Abstract
In mammalian cells, activation of oncogenes usually triggers innate tumor-suppressing defense mechanisms including apoptosis and senescence, which are compromised by additional mutations before cancers are developed. The miR-17-92 gene cluster, a polycistron encoding 6 microRNAs (miRNAs), is frequently overexpressed in human cancers, and has been shown to promote several aspects of oncogenic transformation, including evasion of apoptosis. In the current study, we demonstrate a new role of miR-17-92 in inhibiting oncogenic ras-induced senescence. Further dissection of the miRNA components in this cluster reveals that the miR-17/miR-20a seed family accounts for this anti-senescence activity. miR-17 and miR-20a are both necessary and sufficient for conferring resistance to ras-induced senescence, by directly targeting p21WAF1, a key effector of senescence. By contrast, these components are not essential for the ability of miR-17-92 to evade Myc-induced apoptosis. Moreover, disruption of senescence by miR-17-92 or its miR-17/20a components leads to enhanced oncogenic transformation by activated ras in primary human cells. Taken together with previous reports that miR-17-92 inhibits apoptosis by suppressing Pten via the miR-19 components, our results indicate that this miRNA cluster promotes tumorigenesis by antagonizing both tumor-suppressing mechanisms, apoptosis and senescence, through the activities of different miRNA components encoded in this cluster.
Keywords: oncogene-induced senescence, cancer, ras, microRNA, miR-17-92, miR-17, miR-20a, p21WAF1
Introduction
MicroRNA (miRNA) is a class of evolutionarily conserved non-coding RNAs that suppress the expression of protein-encoding genes (1, 2). Recently, miRNA has been implicated in human cancer (3–6). The miR-17-92 cluster is a polycistron encoding 6 mature miRNAs, belonging to 4 seed families, the miR-17 family (miR-17 and miR-20a), the miR-18 family (miR-18a), the miR-19 family (miR-19a and miR-19b-1), and the miR-92 family (miR-92a-1) (2, 7). Overexpression of miR-17-92 has been detected in various types of human cancer (8, 9). When overexpressed, miR-17-92 promotes cell cycle progression and proliferation (10), inhibits apoptosis (11, 12), and induces tumor angiogenesis (13). Furthermore, transgenic mice with moderate overexpression of miR-17-92 in lymphocytes develop a lymphoproliferative disease (14), whereas deletion of this cluster in mouse disrupts normal B-cell development as a result of premature cell death (12). In addition, miR-17-92 accelerates lymphomagenesis from mouse B cells expressing c-Myc by counteracting c-Myc-induced apoptosis (11). Two recent studies have identified miR-19 as the key oncogenic component of miR-17-92, demonstrating that miR-19 inhibits c-Myc-induced apoptosis and promotes c-Myc-mediated lymphomagenesis by repressing the expression of the Pten tumor suppressor gene, (15, 16).
In contrast to the anti-apoptotic effect of miR-17-92, the role of this cluster in other aspects of tumorigenesis is less well defined. Besides apoptosis, oncogene-induced senescence is another tumor suppressing defense mechanism that needs to be compromised during cancer development. Instead of tumorigenic phenotypes, activation of certain oncogenes such as ras in early-passage primary cells causes a permanent proliferative arrest known as premature senescence (17–19). Induction of senescence by oncogenic ras is mediated by the p38 MAPK (18, 20), and is accompanied by upregulation of inhibitors of cell proliferation, including p16INK4A, p53, p21WAF1, and p14/p19ARF (17, 21, 22). Recent studies show that oncogene-induced senescence occurs in vivo and provides a bona fide barrier to tumorigenesis (23–28).
In the current study, we have demonstrated a new role of miR-17-92 in inhibiting oncogene-induced senescence. Further dissection of miR-17-92 reveals that the miR-17/miR-20a seed family accounts for this anti-senescence activity, by directly targeting p21WAF1; however, these components are not essential for the anti-apoptotic activity of this cluster. Moreover, disruption of senescence by miR-17-92 leads to enhanced oncogenic transformation mediated by activated ras. Therefore, miR-17-92 promotes tumorigenesis by antagonizing both tumor-suppressing mechanisms, apoptosis and oncogene-induced senescence, through the activities of different miRNA seed families encoded by this cluster.
Materials and Methods
Cells culture
BJ and WI38 primary human fibroblasts (ATCC) were maintained in Minimum Essential medium (MEM) supplemented with 10% FCS, non-essential amino acids, glutamine and antibiotics. Grip Tite 293 and 293T (Invitrogen) were grown in Dulbecco’s modified Eagle medium (DMEM) with 10% FCS, sodium pyruvate, glutamine and antibiotics. These cells were authenticated by the suppliers (ATCC and Invitorgen) based on viability, recovery, growth, morphology and isoenzymology, and had been passaged in our laboratory for fewer than 6 months after resuscitation.
Plasmids
The expression vectors for miRNAs and miRNA clusters were constructed by cloning a single copy or multiple (2 or 4) copies of their precursor sequences, together with a short upstream and a short downstream flanking region, into pLV-EF1α-rPuro lentiviral expression vectors (Biosettia) or pWAYP. Maps and sequences for these constructs are available upon request.
