39% [31%;47%]; 61 |
91 / 155 41% [34%;50%]; 59 |
1.02 [0.75;1.37] p=0.84 |
|
| HR for death pos vs. neg p-value |
0.86 [0.64;1.16] p=0.33 |
0.76 [0.56;1.03] p=0.08 |
Interaction p=0.56 |
|
| miR-29b | miR-29b neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
115 / 168 27% [20%;35%]; 51 |
98 / 149 34% [27%;43%]; 46 |
0.90 [0.68;1.19] p=0.47 |
| miR-29b pos (no deaths/no pts) 8-y OS rate; median OS (mo) |
93 / 157 40% [33%;48%]; 64 |
91 / 160 43% [35%;51%]; 63 |
1.03 [0.76;1.40] p=0.84 |
|
| HR for death pos. vs. neg. p-value |
0.90 [0.66;1.23] p=0.52 |
0.79 [0.57;1.09] p=0.15 |
Interaction p=0.53 |
|
| miR-34a | miR-34a neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
116 / 170 29% [22%;37%]; 48 |
92 / 148 38% [30%;46%]; 47 |
0.97 [0.73;1.29] p=0.82 |
| miR-34a pos (no deaths/no pts) 8-y OS rate; median OS (mo) |
94 / 158 39% [31%;47%]; 67 |
97 / 160 39% [32%;48%]; 53 |
0.97 [0.72;1.30] p=0.82 |
|
| HR for death pos. vs. neg. p-value |
0.90 [0.67;1.23] p=0.51 |
0.90 [0.66;1.24] p=0.53 |
Interaction p=1.00 |
|
| miR-34b | miR-34b neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
112 / 164 28% [21%;37%]; 55 |
98 / 149 35% [27%;43%]; 42 |
0.96 [0.72;1.28] p=0.77 |
| miR-34b pos (no deaths/no pts) 8-y OS rate; Median OS (mo) |
94 / 158 39% [32%;47%]; 55 |
88 / 155 43% [35%;51%]; 58 |
0.99 [0.73;1.35] p=0.96 |
|
| HR for death pos. vs. neg. p-value |
0.94 [0.70;1.26] p=0.69 |
0.91 [0.67;1.25] p=0.56 |
Interaction p=0.87 |
|
| miR-34c | miR-34c neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
106 / 163 31% [24%;40%]; 56 |
88 / 146 41% [33%;49%]; 45 |
1.05 [0.78;1.41] p=0.76 |
| mir34c pos (no deaths/no pts) 8-y OS rate; median OS (mo) |
98 / 157 36% [29%;45%]; 54 |
95 / 152 37% [29%;45%]; 52 |
0.85 [0.78;1.41] p=0.29 |
|
| HR for death pos. vs. neg. p-value |
0.92 [0.69;1.24] p=0.60 |
1.13 [0.83;1.55] p=0.43 |
Interaction p=0.34 |
|
| miR-155 | miR-155 neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
111 / 157 27% [20%;35%]; 43 |
98 / 161 39% [31%;47%]; 46 |
1.00 [0.75;1.33] p=1.00 |
| mir155 pos (no deaths/no pts) 8-y OS rate; median OS (mo) |
100 / 172 40% [32%;48%]; 67 |
90 / 147 39% [31%;48%]; 56 |
0.95 [0.71;1.28] p=0.75 |
|
| HR for death pos. vs. neg. p-value |
0.89 [0.66;1.20] p=0.43 |
0.93 [0.68;1.27] p=0.64 |
Interaction p=0.83 |
|
| Let-7a | Let-7a neg (no deaths/no pts) 8-y OS rate; median OS (mo) |
111 / 159 28% [21%;36%]; 45 |
101 / 160 38% [31%;47%]; 45 |
1.06 [0.80;1.40] p=0.69 |
| Let-7a pos (no deaths/no pts) 8-y OS rate; median OS |
100 / 170 39% [31%;47%]; 67 |
89 / 149 39% [31%;47%]; 58 |
0.89 [0.66;1.20] p=0.44 |
|
| HR for death pos. vs. neg. p-value |
0.78 [0.58;1.04] p=0.08 |
0.93 [0.68;1.26] p=0.62 |
Interaction p=0.41 |
Remarks: OS listed in months; 95% confidence interval of HR and 8-y OS rate listed between brackets; Interaction refers to test for interaction of microRNA and treatment.
