Abstract
Yeast Gis1 protein functions as a transcription factor after nutrient limitation and oxidative stress. In this report, we show that Gis1 also regulates the induction of several genes involved in spore wall synthesis during sporulation. Gis1 contains a JmjC domain near its N-terminus. In many proteins JmjC domains provide histone demethylase activity. Whether the JmjC domain of Gis1 contributes to its transcriptional activation is still unknown. Here we show that gis1 point mutations that abolish Fe (II) and α-ketoglutarate binding, known co-factors in other JmjC proteins, are still able to induce transcription normally during glucose starvation and sporulation. Even deletion of the whole JmjC domain does not affect transcriptional activation by Gis1. Moreover, the JmjC domain is not required for the toxicity associated with Gis1 overexpression. The data demonstrate that the JmjC domain is dispensable for transcriptional activation by Gis1 during nutrient stress and sporulation.
Keywords: sporulation, jimonji domain, histone demethylation
Introduction
Upon nitrogen and carbon source starvation, diploid cells of Saccharomyces cerevisiae undergo meiosis and form haploid spores (Zaman, et al., 2008). A special extracellular matrix, the spore wall, is assembled during maturation of the spore. The spore wall enables spores to survive under poor environmental conditions (Smits, et al., 2001). It consists of four distinct layers. The innermost layer is composed mostly of mannan and the next layer of β 1,3-linked glucans (Kreger-Van Rij, 1978). The outer two layers, which are unique to the spore, are composed of chitosan and dityrosine (Pammer, et al., 1992, Briza, et al., 1994). Intriguingly, the order of synthesis of the four layers matches the order of layers in the spore wall: first mannan, then β glucan, then chitosan, and finally dityrosine, which suggests an underlying mechanism of coordinated regulation.
Many genes involved in meiosis and sporulation are transcribed in a series of temporal waves. Individual genes can be classified as early, middle, mid-late, or late genes depending on the timing of their induction (Mitchell, 1994, Vershon & Pierce, 2000). Genes involved in spore wall synthesis generally fall into the middle, mid-late, and late classes (Neiman, 2005). The factors controlling early and middle gene are well understood (Mitchell, 1994, Chu, et al., 1998). However, so far, little is known about factors controlling mid-late and late gene expression. During our previous screen for mutants with spore wall defects, a transcription factor, Gis1, was identified (Coluccio, et al., 2004). The gis1Δ cells lacked the dityrosine layer and some also lacked the chitosan layer. Further tests showed that DIT1::lacZ, a reporter for mid-late gene expression, was not induced in gis1Δ cells. That suggested that Gis1 might function as a novel transcription factor to induce genes required for the synthesis of the spore wall.
Gis1 was first discovered as a multicopy suppressor of the Gal− phenotype of snf1 mig1 srb8/10/11 cells (Balciunas & Ronne, 1999). Subsequent studies established the role of Gis1 as an important transcription activator during nutrient depletion or oxidative stress. It functions in the Ras/cAMP pathway, downstream of Rim15 to induce transcription of several genes, including SSA3 and HSP12 (Pedruzzi, et al., 2000). Further microarray analysis suggested that over 100 genes are regulated by Gis1 during the diauxic transition (Cameroni, et al., 2004). Interestingly, there are reports that Gis1 can also function as a transcriptional repressor (Jang, et al., 1999, Oshiro, et al., 2003). The dual function of Gis1 as either activator or repressor might be explained by its binding partners, promoter context and physiological conditions, such as is the case for Sp3 (Majello, et al., 1997).
GIS1 encodes a protein of 894 amino acids (aa), with two zinc finger domains at its C terminus (aa 828-877). Gis1 also harbors Jumonji N and C domains at its N-terminus, JmjN (aa 11-52) and JmjC (aa 170-324). Notably JmjC domains are found in a large family of histone demethylases (Klose, et al., 2006). Different members of this family have been shown to remove specific methyl groups on histones, including those on histone H3 K4, K9, K36, K27 and R2 (Tsukada, et al., 2006, Yamane, et al., 2006, Agger, et al., 2007, Chang, et al., 2007, Tahiliani, et al., 2007). They act by oxidizing the methyl group to yield formaldehyde and the unmodified lysine or arginine. JmjC containing proteins belong to a super-family of dioxygenases that use α-ketoglutarate and an enzyme-bound Fe (II) ion to oxidize various substrates (Aravind & Koonin, 2001, Chen, et al., 2006). Histone demethylases play a role in several processes including transcriptional activation, gene silencing, heterochromatin establishment and maintenance (Klose & Zhang, 2007). Gis1 belongs to the JHDM3/JMJD2 sub-family, in which members contain JmjN, PHD or Tudor domains, in addition to the JmjC domain. There are two JHDM3/JMJD2 orthologues in yeast, Gis1 and Rph1 (Klose, et al., 2006). Rph1 has demethylase activity for both histone H3 K36me and H3 K9me (Klose, et al., 2007). However, it is still not clear whether Gis1 harbors histone demethylase activity. It was predicted not to have activity based on the fact that it lacked a conserved histidine required for Fe (II) binding (Klose, et al., 2006). Subsequently one group could not demonstrate activity using in vitro enzymatic assays while another group detected weak activity (Klose, et al., 2007, Tu, et al., 2007). Even with little or no histone demethylase activity, the JmjC domain of Gis1 might still be able to regulate gene activity. For instance, the S. pombe JmjC protein, Epe1 has no demonstrable histone demethylase activity and yet plays an important role in allowing transcription of heterochromatic regions (Zofall & Grewal, 2006, Trewick, et al., 2007). Given the close connection between the JmjC domain and epigenetic gene regulation, it is of interest to determine whether the Gis1 JmjC domain contributes to its transcriptional activity.