To construct the LacZ-p21WAF13′UTR reporter, the p21WAF1 3′-UTR sequence was amplified by PCR from the genomic DNA of BJ cells using primers p21-UTR-f (5′-GATCCACCGTCTAGATAATCCGCCCACAGGAAGCCTGCA-3′) and p21-UTR-(5′TGTACATCAAAGCTTTAAAGTCACTAAGAATCATTTATTGAGCACCTGCT-3′), and cloned into pCMV-LacZ (Biosettia). The shRNA constructs for p21WAF1, Pten and Bim were designed based on the single oligonucleotide RNAi technology (Biosettia). The DNA oligonucleotide for each shRNA was cloned into the lentiviral pLV-H1-EF1α-puro vector, following the manufacturer’s protocol, and verified by DNA sequencing. The sequences of oligos used to generate shRNAs are: sh-p21-1, 5′-AAAAGGCTGATCTTCTCCAAGAGTTGGATCCAACTCTTGGAGAAGATCAGCC-3′; sh-p21-2, 5′-AAAAGCCTCTGGCATTAGAATTATTGGATCCAATAATTCTAATGCCAGAGGC-3′; sh-p21-3, 5′-AAAAGGAGGCACTGAAGTGCTTATTGGATCCAATAAGCACTTCAGTGCCTCC-3′; sh-Pten-1162, 5′-AAAAGCGTATACAGGAACAATATTTGGATCCAAATATTGTTCCTGTATACGC-3′; sh-Pten-1364, 5′-AAAAGCTAAGTGAAGATGACAATTTGGATCCAAATTGTCATCTTCACTTAGC-3′; sh-Bim-655, 5′-AAAAGCCTTCAACCACTATCTCATTGGATCCAATGAGATAGTGGTTGAAGGC-3′; sh-Bim-754, 5′-AAAAGGAGACGAGTTTAACGCTTTTGGATCCAAAAGCGTTAAACTCGTCTCC-3′. A scramble sequence (5′-AAAAGCTACACTATCGAGCAATTTTGGATCCAAAATTGCTCGATAGTGTAGC-3′) was used as a negative control.
Retrovirus- and Lentivirus-Based Gene Transduction
Recombinant retroviruses were packaged and transduced into cells as previously described (29). Recombinant lentiviruses were packaged in 293T cells in the presence of helper plasmids (pMDLg, pRSV-REV and pVSV-G) using Lipofectamine 2000 (Invitrogen). 1 × 106 cells were seeded in a 10-cm plate, grown for overnight, infected with at a MOI of 20 in fresh medium containing 8 μg/ml polybrene, and spun for 1 hour at 1600–1800 rpm. Transduced cells were purified with 120 (BJ) or 50 (WI38) μg/ml of hygromycin B, 600 μg/ml of G418, 1.2 μg/ml of Puromycin, or 5 μg/ml of blasticidin.
Western Blot Analysis
Western blot analysis was performed with lysates prepared 7–10 days after transduction of Ras or MKK3/6E from subconfluent cells as described (18). Primary antibodies were purchased from Sigma (actin), Santa Cruz (Ras C-20 and p21WAF1 C-19), Cell Signaling (Pten 1038G6), and Epitomics (Bim Y36). Signals were detected using enhanced chemiluminescence and captured by the FluorChem 8900 Imaging System (AlphaInnotech).
Analysis of Senescence
Senescence in cells was analyzed by measuring the rate of cell proliferation and activity of SA-β-galactosidase, as described previously (18). To quantify SA-β-gal positive cells, at least 200 cells were counted in random fields in each of the duplicated or triplicated wells.
Tumorigenesis Assays
BJ were transduced with appropriated oncogenes and miRNAs at PD22. For soft agar colony formation assay, 2 × 104 of BJ cells at PD26 were resuspended in a medium containing 0.3 % low-melting-point agarose and plated onto a solidified bottom layer medium containing 0.5 % agarose in triplicates in 6-cm plates. Colonies were photographed after 2–4 weeks, stained with 0.02 % Giemsa, and counted. For tumor formation assays, 2 × 106 of BJ cells at PD28 were injected subcutaneously into the flanks of 4-week-old female HSD:athymic nude mice in 100 μl of PBS. Tumor growth was monitored weekly for 10 weeks.
LacZ- p21WAF13′-UTR Reporter Assay
For the LacZ reporter assay, 4 × 103 of Grip Tite 293 cells were seeded the day before transfection, and transfected with 180 ng of miRNA-expressing vector, 20 ng of the LacZ reporter and 10 ng of a CMV-Luciferase reporter (Promega) in 96-well plates in triplicates using Lipofectamine 2000 (Invitrogen). Cells were lysed 24 h after transfection in 100 μl/well of Luciferase Assay Lysis buffer (25 mM Tris-HCl, pH 8.0, 0.1 mM EDTA pH8.0, 10 % glycerol, and 0.1 % Triton X-100), and frozen at −80 C for at least 1 hour. After thawed at room temperature, 10μl of each lysate was mixed with 100μl of Working Solution containing 1.1 mM 3-carboxy-umbelliferyl β-D-galactopyranoside (CUG, Sigma) in 0.1 M sodium phosphate, pH 7.3, 1 mM MgCl2, and 45 mM β-mercaptoethanol in a 96-well black-wall plate. After incubation at room temperature in dark for 30 minutes, 50μl/well of Stop Solution (0.2 M Na2CO3) was added. Signals (390/448 nm) were measured. Firefly luciferase activity was measured with Luciferase Assay System (Promega), and used as an internal normalization control.
miRNA Expression Level Assays
Expression level of the mature miRNA was measured by TaqMan MicroRNA Assays kit (Applied Biosystems) following the manufacturer’s protocol. The RUN48 primers were used as a normalization control.
Myc-induced apoptosis
BJ cells seeded in 6-well plates at 2 × 105 cells/well were transduced first with miR-17-92 or its components or a LacZ control and selected with puromycin, and then with human c-Myc or GFP and selected with blasticidine. Two days post infection, cells were grown in MEM containing 0.5 % of FCS for 2, or 6 days, after which both adherent cells and cells in suspension were harvested, washed with PBS, resuspended and incubated in PBS containing 20μg/ml of propidium iodide at room temperature for 15 minutes, and analyzed by flow cytometry.