Abbreviations: HR: hazard ratio; OS: overall survival; no: number; y: year; pts: patients
Additional prognostic and predictive analyses
As initial analysis of microRNA expression values revealed heterogeneity in the data distribution between the two laboratories, separate prognostic and predictive analyses were performed in both groups to exclude an effect of the standardization procedure on the study outcome and to exclude negative results were not due to inconsistent assay results,
There was no difference in the prognostic effect of any of the seven microRNAs in both groups. There was no difference in the predictive effect of miR-21, miR-34a, miR-34b, miR-34c, nor let-7a, in both groups. There was a borderline difference in the predictive effect of miR-29b in both groups (p=0.03 for comparing the interactions and p=0.04 for comparing deviations from the group averages), but the heterogeneity and trend test did not show anything consistent. There was a difference in the predictive effect of miR-155 in both groups (p=0.007 for comparing the treatment interaction, but the difference was not significant for comparing deviations from the group averages (p=0.12) nor in the trend tests (p=0.07)). As the variations of the hazard ratios in the quartiles within each group were not consistent, the likelihood of a predictive effect is low. Furthermore, for predictive analyses, the treatment effect of chemotherapy was borderline significantly different between both groups and differences in predictive effects of microRNA expression profiles should therefore be interpreted with respect to the average treatment effect in the group. More details regarding these additional analyses are available upon request.
Association of microRNA expression with other IALT-Bio markers
As we did not find neither a significant predictive nor prognostic effect of any of the candidate microRNAs, we subsequently assessed in a secondary analysis the association of microRNAs and other molecular markers relevant in lung cancer. We were able to perform these analyses because data was or became available in the course of the study. It was hypothesized that a combined protein and microRNA expression signature could also potentially be of predictive and/ or prognostic value.
Since TP53 activity is associated with miR-34a/b/c transcription, we looked at an association between TP53 mutations and miR-34a/b/c expression. IALT-Bio investigators previously reported on the prognostic and predictive value of TP53 mutations (exons 5 to 8) and KRAS mutations (codons 12 and 13) in the IALT-Bio cohort (34). TP53 mutations were shown in 46% of patients. After 8 years of follow-up no prognostic value for TP53 mutation status was shown in all cases grouped together. It was recently suggested that patients with non-adenocarcinoma NSCLC and TP53 wild type might benefit from adjuvant cisplatin-based chemotherapy, whereas it might be less beneficial in patients with mutated TP53 (34). Nevertheless, there was no global association between TP53 mutations and miR-34a status (positivity: p= 0.70; trend: p=0.33). The association between TP53 mutations and miR-34a positivity did not vary with histology (p=0.23). Equally, there was no global association between TP53 mutations and miR-34b or miR-34c (positivity: p=0.91, trend: p=0.49; positivity: p=0.57, trend: p=0.35, respectively) and this did not vary with stratification for histology (p=0.47; p=0.78, respectively).
Next we looked at potential associations between miR-21 expression and EGFR mutation status, as increased miR-21 expression was detected previously in EGFR mutant tumors, EGFR being one of the key markers in NSCLC biology (22). Only 20 tumors had EGFR mutations and it was therefore not possible to perform a sub analysis by histology. We did not detect a global association between occurrence of EGFR mutations and miR-21 expression (positivity: p=0.73, trend: p=0.29).
In the aforementioned study, KRAS mutation was suggested to be a biomarker of poor prognosis in patients with non-adenocarcinoma (34). Since KRAS is one of the known targets of let-7a, we investigated the association of KRAS mutations and let-7a expression. We found that there is a global borderline association between KRAS mutations and let-7a (positivity: p=0.10, trend: p=0.03). The association between KRAS mutations and let-7a positivity did not vary with histology (p=0.42).