In this report, we show that GIS1 is required for induction of genes involved in spore wall synthesis during sporulation, including DIT1, SPS100 and SHC1. However, transcriptional activation by Gis1 during either sporulation or glucose limitation is not impaired in gis1 mutants with abolished co-factor binding sites in the JmjC domain. Even deletion of whole JmjC domain does not affect normal transcriptional activation by Gis1. Thus our data indicate that the JmjC domain is dispensable for transcriptional activation by Gis1.
Materials and methods
Plasmids
The wild type GIS1 ORF plus 440 bp of upstream and 237 bp of downstream sequence was cloned into the Spe I site of pRS316 (Sikorski & Hieter, 1989) to yield pYY31. All the plasmids described below contain the same upstream sequence and all the plasmids except the ones with a FLAG tag contain the same downstream sequence. GIS1 point mutations in pYY31 were generated using the QuikChange II mutagenesis kit (Stratagene). Plasmid pYY32 has the gis1-H204A mutation, pYY40 has H204A-D206A and pYY41 has Y292H. The JmjC region encoding amino acids 170-324 of Gis1 was deleted by PCR to generate a Gis1ΔJmjC fragment. The zinc finger region encoding amino acids 828-894 of Gis1 was deleted by PCR to generate a Gis1ΔZnF fragment. These fragments were cloned into the Spe I site of pRS316 and a FLAG tag was fused to the C terminus of the ORF by PCR to generate wild-type GIS1 (pYY53), GIS1ΔJmjC (pYY54) and GIS1ΔZnF (pYY55). For GIS1 overexpression, wild type or various mutant GIS1 genes were cloned into Spe I site of pRS424 (Christianson, et al., 1992) to generate wild-type GIS1 (pYY86), gis1ΔJmjC (pYY87), gis1ΔZnF (pYY88), gis1-H204A D206A (pYY93), and gis1-Y292H (pYY94). All these overexpression constructs have a FLAG tag fused to the C terminus of the ORF.
Transcriptional induction during sporulation
GIS1, gis1ΔJmjC or gis1ΔZnF were cloned into the Spe1 site of the integration plasmid pRS306 (Sikorski & Hieter, 1989) to yield pYY49, pYY50 and pYY51, respectively. These plasmids were linearized at the StuI site within URA3 and transformed into a homozygous gis1Δ diploid strain (SK1 background) to yield strains YYY73 (vector), YYY74 (wild-type GIS1), YYY75 (gis1ΔJmjC) and YYY76 (gis1ΔZnF). Correct integration of the plasmids at the URA3 locus was confirmed by PCR. Cells were grown in YPD medium for one day and diluted into YP acetate medium to OD600 = 0.1. Cells were grown to OD600 = 0.8 and half the cells were collected for pre-sporulation samples. The rest of the cells were washed once with H2O, and then incubated in 2% potassium acetate for an additional 10 hr to induce sporulation, at which time cells were collected. A 10 hr time point was chosen because in the SK-1 background DIT1 transcription is close to a maximum at that time (Chu, et al., 1998, Primig, et al., 2000). RNA was extracted from the cells using a RiboPure-Yeast kit (Ambion) and converted to cDNA by Superscript II Reverse Transcriptase (Invitrogen). The expression level of genes was evaluated by RT-PCR using the following primers: 5′ CATCTTGCCAGCGTTTCC 3′ and 5′ TCGTGCAATT TGGTCGTG 3′ for DIT1; 5′ AGTTCGGACGCGGAAGGTAG 3′ and 5′ GGTCCCGGAGGAGGCATTAG 3′ for SPS100; 5′ CGGCAGACGG TAATTCCGAG 3′ and 5′ GGTTTTCCATCTTGGTAGCA AGC 3′ for SHC1; 5′ TTGGTATTGATTTGGGAACA 3′ and 5′ GGAAGTGCTTGGCATCTGTC 3′ for SSA3. As input controls, the levels of 18s rRNA and ACT1 RNA were evaluated by RT-PCR using the following primers: 5′ GCCGATGGAAGTTTGAGG 3′ and 5′ TACTAGCGACGGGCGGTGT 3′ for rRNA and 5′ACTGAATTAACAATGGATTCTG 3′ and 5′ CATGATACCTTGGTGTCTTG 3′ for ACT1 RNA. Samples were taken after the indicated number of PCR cycles in the linear range of amplification. PCR products were subjected to agarose gel electrophoresis and visualized by ethidium bromide staining. Control PCR reactions on the RNA samples before reverse transcription gave no signal, showing that the RNA samples were free of DNA.