Results
The mir-17-92 cluster disrupts oncogenic ras-induced senescence
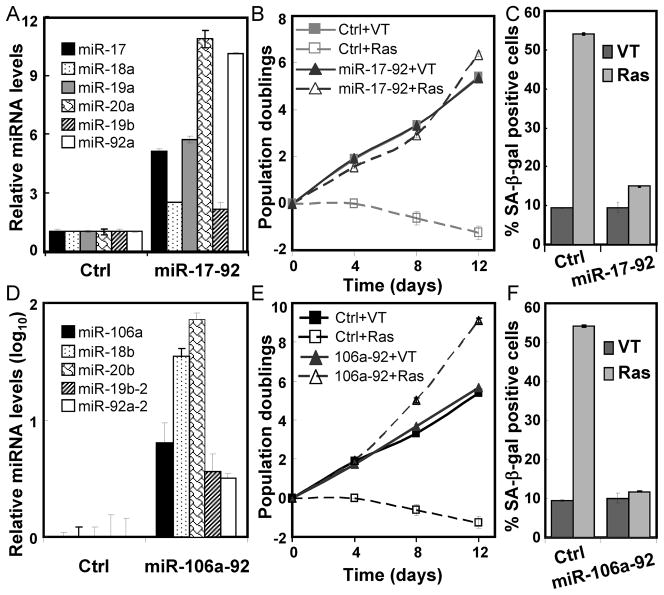
The involvement of miR-17-92 in cancer prompted us to explore whether it inhibits oncogene-induced senescence (30). When the miR-17-92 cluster was transduced into BJ primary human fibroblasts via a lentivirus, increased expression levels of all 6 mature miRNAs were detected, ranging from 2- to 11-fold over the endogenous levels (Fig. 1A). Transduction of an activated ras allele, Ha-rasV12, caused a proliferative arrest (Fig. 1B) and increased expression of senescence markers senescence-associated (SA)-β-galactosidase (Fig. 1C) and MCL1 (24) (Fig. S1) in control BJ cells, indicating senescence induction by ras. In contrast, BJ cells expression miR-17-92 were resistant to ras-induced proliferative arrest and expression of SA-β-gal and MCL1 (Fig. 1B–C, S1). Thus, the miR-17-92 cluster can disrupt oncogenic ras-induced senescence when ectopically expressed in primary BJ fibroblasts. Increased expression of miR-17-92 also compromised oncogenic ras-induced senescence in WI38 lung fibroblasts (Fig. S2A–B), suggesting that the anti-senescence activity of the miR-17-92 cluster is not restricted in BJ foreskin fibroblasts, but operates in primary fibroblasts derived from multiple origins.
Figure 1. The miR-17-92 and miR-106a-363 miRNA clusters disrupt oncogenic ras-induced senescence.
(A) The expression level of each miRNA encoded by miR-17-92 in BJ cells transduced with a control (Ctrl) or miR-17-92-expressing (miR-17-92) lentivirus. The signals obtained with miRNA-specific primers were normalized to that with the RUN48 primers. The relative miRNA level was calculated by dividing the miRNA level in miR-17-92-transduced cells by that in the control cells.
(B) The population doublings of BJ cells transduced with miR-17-92 or vector (Ctrl) and Ha-RasV12 (Ras) or vector (VT) were monitored for 12 day starting on day-5 post ras transduction.
(C) The percentage of SA-β-gal positive cells in BJ cell populations described in (B) on day-12 post ras transduction.
(D) The expression level (in log10 scale) of each miRNA encoded by miR-106a-92 in BJ cells transduced with a control (Ctrl) or miR-106a-92-expressing (miR-106a-92) lentivirus. The signals obtained with miRNA-specific primers were normalized to that with the RUN48 primers. The relative miRNA level was calculated by dividing the miRNA level in miR-106a-92-transduced cells by that in the control cells.
(E) The population doublings of BJ cells transduced with miR-106a-92 or vector (Ctrl) and Ha-RasV12 (Ras) or vector (VT) were monitored for 12 day starting on day-5 post ras transduction.
(F) The percentage of SA-β-gal positive cells in BJ cell populations described in (E) on day-12 post ras transduction.
(A–F) Values are mean ± SD for triplicates.
Members of the miR-17-92 cluster have homologues in 2 other miRNA clusters, one of which is the miR-106a-363 cluster on the X chromosome (6). The miR-106a-363 cluster encodes miR-106a and miR-20b, miR-18b, miR-19b-2, and miR-92a belonging to the same miRNA family as miR-17 and miR-20a, miR-18a, miR-19a and miR-19b-1, and miR-92a-1 on the miR-17-92 cluster, respectively. Expression of the major part of the miR-106a-363 region (miR-106a-92) containing miRNAs homologous to those in the miR-17-92 cluster (Fig. 1D) also rendered BJ cells refractory to oncogenic ras-induced senescence (Fig. 1E, 1F). Therefore, these two homologous miRNA clusters can both disrupt oncogene-induced senescence when expressed in primary human cells.
The miR-17/20a components are required for the anti-senescence activity of miR-17-92
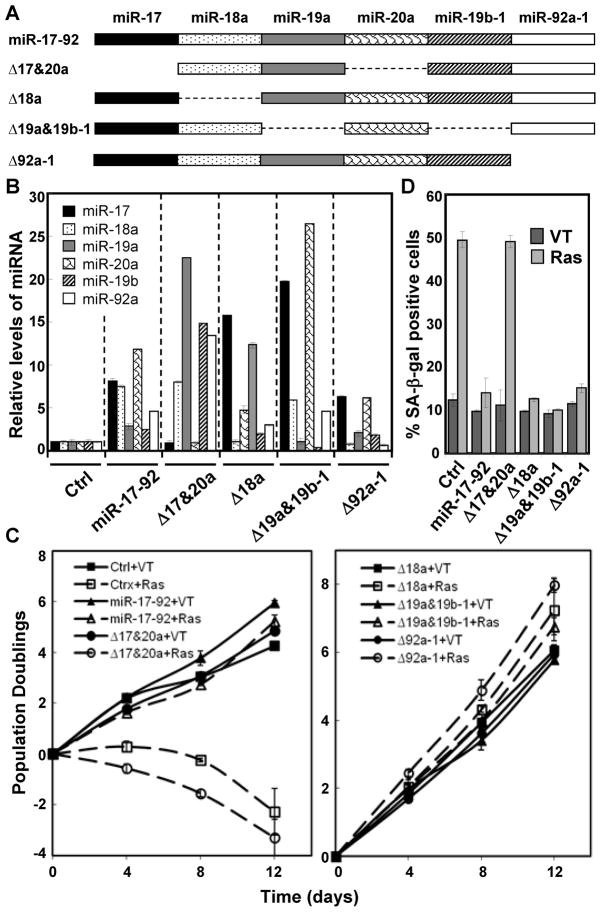
The miR-17-92 cluster encodes 6 miRNAs belonging to 4 different seed families, each with a unique sequence at positions 2–7. To dissect the contribution of these miRNA components to the anti-senescence activity, miR-17-92 mutants were constructed, each containing deletion of a single seed family (Fig. 2A). When transduced into BJ cells, these mutants displayed expected expression patterns for the miRNA components, with abrogation of the expression of the mutated, but not the other, miRNAs (Fig. 2B). Deletion of the miR-17/miR-20a (Δ17&20a) family abolished the ability of miR-17-92 to confer resistance to ras-induced senescence (Fig. 2C, left panel, 2D and S1). On the other hand, miR-17-92 mutants with deletion of the other seed families (Δ18a, Δ19a&19b-1 orΔ92a-1) were fully capable of blocking oncogenic ras-induced senescence (Fig. 2C, right panel and 2D). Similar observations was made in WI38 cells (Fig. S2A–B). These data demonstrate that the miR-17/miR-20a seed family is essential for the miR-17-92 cluster to prevent ras-induced senescence, while the other seed families are dispensable.