Finally we assessed a potential association between miR-29b and p16 expression. De novo DNA methyltransferase 3A and 3B are targets of miR-29b and p16 expression is known to be silenced in lung cancer due to hypermethylation. However, we found that there is no global association between p16 expression as assessed by immunohistochemistry and miR-29b (positivity: p=0.14, trend: p=0.10). The association between p16 IHC positivity and miR-29b positivity did not vary with histology (p=0.86).
Analyses using tumor adjacent normal tissue
In total, 79 samples have measurements of a least one matched tumor and normal microRNA. The median expression ratio for tumor and normal tissues (T/N) was, as expected, positive for the “oncomiRs” miR-21 (0.95±1.33 (±SD)) and miR-155 (0.05±1.13), indicating a higher expression in the tumor tissue compared to the normal tissue. Relative T/N expression was, as expected, negative for the “tumor suppressor miRs” miR-29b (−0.68±1.33), miR-34a (−0.40±1.38), miR-34b (−0.37±2.88), miR-34c (−0.51±2.73), and let-7a (−0.61±1.40).
In situ hybridization
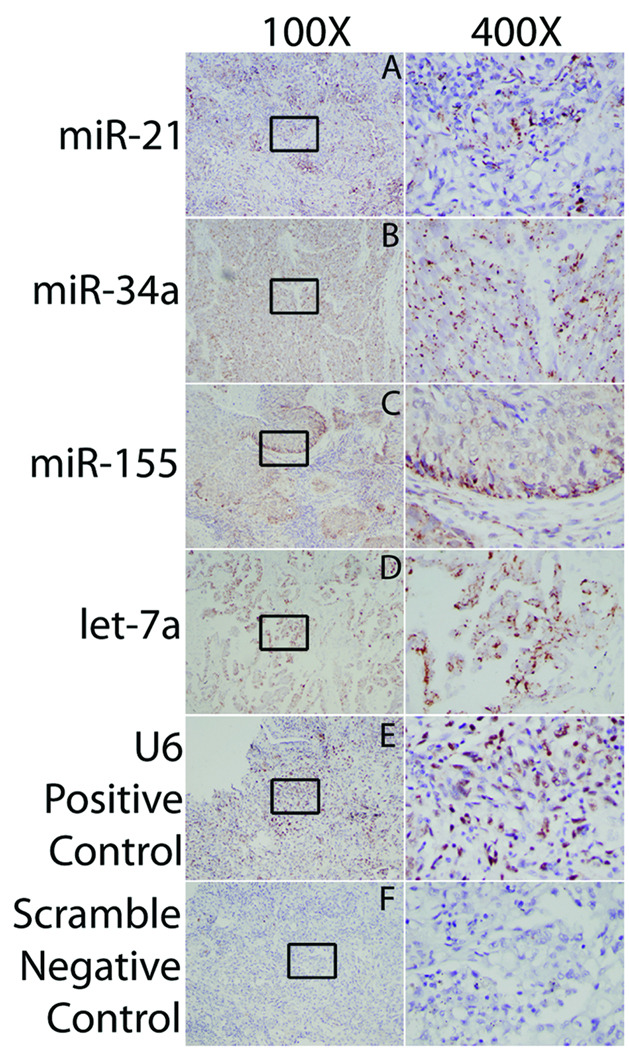
We performed in situ hybridization (ISH) of the seven tested microRNAs. For the IALT-Bio cohort, none of the tested microRNAs were neither of prognostic nor predictive. MicroRNA-21 was the only microRNA with a suggested deleterious prognostic effect of lower values. Figure 2 shows a representative pattern of in situ miR-21 staining in a non-small cell lung cancer adenocarcinoma case with high miR-21 expression according to the qRT-PCR assay. In general we observed that staining was exclusively present in the cytoplasm for all probes except, as expected, for the experimental control, endogenous small nucleolar RNA U6, which showed nuclear expression. As for miR-155 staining, we frequently observed that the staining was localized to the outer end/leading edge of each nodule of squamous cells within the tumor. Additionally, Figure 2 shows miR-34a and let-7a positive cases (data for miR-29b, miR-34b/c not shown).