Transcriptional induction during post-diauxic transition
pRS316-based plasmids expressing wild type or various GIS1 mutants, were transformed into a gis1Δ strain (BY4741 background). Cells were grown in SC-Ura medium with 2% glucose until exponential phase and the culture split in half. One half of the cells were collected. The rest were washed with H2O, grown in SC-Ura medium with 0.1% glucose for an additional 4 hr to induce the diauxic transition, at which time cells were collected. RNA was extracted and converted to cDNA. The expression levels of SSA3, DIT1 and SPS100 were evaluated by RT-PCR. The samples were analyzed after the indicated number of PCR cycles as described above.
Gis1 overexpression
Multi-copy pRS424-based plasmids overexpressing wild type or mutant GIS1 were transformed into strain W303-1a. Cells were collected from the original tranformation plates and spotted in 3-fold dilutions onto an SC-Trp plate. Plates were incubated at 30 °C for 2 days and the growth of cells recorded each day.
Westerns
Yeast cell extracts were prepared as described previously (Krishnamoorthy, et al., 2006). Proteins were transferred to a PVDF membrane and blotted with mouse anti-FLAG antibody (Sigma F1804) to detect Gis1 and with rat-anti-tubulin antibody to detect tubulin as a loading control.
Results
GIS1 induces genes for spore wall synthesis during sporulation
To identify genes regulated by GIS1 during sporulation, we carried out a microarray analysis of transcripts during sporulation in wild type and gis1Δ diploid strains (data not shown). The three genes with poor induction in gis1Δ cells were chosen for further analysis by RT-PCR (Fig. 1). Consistent with our previous report (Coluccio, et al., 2004), the induction of DIT1 during sporulation was severely impaired in gis1Δ cells. Besides DIT1, a new target of GIS1, called SPS100, was identified. The protein encoded by SPS100 is highly glycosylated and is a component of the glycoprotein matrix of the spore wall (Law & Segall, 1988). During sporulation, SPS100 was induced in wild-type cells. However, in the gis1Δ mutant, SPS100 was not induced, and its expression was reduced somewhat compared to vegetative growth. GIS1 also contributes very slightly to the induction of SHC1 during sporulation (Fig. 1). This gene encodes the activator of CHS3 (chitin synthase III) and is required for the synthesis of the chitosan layer of the spore wall (Sanz, et al., 2002).
Fig. 1.
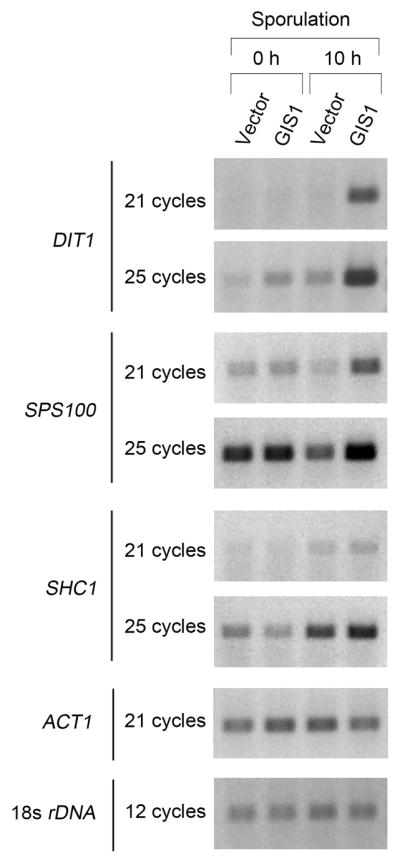
Gis1 is required for induction of genes for spore wall synthesis during sporulation. Homozyous gis1Δ diploid cells were transformed with an integration plasmid, either pRS306 vector or pRS306-GIS1. RNAs were extracted from the cells growing in pre-sporulation medium or after 10 hours in sporulation medium. RT-PCR was performed to check the levels of DIT1, SPS100 and SHC1 RNA. The levels of 18s rDNA and ACT1 RNAwere also measured to serve as an internal controls. Samples were collected after the indicated number of PCR cycles in the linear range of amplification.