Figure 2. The miR-17 and miR-20a components are essential for miR-17-92-mediated resistance to oncogenic ras-induced senescence.
(A) Schematic diagrams of miR-17-92 deletion mutants used in the study.
(B) The expression level of each miRNA encoded by miR-17-92 in BJ cells transduced with vector (Ctrl), or wild type or indicated mutants of miR-17-92. The signals obtained with miRNA-specific primers were normalized to that with the RUN48 primers. The relative miRNA level was calculated by dividing the miRNA level in miR-17-92-transduced cells by that in the control cells.
(C) The population doublings of BJ cells transduced with wild type or indicated mutants of miR-17-92 or vector (Ctrl) and Ha-RasV12 (Ras) or vector (VT) were monitored for 12 day starting on day-5 post ras transduction.
(D) The percentage of SA-β-gal positive cells in the BJ cell population described in (C) on day-12 post ras transduction.
(B–D) Values are mean ± SD for triplicates.
We previously demonstrated that oncogenic ras-induced senescence is mediated by the p38 MAPK, and that premature senescence can be induced by constitutively active forms of MKK3 (MKK3E) and MKK6 (MKK6E), the upstream kinases of p38 (18). The miR-17-92 cluster also conferred resistance to MKK3E-induced senescence in BJ cells (Fig. S2C–D), suggesting that these miRNAs disrupted senescence by targeting a signaling step downstream of p38.
A requirement of the miRNAs belonging to the same family as miR-17/miR-20a was also observed for the anti-senescence activity of the homologous miR-106a-363 cluster. Disruption of ras-induced senescence by miR-106a-92 in the absence of miR-363 (Fig. 1E, 1F, S3) indicates that miR-363 is dispensable. In addition, deletion of miR-106a and miR-20b, but not miR-18b, miR-19b-2 or miR-92a, abolished miR-106a-92-conferred resistance to senescence (Fig. S3), suggesting that miR-106a/20b are the only components essential for the senescence bypass by the miR-106a-363 cluster.
miR-17 and miR-20a are dispensable for the anti-apoptotic activity of the miR-17-92 cluster
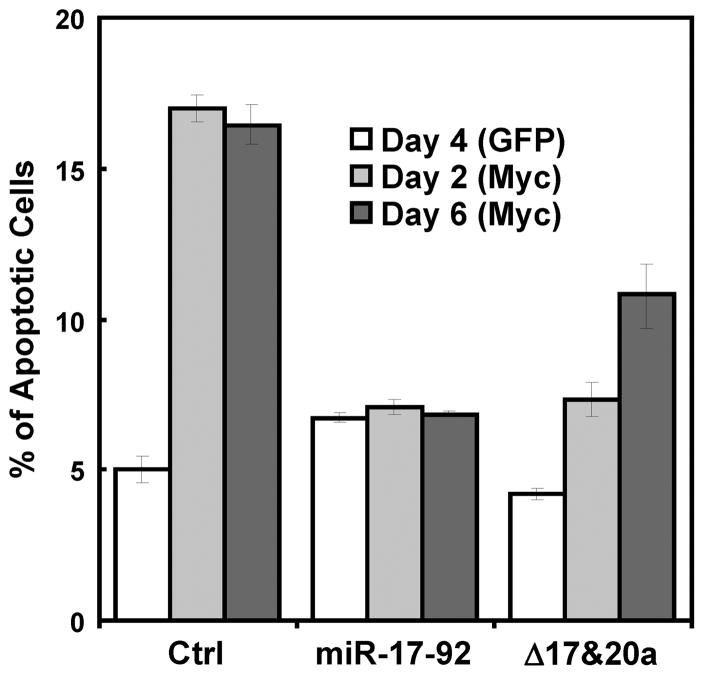
To further dissect the oncogenic function of miR-17-92, we determined the requirement of the miR-17/20a components for the anti-apoptotic activity. It has been shown that the miR-19 components mediate the evasion of Myc-induced apoptosis in B lymphocytes and acceleration of Myc-induced lymphomagenesis (15, 16). To avoid the cell type-specific effects, we analyzed the anti-apoptotic activity of miR-17-92 in BJ fibroblasts, the same cell type in which its anti-senescence activity was dissected. Myc was reported to induce apoptosis in growth factor-deprived fibroblasts (31). We thus examined whether the miR-17-92 cluster also counteracted Myc-induced apoptosis in this setting. Compared to the BJ with a GFP control, cells transduced with the human c-Myc oncogene displayed an increased rate of cell death after 2–6 days of serum starvation, as measured by the percentage of cells that failed to exclude propidium iodide (Fig. 3). By contrast, in BJ cells transduced with miR-17-92 or mutant miR-17-92 lacking the miR-17 and miR-20a components, Myc-dependent apoptosis was greatly reduced compared to the cells transduced with a vector control (Fig. 3). Therefore, like in mouse B-lymphocytes, the miR-17-92 cluster disrupts Myc-induced apoptosis in growth factor-deprived primary human fibroblasts, and this anti-apoptotic function of miR-17-92 does not absolutely require the miR-17/20a components. This finding, in combination with the previous reports that miR-17-92 blocks Myc-induced apoptosis through miR-19 in B-cells (15, 16), indicate that the anti-senescence and anti-apoptotic activities of miR-17-92 rely on separate components encoded by this cluster.