Fig 2. In situ hybridization for microRNAs in lung tumors.
Lung tumors were hybridized with biotin-labeled microRNA (miR) probes that were detected using a biotinyl tyramide-based system and Vector NovaRed as a substrate (brown/red). Tissues were counterstained with Mayer’s hematoxylin (blue). Tissue sections were sourced from both the IALT cohort, and the University of Maryland. Representative slides with positive staining cells are shown for various cancer types, but was not limited to these specific types. Positively staining cells are shown for (A) miR-21 in adenocarcinoma, (B) miR-34a in a large cell carcinoma with neuroendocrine features, (C) miR-155 in squamous cell carcinoma (SCC), and (D) let-7a in adenocarcinoma. Staining was limited to the cytoplasm, as expected, and was diffusely localized within tumor types except for miR-155 in SCC. Mir-155 was expressed in cells on the edge of squamous cell nodules within SSCs. (E) U6 and (F) Scramble probes were used as positive and negative controls, respectively.
Discussion
Based on preclinical studies as well as translational studies using clinical specimens, microRNA expression levels have been suggested to be biomarkers for prognosis as well as treatment outcome in numerous malignancies (35–37). Expression of seven candidate microRNAs was hypothesized to predict outcome of adjuvant chemotherapy in NSCLC, aiming to establish routine markers for selection of patients for adjuvant chemotherapy in order to improve treatment outcome.
We tested this hypothesis in a very large NSCLC cohort of 639 cases, randomized for treatment, using extensive quality and methodological control measures. We disposed of only one tumor section per case and made a selection of seven biologically relevant microRNAs to be analyzed for associations with clinicopathological covariates, prognosis and treatment outcome.
Interestingly, microRNA expression did correlate with histology for five out of seven microRNAs assessed: miR-21, miR-29b, miR-34a/b/c. Recently, a study on microRNA expression profiling was reported using 290 FFPE specimens from the EAGLE study, a population-based case control study in lung cancer (38). A microRNA signature, including miR-29b and let-7a, was identified that strongly differentiated histological subtypes, i.e. adenocarcinoma and squamous cell carcinoma in male smoker patients. Interestingly, cigarette smoking intensity showed an inverse correlation with let-7 expression in female adenocarcinoma patients and a positive correlation with miR-21 expression in male squamous cell carcinoma patients. Furthermore, only in the group of male smokers with squamous cell carcinoma, a prognostic miR signature could be established, which included miR-34c-5p and miR-34a (38). Unfortunately we did not dispose of smoking status data for IALT cases to confirm these relationships.
We found that positive expression of miR-21, miR-29b, miR-34a and miR-155 was associated with intense lymphoid infiltration, or presence of lymphatic invasion (miR-34a only). In this regard, it was previously shown that miR-21 expression is associated with an increased rate of lymph node metastasis in breast cancer and colorectal cancer patients (39;40).
Overall, we were not able to show a prognostic value of any of the seven microRNAs in this patient cohort. Upregulated miR-21 expression has been previously associated with worse outcome in NSCLC cases (23;24;41). However, the study by Markou et al. consisted of few cases, 48, and it made use of matched tumor and normal tissues (24). Consequently, results reported were based on the tumor tissue/ normal tissue (T/N) expression ratio. In contrast, for our main analysis we made use of tumor tissue expression values. Additionally, the study by Raponi et al. made use of a microRNA array approach (23). Another characteristic of our study was that we disposed only of FFPE tissue sections instead of frozen tumor specimens.
We did establish an association between let-7a expression and KRAS mutation status, albeit only in the multivariate analysis on the means and not the dichotomized expression values. The significance of this finding is unclear. So far it has been reported that the KRAS-LCS6 polymorphism results in upregulation of the KRAS gene and concomitant downregulation of let-7 (42). However, this polymorphism was not found to be associated with KRAS mutations (43).
We could not confirm an association between EGFR mutation status and miR-21 expression, as reported previously (22).