Gis1 has been reported to bind to either a PDS (T(T/A)AG3AT) or STRE (AG4) element (Pedruzzi, et al., 2000, Roosen, et al., 2005, Badis, et al., 2008). The promoters of DIT1, SPS100 and SHC1 all contain the STRE element, AG4, suggesting that Gis1 may act directly on the promoters of these genes. Our results show that Gis1 regulates the induction of some of the genes for spore wall synthesis during sporulation.
JmjC domain is dispensable for Gis1 to induce SSA3 during post-diauxic transition
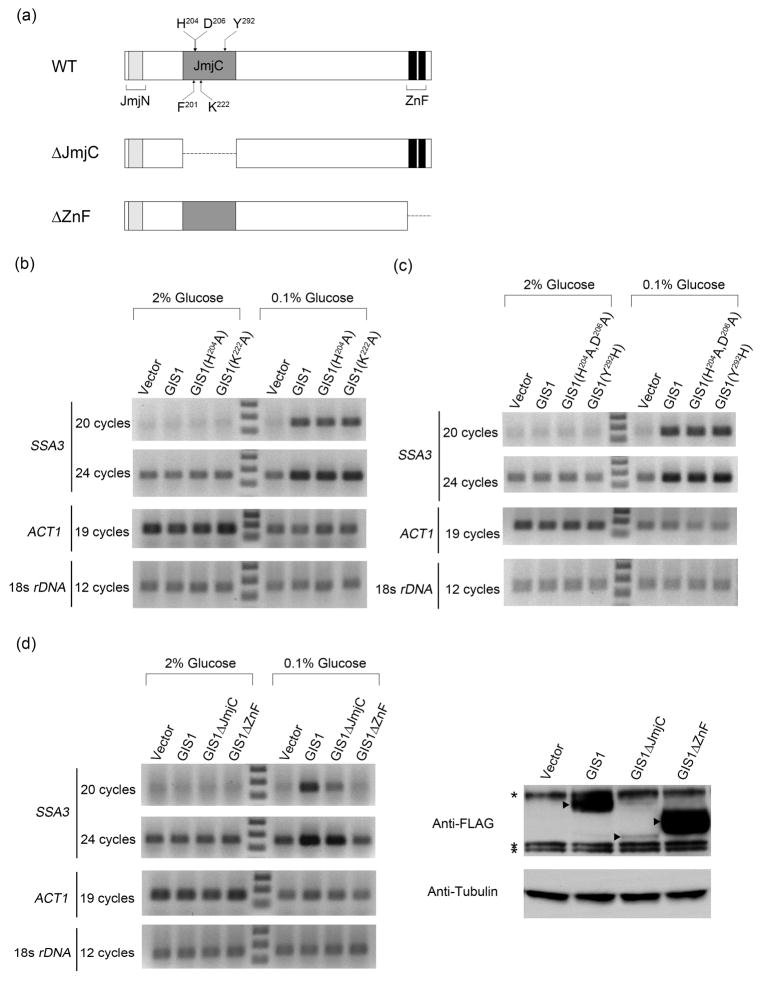
It is known that SSA3 is strongly induced after cells undergo a diauxic transition as they are starved for glucose, and this induction depends on GIS1 (Pedruzzi, et al., 2000). We wanted to determine if the JmjC domain of Gis1 and its putative histone demethylase activity are required for this induction. To address this question, several co-factor binding residues of Gis1 were mutated to alanine separately or in combination to abolish the potential enzymatic activity. As mentioned in the Introduction, Fe (II) and α-KG are the necessary co-factors for the histone demethylation reaction catalyzed by JmjC-domain-containing proteins (Chen, et al., 2006). The structure of JMJD2A, a histone demethylase with a JmjC domain, identified three residues within the JmjC domain that bind Fe (II) and two that bind to α-ketoglutarate (α-KG). These residues are conserved in most JmjC-containing proteins. Based on the alignment with other members of JHDM3A/JMJD2 family, Gis1 contains two conserved residues for Fe (II) binding (H204, D206) and two for α-KG binding (F201, K222), However, the third Fe (II)-binding residue, a histidine, is missing in Gis1 and instead is a tyrosine (Y292) (Fig. 2a). Therefore Gis1 has been predicted not to be able to bind Fe (II) and thus not have histone demethylase activity (Klose, et al., 2006). Whether this substitution ablates the histone demethylase activity of Gis1 is still not settled, since both negative and positive results have been reported (Klose, et al., 2007, Tu, et al., 2007). To assess transcriptional activity of the GIS1 mutants, we checked the GIS1-dependent induction of SSA3 after shifting cells from 2% to 0.1% glucose for 4 hours. In cells with wild-type GIS1, SSA3 was induced upon glucose starvation and as expected, the induction was totally abolished in a gis1Δ mutant (Fig. 2b). However, SSA3 was induced normally in cells expressing gis1-H204A or gis1-K222A as the only source of GIS1 (Fig. 2b). The data suggest that mutation of either the conserved Fe (II) or α-KG binding site doesn’t affect the transcriptional activity of Gis1. To confirm this conclusion, a double mutant (gis1-H204A-D206A) that removed both Fe (II) binding sites was constructed. SSA3 was also induced normally in this mutant (Fig. 2c). As stated previously, both Fe (II) andα-KG are necessary for the histone demethylase activity of JmjC proteins. Since GIS1 mutants predicted to abolish Fe (II) and α-KG binding still induced transcription normally, it suggests that any histone demethylase activity of Gis1 is dispensable for the transcriptional activation.