Figure 3.
The miR-17/20a components are not essential for the ability of miR-17-92 to disrupt Myc-induced apoptosis. BJ cells were transduced miR-17-92, mutant miR-17-92 lacking miR-17/20a (Δ17&20a) or vector (Ctrl), and c-Myc (Myc) or GFP control (GFP), and serum-starved for 2 or 6 days. The percentage of apoptotic cells were determined by flow cytometry after staining with propidium iodide. Values are mean ± SD for triplicates.
miR-17 and miR-20a are sufficient for the disruption of oncogenic ras-induced senescence
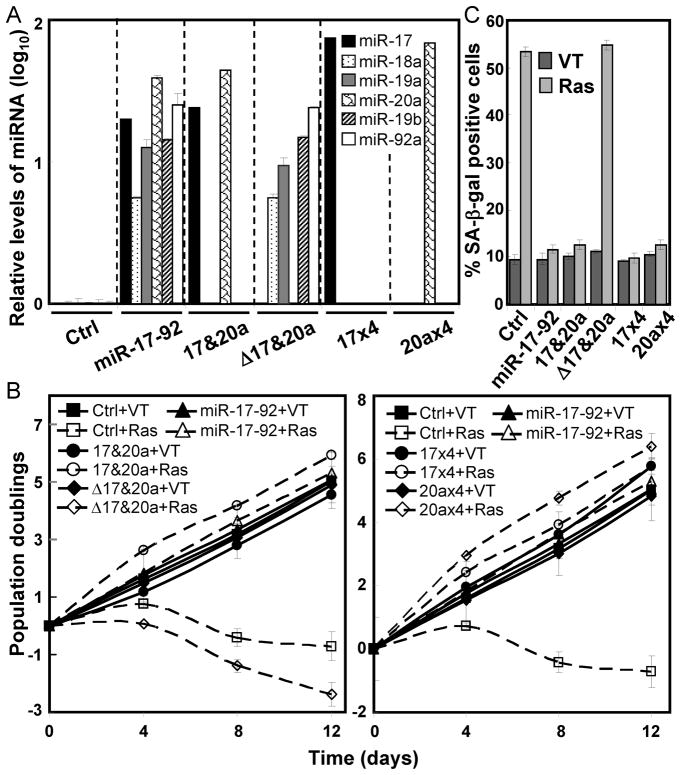
We next investigated whether the miR-17/miR-20a family by itself was sufficient for overcoming oncogenic ras-induced senescence. In BJ cells transduced with miR-17 and miR-20a only (Fig. 4A), oncogenic ras failed to induce proliferative arrest (Fig. 4B) or accumulation of SA-β-gal (Fig. 4C), indicating miR-17 and miR-20a are sufficient to confer the anti-senescence activity of miR-17-92. In contrast, none of the other miRNAs in the cluster was sufficient for bypassing senescence, since cells transduced with the Δ17&20a mutant, which mediated the expression of miR-18a, miR-19a, miR-19b-1 and miR-92a-1 at levels similar to the wild type miR-17-92 (Fig. 4A), became senescent upon ras activation (Fig. 4B, left panel, 4C and S1).
Figure 4. The miR-17/20a components of miR17-92 are sufficient to confer resistance to oncogenic ras-induced senescence.
(A) The expression level (in log10 scale) of each miRNA encoded by miR-17-92 in BJ cells transduced with vector (Ctrl), wild type miR-17-92, miR-17 and miR-20a (17&20a), a mutant miR-17-92 lacking miR-17 and miR-20a (Δ17&20a), or 4 tandem copies of miR-17 (17×4) or miR-20a (20a×4). The signals obtained with miRNA-specific primers were normalized to that with the RUN48 primers. The relative miRNA level was calculated by dividing the miRNA level in miRNA-transduced cells by that in the control cells.
(B) The population doublings of BJ cells transduced with vector (Ctrl), wild type miR-17-92, miR-17 and miR-20a (17&20a), a mutant miR-17-92 lacking miR-17 and miR-20a (Δ17&20a) or 4 tandem copies of miR-17 (17×4) or miR-20a (20a×4), and Ha-RasV12 (Ras) or vector (VT) were monitored for 12 day starting on day-5 post ras transduction.
(C) The percentage of SA-β-gal positive cells in BJ cell populations described in (B) on day-12 post ras transduction. (A–C) Values are mean ± SD for triplicates.
miR-17 and miR-20a share identical seed sequences and high overall sequence homology, and thus likely repress the expression of the same genes and have similar biological functions. Indeed, BJ cells transduced with lentiviruses encoding 4 copies of miR-17 (17×4) or miR-20a (20a×4) were resistant to ras-induced senescence (Fig. 4B, right panel, 4C and S1), indicating that miR-17 or miR-20a alone is sufficient to overcome senescence. We further examined the dosage effect of miR-17 and miR-20a on senescence. Lentiviral vectors were constructed that encode either a single copy or 2 or 4 tandem copies of miR-17 or miR-20a. These constructs resulted in copy number-dependent expression of miR-17 and miR-20a when transduced into BJ cells (Fig. S4A, S4B). The expression levels derived from the constructs containing 4 copies of the miRNAs were comparable to the levels mediated by the wild type miR-17-92 cluster, whereas those from the 1- and 2-copy vectors were consistently lower. Corresponding to the expression levels, only the 4-copy vectors conferred complete resistance to ras-induced senescence as the miR-17-92 cluster (Fig. S4C–E). Two tandem copies of miR-17 or miR-20a partially abrogated ras-induced senescence; and a single copy of these miRNAs did not block senescence at all. These results suggest that miR-17 and miR-20a, when expressed alone, can prevent senescence induction in a dosage-dependent manner.
miR-17 and miR-20a alone or in combination, but not the other miRNAs in miR-17-92, were also sufficient to block ras-induced senescence in WI38 human lung fibroblasts (Fig. S2A–B). In addition, miR-17 and miR-20a prevented MKK3E-induced senescence (Fig. S2C–D). These results confirm that the miR-17/20a components mediate the anti-senescence function in multiple cell strains, by targeting signaling downstream of p38.