Additionally, we could not show a predictive effect of any of the seven microRNAs on treatment outcome, which consisted of cisplatin-based adjuvant chemotherapy. Drugs combined with cisplatin among the 639 patients were either etoposide (54%), vinorelbine (33%), vinblastine (8%) or vindesine (6%) (8). We previously showed that high microRNA-21 expression is related to fluoropyrimidine (5-FU) resistance in colorectal cancer as well as gemcitabine resistance in pancreatic cancer and the predictive value of microRNA expression may be chemotherapy and/ or tumor specific (21;44).
As the prognostic and/ or predictive value of microRNAs might be dependent on the expression of more than one microRNA, we performed cluster analysis using standardized values of the 7 microRNAs. However, no combination of expression profiles of the seven microRNAs was found to be prognostic or predictive (data not shown). Finally, in situ hybridization was performed using specimens from this cohort as well as specimens obtained from the University of Maryland. Treatment conditions for microRNA in situ hybridization vary per probe and histology. Conditions had to be optimized using serial sections of an independent test cohort for which qRT-PCR data was available. For the IALT cases, a disadvantage was that we were completely blinded to origin of cases, specimens being collected from a worldwide multicenter study, with non standardized fixation and storage conditions. Fixation and procurement variations all affect the efficacy of in situ hybridization to a much bigger extent than microRNA isolation and qRT-PCR (45;46). Nevertheless, in standardized test cases as well as IALT-Bio cases, in situ expression of microRNAs could be demonstrated.
In conclusion, many reports exist on microRNA expression profiling and the prognostic and/or treatment predictive impact of microRNA expression in NSCLC. However, many studies are limited by small sample size and by not being randomized. This study constitutes the largest ever group of NSCLC patients analyzed for the prognostic and predictive value of microRNA expression. The seven target microRNAs chosen for evaluation, miR-21, miR-29b, miR-34a/b/c, miR-155 and let-7a, were neither prognostic nor predictive in this patient cohort. Further studies, e.g. making use of a microRNA array approach, are warranted in NSCLC, in order to identify the prognostic or predictive value of expression of other microRNAs which were not included in this study.
Supplementary Material
Acknowledgments
Financial support: this work was in part supported by an unrestricted research grant from Eli Lilly and grants from the Programme Hospitalier de Recherche Clinique 2005, as well as Cancéropôle Rhône-Alpes.
References
- 1.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 2.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Li W, Yang Y, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 6.Fujita Y, Kojima K, Hamada N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 7.Filipits M, Pirker R, Dunant A, et al. Cell cycle regulators and outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer: the International Adjuvant Lung Cancer Trial Biologic Program. J Clin Oncol. 2007;25:2735–2740. doi: 10.1200/JCO.2006.08.2867. [DOI] [PubMed] [Google Scholar]
- 8.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Rosell R, De LM, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 10.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 11.Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol. 2010;28:35–42. doi: 10.1200/JCO.2009.23.2272. [DOI] [PubMed] [Google Scholar]
- 13.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 14.Filipits M, Haddad V, Schmid K, et al. Multidrug resistance proteins do not predict benefit of adjuvant chemotherapy in patients with completely resected non-small cell lung cancer: International Adjuvant Lung Cancer Trial Biologic Program. Clin Cancer Res. 2007;13:3892–3898. doi: 10.1158/1078-0432.CCR-06-2446. [DOI] [PubMed] [Google Scholar]
- 15.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 17.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 19.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 21.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 24.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 25.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 28.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 29.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 31.Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumamoto K, Spillare EA, Fujita K, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 34.Ma X, Vataire AL, Sun H, et al. TP53 and KRAS muations as markers of outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small cell lung cancer (NSCLC): the International Adjuvant Lung Cancer Trial (IALT) Biological Program. Ann Oncol. 2008;19:viii61. [Google Scholar]
- 35.Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 36.Hu X, Schwarz JK, Lewis JS, et al. A MicroRNA Expression Signature for Cervical Cancer Prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 41.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 42.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69:51–53. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84–88. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.