Fig. 2.
The Gis1 JmjC domain is dispensable for the induction of SSA3 during glucose limitation. (a) Schematic representation of Gis1 domains and deletion mutants used. The predicted residues necessary for Fe (II) binding sites, F201 and K222; and those for α-ketoglutarate (α-KG) binding, H204 and D206 are indicated. Y292, which is a conserved Fe (II)-binding histidine in most JHDM3/JMJD2 family members, is also highlighted. (b–d) SSA3 induction after glucose limitation. Cells were transformed with a plasmid expressing wild type or the indicated GIS1 mutant. RNAs were extracted from cells growing in 2% glucose or in 0.1% glucose for 4 hours. RT-PCR was performed to check the levels of SSA3. Cells from the 0.1% glucose cultures were also used for western analysis (d). Arrows indicate wild type or mutant Gis1 proteins and asterisks indicate cross-reacting bands recognized by the anti-FLAG antibody.
We also mutated Gis1 Y292 to H, thus creating a protein that contained all the conserved Fe (II) and co-factor binding residues present in most JmjC domains. If Gis1 lacks demethylase activity because of Y292, this change might restore activity to the protein and thus affect its transcriptional activity. Induction of SSA3 in gis1-Y292H cells was the same as that seen for wild type (Fig. 2c) and therefore this mutation had no obvious effect on transcriptional activity.
Epe1, a JmjC-domain containing protein in S. pombe, lacks one conserved Fe (II) binding site and has no detectable histone demethylase activity (Trewick, et al., 2007). And yet the ability of Epe1 to negatively regulate heterochromatin formation and counteract silencing requires the JmjC domain. This indicates that the JmjC domain can regulate gene activity without histone demethylase activity (Zofall & Grewal, 2006, Trewick, et al., 2007). To check whether the JmjC domain of Gis1 can function in the similar manner, we constructed a gis1ΔJmjC mutant, in which the whole JmjC domain (aa 170-324) was deleted. As a control, we also created a deletion lacking the zinc finger DNA binding domain, gis1ΔZnF (Fig. 2a). The induction of SSA3 was totally abolished in the cells expressing gis1ΔZnF, indicating that the zinc finger is necessary for transcriptional activation (Fig. 2c). The Gis1ΔJmjC protein turned out to be very unstable, since the protein level was very low compared to wild-type Gis1 (Fig. 2d, right panel). Nevertheless, gis1ΔJmjC cells still induced the transcription of SSA3, albeit to a lesser extent than wild type (Fig. 2c). Given that the reduction of the protein level of Gis1ΔJmjC was much greater than the reduction in transcriptional activity, it suggests that the Gis1ΔJmjC protein still has normal ability to induce transcription of SSA3.