Taken together, our results demonstrate that miR-17 and miR-20a are both necessary and sufficient for disrupting oncogenic ras-induced senescence, and thus are the key components mediating the anti-senescence activity of miR-17-92. While this manuscript was in preparation, an independent study was published, in which miR-17 and miR-20a were identified from a screen designed to isolate miRNAs that could compromise oncogenic ras-induced senescence in human mammary epithelial cells (HMECs) (32). This finding reinforces our conclusion that the anti-senescence effect of the miR-17-92 cluster is attributed to the miR-17/20a components.
miR-17 and miR-20a overcome ras-induced senescence at least partly by inhibiting the induction of p21WAF1 expression
To determine the mechanism underlying the anti-senescence activity of miR-17-92, we analyzed the relevance of the known miR-17-92 targets in oncogene-induced senescence. miR-17-92 represses the expression of multiple direct targets, including the E2F transcription factors, Pten, Bim and p21WAF1 (12, 14, 33–38). Among these targets, the suppression of Pten has been specifically attributed to the miR-19 components (15, 16), while that of E2F to miR-17 and miR-20a (34). In addition, it has been demonstrated that expression of p21 WAF1 can be repressed by the miR-17/20a family (32, 37, 38), although it is unclear whether the other components of miR-17-92 also contribute to p21WAF1 suppression.
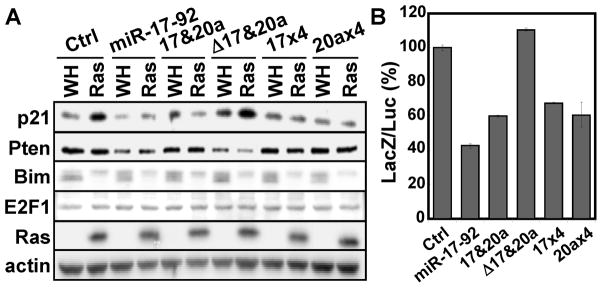
Since miR-17 and miR-20a are the key mediators of the anti-senescence activity of miR-17-92, we focused on the targets of this miRNA family. Inactivation of E2F promotes, rather than blocks, oncogene-induced senescence (22). Therefore, we reason that suppression of p21WAF1, an important senescence mediator (19), by miR-17/20a may contribute to the anti-senescence activity of the miR-17-92 cluster. Indeed, oncogenic ras induced p21WAF1 expression, and this induction was greatly reduced in cells expressing the miR-17-92 cluster, miR-17 and miR-20a, miR-17 alone, or miR-20a alone (Fig. 5A). The ability of miR-17-92 to disrupt p21WAF1 induction depended on the miR-17/20a components, as miR-17-92 containing mutations in miR-17 and miR-20a (Δ17&20a) failed to reduce p21WAF1 expression in BJ cells with activated ras (Fig. 5A). Therefore, miR-17 and miR-20a are the miR-17-92 components that are both necessary and sufficient for mediating the abrogation of ras-induced p21WAF1 expression.
Figure 5. miR-17-92 directly suppresses the expression of p21WAF1 through the miR-17/20a components.
(A) Western blot analysis of BJ cells transduced with vector (Ctrl), wild type miR-17-92, miR-17 and miR-20a (17&20a), a mutant miR-17-92 lacking miR-17 and miR-20a (Δ17&20a), or 4 tandem copies of miR-17 (17×4) or miR-20a (20a×4), and Ha-RasV12 (Ras) or vector (WH) on day-8 post ras transduction.
(B) Activity of the LacZ reporter containing a 3- UTR of p21WAF1 in the presence of vector (Ctrl), wild type miR-17-92, miR-17 and miR-20a (17&20a), a mutant miR-17-92 lacking miR-17 and miR-20a (Δ17&20a), or 4 tandem copies of miR-17 (17×4) or miR-20a (20a×4). The LacZ activity in each sample was normalized to that of a luciferase internal control reporter. The percentage of suppression was calculated by dividing the LacZ activity in miRNA-transduced cells by that in the control cells. Values are mean ± SD for triplicates.
It has been shown that miRNAs belonging to the miR-17/20a family (miR-106a, -106b, -20b, and -17-5) suppress the expression of p21WAF1 by directly binding to its 3′-UTR, which contains 2 sites with sequences complementary to the seed region of miR-17/20a (32, 37, 38). To confirm that the miR-17-92 cluster directly target p21WAF1 through miR-17 and miR-20a, we analyzed a LacZ reporter fused to human p21WAF1 3′-UTR (Fig. 5B). The reporter expression was significantly repressed by miR-17-92, miR-17 and miR-20a, miR-17 alone, or miR-20a alone, but not by the Δ17&20a mutant. Therefore, miR-17 and miR-20a, but not any of the other components, mediate the p21WAF1-inhibitory activity of the miR-17-92 cluster by directly targeting the 3′-UTR of p21WAF1. Consistent with the importance of p21WAF1 in senescence (39, 40), silencing of p21WAF1 expression by shRNA led to disruption of ras-induced senescence (Fig. S5). This finding further confirms the contribution of the miR-17/20a-mediated suppression of p21WAF1 in the anti-senescence activity of the miR-17-92 cluster.
In contrast to p21WAF1, the other known targets of miR-17-92 showed different expression patterns in response to various miR-17-92 constructs (Fig, 5A). Consistent with previous reports (15, 16), Pten expression was suppressed only by the miR-17-92 constructs containing functional miR-19 (wild type andΔ17&20a), but not by miR-17 and/or miR-20a. Despite previous findings that Bim and E2F1 were miR-17-92 targets (12, 14, 35), we detected no significant suppression of these proteins by any miR-17-92 constructs in our system, possibly due to different experimental conditions. Thus, Pten, Bim and E2F1 are not specific targets of miR-17 or miR-20a under these conditions. Furthermore, silencing of Pten or Bim expression by shRNA (Fig. S6A) failed to block ras-induced senescence (Fig. S6B, S6C). These data further demonstrate that the anti-senescence function of the miR-17-92 cluster does not involve other miR-17-92 targets such as Pten, Bim and E2F1, but specifically relies on miR-17 and miR-20a that suppress the induction of p21WAF1 by oncogenic ras.
The miR-17-92 cluster and its miR-17/20a components enhance oncogenic ras-mediated cellular transformation
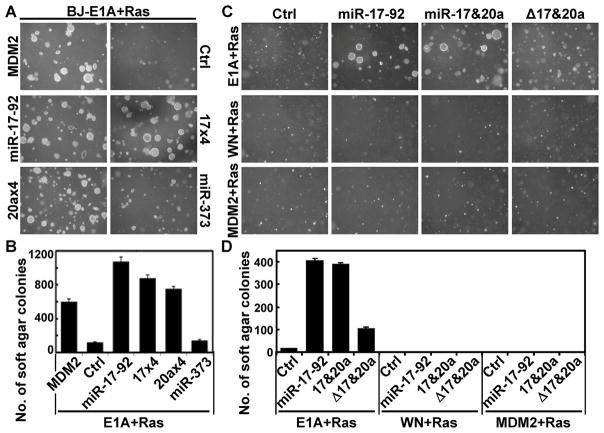
Oncogene-induced senescence is a tumor-suppressing mechanism that needs to be disrupted during cancer development (23, 41). We thus analyzed the contribution of miR-17-92-mediated senescence bypass to oncogenic transformation. Primary human cells, such as BJ fibroblasts, can be fully transformed by the combination of an adenovirus-encoded oncogene E1A, a cellular oncogene MDM2 that inhibits p53, and oncogenic ras (28, 42). Since miR-17-92 suppresses the induction of p21WAF1, a key effector of the p53 function, we tested whether it could replace MDM2 in this human cell transformation model. As expected, BJ cells transduced with E1A, MDM2 and Ha-RasV12 formed colonies in semi-solid soft agar medium (Fig. 6A, 6B), and developed tumors when injected subcutaneously into nude mice (Table 1). The combination of E1A, miR-17-92, and Ha-RasV12 also resulted in efficient anchorage-independent growth in soft agar and tumor development in nude mice, while E1A, Ha-RasV12 and the vector control for miRNA failed to do so (Fig. 6A, 6B, 6C, 6D and Table 1). Therefore, miR-17-92 is able to replace the p53-inhibitory function of MDM2, and promote tumorigenesis in cooperation with E1A and oncogenic ras.
Figure 6. miR-17-92 and its miR-17/20a components promote anchorage-independent growth in cooperation with E1A and activated ras.
(A) Representative fields of soft-agar colonies formed by BJ cells transduced with E1A, Ha-RasV12, and MDM2, miR-17-92, 4 tandem copies of miR-17 (17×4) or miR-20a (20a×4), miR-373 or vector (Ctrl), 4 weeks after seeding in 6-cm plates.
(B) Quantification of the number of colonies in the experiment described in (A), after staining with Giemsa.
(C) Representative fields of soft-agar colonies formed by BJ cells transduced with indicated combinations of oncogenes and miRNA 4 weeks after seeding in 6-cm plates. Vector (Ctrl), wild type miR-17-92 (miR-17-92), miR-17 and miR-20a (17&20a), or a mutant miR-17-92 lacking miR-17 and miR-20a (Δ17&20a) were transduced into BJ cells expressing E1A and Ha-RasV12, vector (WN) and Ha-RasV12, or MDM2 and Ha-RasV12.
(D) Quantification of the number of colonies in the experiment described in (C), after staining with Giemsa.
(B, C) Values are mean±SD for triplicates.
Table 1.
miR-17-92 and its miR-17/20a components cooperate with E1A and activated ras to promote formation of subcutaneous tumors in nude mice. 2 × 106 of BJ cells transduced with indicated combinations of oncogenes and miRNA were injected subcutaneously into nude mice. The number of tumors arising/number of injections within 10 weeks after injection is shown
| Cell lines | Tumor Frequency |
|---|---|
| E1A+Ras+Ctrl | 0/6 |
| E1A+Ras+miR-17-92 | 6/6 |
| E1A+Ras+17&20a | 6/6 |
| E1A+Ras+Δ17&20a | 0/6 |
| E1A+Ras+17×4 | 6/6 |
| E1A+Ras+20a×4 | 6/6 |
| E1A+Ras+miR-373 | 1/6 |
| WN+Ras+Ctrl | 0/6 |
| WN+Ras+miR-17-92 | 0/6 |
| WN+Ras+17&20a | 0/6 |
| WN+Ras+Δ17&20a | 0/6 |
| MDM2+Ras+Ctrl | 0/6 |
| MDM2+Ras+miR-17-92 | 0/6 |
| MDM2+Ras+17&20a | 0/6 |
| MDM2+Ras+Δ17&20a | 0/6 |
Furthermore, consistent with their ability to overcome ras-induced senescence, miR-17 and miR-20a together (Fig. 6C, 6D, and Table 1), or miR-17 or miR-20a alone (Fig. 6A, 6B, and Table 1), also transformed BJ cells together with E1A and Ha-RasV12. In contrast, cells expressing theΔ17&20a mutant of miR-17-92 in combination with E1A and Ras only formed a relatively low number (Fig. 6D) of small (Fig. 6C) colonies on soft agar as compared to the wild type miR-17-92, and failed to develop tumors in nude mice (Table 1). Thus, like in the case of resistance to oncogene-induced senescence, miR-17 and miR-20a are the major components that contribute to the transforming activity of miR-17-92 in the presence of E1A and activated Ras. In addition, miR-373, a miRNA shown to disrupt ras-induced senescence by targeting LATS2 (43), was unable to support anchorage-independent growth together with E1A and ras, and cells expressing miR-373, E1A and ras only formed one tumor out of the 6 injections. Thus, miR-373 failed to mediate efficient cell transformation with E1A and ras, suggesting that disruption of senescence is not sufficient to cause oncogenic transformation in this system. Therefore, it is likely that by directly suppressing p53, miR-17-92 and MDM2 contribute to oncogenic transformation in this model via a senescence-dependent, as well as a senescence-independent, mechanism.