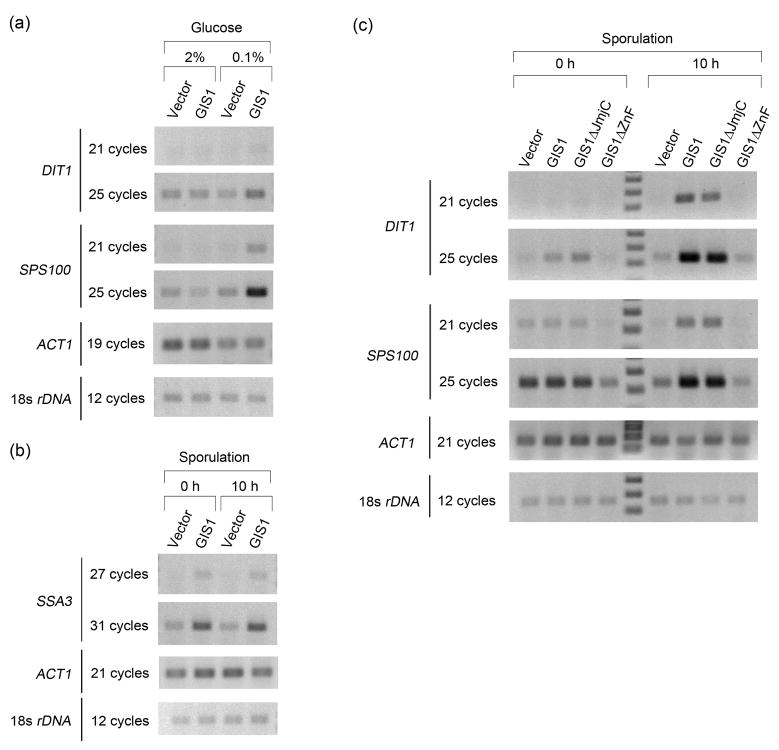
JmjC domain is dispensable for GIS1 induction of DIT1 and SPS100 during sporulation
To assess Gis1-dependent transcriptional activity in different processes, we checked the transcription of the sporulation induced genes, DIT1 and SPS100, in vegetative cells after a shift to 0.1% glucose medium and that of SSA3 during sporulation. As shown in Fig. 3a, in vegetative cells switched to low glucose, DIT1 was induced slightly and there was a large induction of SPS100. Induction of both genes was Gis1-dependent, which suggests that the DIT1 and SPS100 regulatory regions respond to Gis1 in both vegetative cells and during sporulation. To check on the induction of SSA3 during sporulation we compared its expression in pre-sporulation medium (YP acetate) with 10 hours in sporulation medium. GIS1 is responsible for a low basal level of transcription of SSA3 in pre-sporulation medium (Fig. 3b); however, this level is much lower than that seen for glucose-repressed cells during vegetative growth (Fig. 2b and 2d). Importantly, transcription of SSA3 was not increased during sporulation, either with or without GIS1 (Fig. 3b). This implies that Gis1 uses a somewhat different mechanism to induce transcription during nutritional stress versus during sporulation. Given such a difference, it was of interest to investigate whether JmjC domain was required for the transcriptional activation by GIS1 during sporulation.
Fig. 3.
Comparison of induction by Gis1 during glucose limitation and sporulation. (a) DIT1 and SPS100 were induced by Gis1 in haploid cells after glucose limitation. (b) SSA3 was not induced during sporulation of diploid cells. (c) DIT1 and SPS100 induction analysis during sporulation. Plasmids expressing wild type or the indicated GIS1 mutant were integrated into the genome of a homozygous gis1Δ diploid strain. RNAs were extracted from the cells growing in pre-sporulation medium or after 10 hours in sporulation medium and RT-PCR was performed to check the levels of DIT1 and SPS100.
The genes for wild-type GIS1, gis1ΔZnF, gis1ΔJmjC or vector were integrated into the genome of a gis1Δ diploid and transcriptional induction of DIT1 and SPS100 during sporulation was assessed (Fig. 3c). As expected, the induction of both genes was abolished in the cells expressing gis1ΔZnF, reflecting the indispensable role of the DNA binding domain for induction. Interestingly, the level of SPS100 in gis1ΔZnF was even lower than that in gis1Δ cells, which might be explained by a dominant negative effect of Gis1ΔZnF protein. The induction of DIT1, but not SPS100, with Gis1ΔJmjC was slightly lower than with wild-type Gis1. The result for DIT1 was most likely due to the low protein level of Gis1ΔJmjC (Fig. 2d). Taken together, our data suggest that the JmjC domain is not required for the transcriptional activation by Gis1 during sporulation.
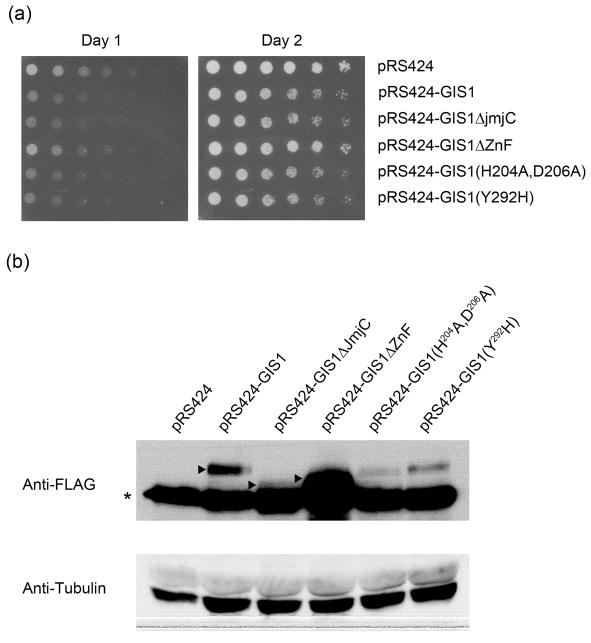
JmjC domain is dispensable for the growth inhibition caused by overexpression of GIS1
GIS1 is negatively regulated by the cAMP-dependent protein kinase in the Ras/cAMP pathway (Pedruzzi, et al., 2000). Overexpression of GIS1 under a strong promoter, like GAL1, inhibits the growth of wild-type cells, and derepresses the cAPK-repressible genes (Pedruzzi, et al., 2000). Since the growth inhibition caused by overexpression of GIS1 reflects the capacity of Gis1 to antagonize cAPK-repressed transcription, it provides an alternative way to check the integrity of GIS1-dependent transcriptional activation. As shown by Fig. 4A, overexpression of Gis1 from an ADH1 promoter on a multicopy plasmid caused a slight growth defect in wild-type cells. No growth defect was observed by overexpressing gis1ΔZnF. This indicates that the zinc finger is necessary for this effect and demonstrates that the growth defect is due to the DNA binding ability of Gis1 and likely to its transcriptional activity. Overexpression of the point mutants, gis1-H204A, gis1-H204A-D206A, gis1-Y292H, or gis1ΔJmjC led to the same growth defect as seen for GIS1. Interestingly, the protein levels of the Gis1 mutants, except gis1ΔZnF, were always lower than that of wild type (Fig. 4b). Since the mutants showed the same toxicity as wild type, it suggests some mechanism exists to antagonize the excess toxicity caused by Gis1.
Fig. 4.
JmjC domain is dispensable for the growth inhibition caused by overexpression of GIS1. (a) Cells were transformed with plasmids overexpressing wild type GIS1 or the indicated mutants. Transformants were resuspended in H2O and 3-fold dilutions spotted onto YPD medium. Growth of cells at 30° was checked after one and two days. (b) Western analysis was performed to check the expression levels of wild type and mutant Gis1 protiens. Arrows indicate the Gis1 proteins and asterisks indicate cross-reacting bands recognized by anti-FLAG antibody.
Discussion
We report here three genes DIT1, SPS100 and SHC1, whose expression is regulated by GIS1 during sporulation. DIT1 is the canonical mid-late gene while SHC1 and SPS100 have been classified as late genes (Law & Segall, 1988, Briza, et al., 1994), (Primig, et al., 2000). Thus Gis1 appears to regulate genes only late in the sporulation process. To our knowledge Gis1 is the first transcriptional activator identified that works specifically at these later times in sporulation. In addition, all three of these target genes are involved in spore wall synthesis. DIT1 is required for the production of a precursor of the dityrosine layer, SHC1 participates in the synthesis of the chitosan layer, and Sps100 is likely a spore wall component (Law & Segall, 1988, Briza, et al., 1994, Sanz, et al., 2002). This is consistent with our previous observations that gis1Δ cells have defects in the chitosan and dityrosine layers (Coluccio, et al., 2004).
The transcriptional level of GIS1 does not change significantly during sporulation (Chu, et al., 1998). This suggests that Gis1 is activated through a post-transcriptional mechanism relatively late in sporulation. During the post-diauxic transition, Gis1 is activated by the Rim15 kinase in the Ras/cAMP pathway (Pedruzzi, et al., 2000). The Ras/cAMP pathway is also linked with triggering of meiosis (Mitchell, 1994), but whether it is still active during the mid-late and late stage of sporulation is unknown. As shown in Fig. 3, DIT1 and SPS100 could be induced by Gis1 during both sporulation and the post-diauxic transition, suggesting that the pathway to activate Gis1 is somewhat similar in both processes. Based on this idea, the Ras/CAMP pathway might contribute to the activation of Gis1 during sporulation. On the other hand, SSA3 is strongly induced by Gis1 during the post-diauxic transition, but is not induced during sporulation. A repressive mechanism might exist at the SSA3 locus during sporulation that prevents Gis1-dependent induction, which is suggested by the low level of SSA3 after 10 hours in sporulation medium (Fig. 3b). The detailed mechanism by which Gis1 is activated during sporulation will be an interesting subject for future investigation.
In this report, we showed that the entire JmjC domain is dispensable for transcriptional activation by Gis1. This indicates that other regions of Gis1 are required for activity. As shown in Fig. 2, the zinc finger is required for transcriptional activation, most likely because that is the domain that binds to DNA of target genes. While this manuscript was in preparation, a recent paper reported that Gis has two transcriptional activation domains, one adjacent to the zinc finger domain and the other a coiled-coiled domain C-terminal to the JmjC domain (Zhang & Oliver, 2010).
Since the JmjC domain is conserved during evolution, it is likely to contribute to other functions of Gis1 besides transcriptional activation. Our data showed that the JmjC domain was important for stability of Gis1. Point mutations in the domain reduced the amount of Gis1 (Fig. 4b), while deletion of the JmjC domain cut the protein level dramatically (Fig. 2d, 4b). A recent report showed that the transcriptional activity of Gis1 is negatively regulated by proteosome-mediated limited proteolysis (Zhang & Oliver, 2010). Thus the JmjC domain might regulate the function of Gis1 by affecting its degradation.