Moreover, miR-17-92 and its components failed to transform BJ cells together with activated Ras or with MDM2 and activated Ras in the absence of E1A (Fig. 6C, 6D), indicating that the essential role of E1A in this system cannot be replaced by miR-17-92. Therefore, the major oncogenic activity of miR-17-92 and its miR-17/20a components in this cellular transformation model specifically overlaps with that of MDM2, but not that of E1A. By inactivating the p53-p21WAF1 pathway, these miRNAs functionally mimic MDM2 in both senescence resistance and oncogenic transformation.
Discussion
The oncogenic miR-17-92 cluster is frequently overexpressed in human cancer (6, 7). By demonstrating the role of miR-17-92 in suppressing oncogene-induced senescence, the current study adds a new aspect to the tumorgenic activity of this pleiotropic miRNA cluster. Based our findings, miR-17-92 is capable of disrupting both apoptosis and senescence, two important tumor-suppressing defense mechanisms triggered upon oncogene activation. Furthermore, the anti-senescence activity of miR-17-92 is mediated by the miR-17/20a components, which, in contrast, are dispensable for the anti-apoptotic function of this cluster.
The miR-17-92 cluster and its miR-17/20a components confer resistance to oncogene-induced senescence at least partly by directly targeting p21WAF1, a key effector of senescence induction. Although the current study focuses on ras-induced senescence, miR-17-92 may also impact other types of senescence, such as replicative and oxidative stress-induced senescence, that rely on activation of the p53-p21WAF1 pathway. However, our results do not rule out the possible involvement of other direct targets of miR-17/20a, which, when downregulated together with p21WAF1, may synergistically abrogate ras-induced senescence. Nevertheless, at least some of the miR-17-92 targets, including Pten, Bim and E2F1, are unlikely to contribute to the anti-senescence activity. By contrast, previous reports showed that direct suppression of Pten expression by miR-19 mediates the anti-apoptotic function of miR-17-92 (15, 16). Therefore, the miR-17-92 cluster achieves its various oncogenic functions through distinct miRNA components that suppress the expression of different direct targets.
Interestingly, although miR-17-92 compromised ras-induced senescence in both BJ and WI38 fibroblasts, the block of senescence seemed to be partial in WI38 cells. It was reported that the p16INK4A levels is much higher in WI38 than in BJ cells, and that high p16INK4A levels prevent efficient disruption of senescence upon p53 inactivation (44). Thus, complete abrogation of ras-induced senescence in WI38 cells may require simultaneous inactivation of the p16INK4A and p53-p21WAF1 pathways.
miR-106b-25 and miR-106a-363 are 2 paralogs of the miR-17-92 cluster in mammals (7). Like miR-17-92, the miR-106a-363 cluster is also oncogenic (45). We demonstrate that miR-106a-363 also confers resistance to oncogenic ras-induced senescence, through its miR-106a/20b components that belong to the same family as miR-17a/20a from miR-17-92. Therefore, at least 2 of these paralogous miRNA clusters have anti-senescence activity. These findings thus provide a mechanism underlying at least part of the oncogenic activity of miR-106a-363.
Using a cell transformation model, we showed that the miR-17-92 cluster replaced MDM2, and cooperated with E1A and activated ras to transform primary human fibroblasts. Furthermore, the transforming activity of miR-17-92 in this model was fully recapitulated by its miR-17/20a components that disrupt senescence, but not by the other components including miR-19 that mediates resistance to apoptosis. This result indicates that in the presence of E1A and activated ras, senescence, rather than apoptosis, is the main rate-limiting factor for cellular transformation, and that inactivation of the p53-p21WAF1 pathway by either MDM2 or miR-17/20a is sufficient to compromise this barrier.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH (CA106768, CA131231 and RR025744 to P.S. and AI041637 and AI068896 to J.H.), an international collaborative grant from NSF China (30828019, P.S. and J.H.), 973 program (2009CB522200, J.H.), and NSFC (30830092, J.H.). This Scripps manuscript number is 20768.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi XB, Tepper CG, vere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–38. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- 4.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–56. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Dillhoff M, Wojcik SE, Bloomston M. MicroRNAs in Solid Tumors. J Surg Res. 2008 doi: 10.1016/j.jss.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 11.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–9. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 20.Iwasa H, Han J, Ishikawa F. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 2003;8:131–44. doi: 10.1046/j.1365-2443.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22:3497–508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 23.Narita M, Lowe SW. Senescence comes of age. Nat Med. 2005;11:920–2. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- 24.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 25.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 26.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, et al. PRAK Is Essential for ras-Induced Senescence and Tumor Suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 29.Sun P, Dong P, Dai K, Hannon GJ, Beach D. p53-independent role of MDM2 in TGF-beta1 resistance. Science. 1998;282:2270–2. doi: 10.1126/science.282.5397.2270. [DOI] [PubMed] [Google Scholar]
- 30.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 31.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–28. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 32.Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010 doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- 33.Novotny GW, Sonne SB, Nielsen JE, Jonstrup SP, Hansen MA, Skakkebaek NE, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007;14:879–82. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 34.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 35.Sylvestre Y, De GV, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 36.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 37.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de MI, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Wei W, Jobling WA, Chen W, Hahn WC, Sedivy JM. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol Cell Biol. 2003;23:2859–70. doi: 10.1128/MCB.23.8.2859-2870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voorhoeve PM, Agami R. The tumor-suppressive functions of the human INK4A locus. Cancer Cell. 2003;4:311–9. doi: 10.1016/s1535-6108(03)00223-x. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Sun P. The pathways to tumor suppression via route p38. Trends Biochem Sci. 2007;32:364–71. doi: 10.1016/j.tibs.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Seger YR, Garcia-Cao M, Piccinin S, Cunsolo CL, Doglioni C, Blasco MA, et al. Transformation of normal human cells in the absence of telomerase activation. Cancer Cell. 2002;2:401–13. doi: 10.1016/s1535-6108(02)00183-6. [DOI] [PubMed] [Google Scholar]
- 43.Voorhoeve PM, le SC, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv Exp Med Biol. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 44.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–22. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.