As mentioned above, Gis1 has also been reported to function in repression of two genes, the damage-responsive gene PHR1 (Jang, et al., 1999) and DPP1. The latter gene is normally induced during inositol induction and in stationary phase (Oshiro, et al., 2003). It was reported that induction of a lacZ reporter gene fused to the DPP1 promoter was greatly reduced in a gis1 mutant. Another paper reported that a Gis1 JmjC domain-LexA fusion could repress transcription when recruited to a promoter with LexA binding sites (Tronnersjo, et al., 2007). Thus it seemed possible that the JmjC domain might be required for the repressive activity of Gis1. We checked repression of DPP1 carefully under various growth conditions using RT-PCR and saw no difference between wild-type and gis1Δ cells (data not shown). As mentioned, the previous report (Oshiro, et al., 2003) used a lacZ reporter gene so that may explain the discrepancy with our negative results for DPP1. We also did a microarray experiment comparing wild-type and gis1Δ cells growing exponentially and also after 4 hours in low glucose. We did not observe any genes whose expression increased significantly in gis1Δ cells under either growth condition, and thus could not test if the JmjC domain was necessary for repression.
Supplementary Material
Acknowledgments
We thank Evelyn Prugar and Chia-Lin Wang for help with the experiments and Tim Hughes for information about Gis1 DNA binding sites. The research was supported by NIH grants P01 GM088297 to A.M.N. and R.S. and GM55641 to R.S.
References
- 1.Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badis G, Chan ET, van Bakel H, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balciunas D, Ronne H. Yeast genes GIS1–4: multicopy suppressors of the Gal- phenotype of snf1 mig1 srb8/10/11 cells. Mol Gen Genet. 1999;262:589–599. doi: 10.1007/s004380051121. [DOI] [PubMed] [Google Scholar]
- 5.Briza P, Eckerstorfer M, Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci U S A. 1994;91:4524–4528. doi: 10.1073/pnas.91.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameroni E, Hulo N, Roosen J, Winderickx J, De Virgilio C. The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle. 2004;3:462–468. [PubMed] [Google Scholar]
- 7.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Zang J, Whetstine J, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 11.Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang YK, Wang L, Sancar GB. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol Cell Biol. 1999;19:7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 14.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 15.Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol Cell Biol. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreger-Van Rij NJ. Electron microscopy of germinating ascospores of Saccharomyces cerevisiae. Arch Microbiol. 1978;117:73–77. doi: 10.1007/BF00689354. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamoorthy T, Chen X, Govin J, et al. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 2006;20:2580–2592. doi: 10.1101/gad.1457006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law DT, Segall J. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol Cell Biol. 1988;8:912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majello B, DeLuca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. Journal of Biological Chemistry. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshiro J, Han GS, Iwanyshyn WM, Conover K, Carman GM. Regulation of the yeast DPP1-encoded diacylglycerol pyrophosphate phosphatase by transcription factor Gis1p. J Biol Chem. 2003;278:31495–31503. doi: 10.1074/jbc.M305452200. [DOI] [PubMed] [Google Scholar]
- 23.Pammer M, Briza P, Ellinger A, Schuster T, Stucka R, Feldmann H, Breitenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992;8:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- 24.Pedruzzi I, Burckert N, Egger P, De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000;19:2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primig M, Williams RM, Winzeler EA, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 26.Roosen J, Engelen K, Marchal K, et al. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol. 2005;55:862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanz M, Trilla JA, Duran A, Roncero C. Control of chitin synthesis through Shc1p, a functional homologue of Chs4p specifically induced during sporulation. Mol Microbiol. 2002;43:1183–1195. doi: 10.1046/j.1365-2958.2002.02812.x. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smits GJ, van den Ende H, Klis FM. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- 30.Tahiliani M, Mei P, Fang R, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 31.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tronnersjo S, Hanefalk C, Balciunas D, Hu GZ, Nordberg N, Muren E, Ronne H. The jmjN and jmjC domains of the yeast zinc finger protein Gis1 interact with 19 proteins involved in transcription, sumoylation and DNA repair. Mol Genet Genomics. 2007;277:57–70. doi: 10.1007/s00438-006-0171-3. [DOI] [PubMed] [Google Scholar]
- 33.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 34.Tu S, Bulloch EM, Yang L, et al. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vershon AK, Pierce M. Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- 36.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Oliver SG. The transcription activity of Gis1 is negatively modulated by proteasome-mediated limited proteolysis. J Biol Chem. 2010;285:6465–6476. doi: 10.1074/jbc.M109.073288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.