Abstract
Development of targeted therapeutics for hepatocellular carcinoma (HCC) remains a major challenge. The ubiquitination modulator COP1 regulates p53 activity by ubiquitination and it is frequently overexpressed in human HCC. In this study we tested the hypothesis that COP1 blockade by siRNA-mediated inhibition could affect the course of HCC progression. The COP1 isoform COP1-1 was selected as the most effective target for siRNAs in terms of growth inhibition and apoptotic induction in several HCC cell lines. Growth inhibition occurred in HCC cells that retained wild-type p53 or expressed mutant p53 (Y220C or R249S), whereas p53 null Hep3B cells were resistant. Microarray expression analysis revealed that the anti-proliferative effects of COPI-1 blockade were driven by a common subset of molecular alterations including a p53-associated functional network. In an orthotopic mouse xenograft model of HCC, systemic delivery of a modified COP1 siRNA by stable nucleic-acid-lipid particles (SNALP) suppressed neoplastic growth in liver without unwanted immune responses. Our findings offer a first proof of principle that COP1 can be a promising target for systemic therapy of HCC.
Introduction
HCC is the third most lethal neoplasm causing an estimated 600,000 deaths annually (1). In the United States the incidence of HCC has doubled over the past two decades, with only 30–40% of patients being eligible for curative treatments due to the late diagnosis, underlying liver disease and lack of effective treatment options (2–4). HCCs are phenotypically and genetically heterogeneous tumors driven by diverse molecular mechanisms (5). However, HCCs exhibit certain common traits selected through genomic and epigenetic alterations (6,7). Identification of both common and subclass specific genomic alterations may provide an opportunity for treatment of HCC through targeted therapy (8).
We have previously observed that COP1, an E3-ubiquitin ligase also known as RFWD2, is generally overexpressed in human HCC and could accurately predict patient survival (9). Even though the overall biological role of the mammalian COP1 is yet to be defined, several functions have been elucidated (10). In particular, COP1 has been shown to act as a negative regulator of p53 via ubiquitination (11). Given the significance of p53 and the altered expression of COP1 in human cancer, we have tested whether the targeting of COP1 could affect the course of HCC progression. Here we report that siRNA-mediated knockdown of COP1 inhibited proliferation and induced apoptosis in HCC cells through common molecular alterations. We also show that systemic silencing of COP1 effectively suppressed human HCC cell growth in orthotopic xenograft mouse model, suggesting that COP1 is a promising target for systemic HCC therapy.
Materials and Methods
Cell lines and siRNA treatment
PLC, Hep3B, and HepG2 obtained from the American Type Culture Collection (ATCC), Huh7 from Riken Cell Bank (deposited by Dr. Nam-Ho Huh) and Huh1 from Health Science Research Resource Bank were passaged for fewer than 6 months. ATCC performed cell line authentication using DNA fingerprinting by short tandem repeat analysis. Riken and Health Science Research Resource cell banks did not provide information on method of authentication. All cell lines were karyotyped upon receipt for future reference. All native siRNA duplexes used for in vitro studies were chemically synthesized by Ambion. Cells were transiently transfected with 15 nM control siRNA (Negative Control #1) or COP1-specific siRNA complexed with Lipofectamine 2000 (Invitrogen). 2’OMe-modified siRNA COP1-4/7 and β gal478 were synthesized and annealed by Integrated DNA Technologies, and formulated into SNALP suitable for in vivo delivery to liver as described (12–14). A list of siRNAs is provided in Supplementary Table 1. Vybrant MTT Cell Proliferation Assay (Invitrogen) and ApoStrand ELISA Apoptosis Detection Kit (Biomol International) were used to evaluate the biological effects of siRNA treatment. qRT-PCR and immunoblotting were performed using standard methods (Supplementary Material and Methods).
Cytokine ELISA
The production of cytokines in culture supernatant of mouse Flt3L dendrocytes or in mouse serum was measured by sandwich ELISA kits for IFN-α , IFN-β (PBL Biomedical Laboratories) and IL-6 (BD Biosciences).
Systemic administration of SNALP-formulated siRNA in vivo.
Animals were housed in an AAALAC facility and cared for in accordance with the guidelines from the Animal Care and Use Committee at the National Cancer Institute, NIH. Huh7-luc+ (5 × 105) or HepG2-luc+ (7 × 105) cells were injected into the splenic pulp of 6-week-old male SCID/Beige mice (Charles River). SNALP-formulated siRNAs (2 mg/kg) were injected into the lateral tail vein four times with a 3-day interval. Tumor growth was monitored by bioluminescence imaging for 4-weeks with 3–4 day intervals using an IVIS Imaging System (Supplementary Material and Methods).
Microarray experiments
Microarray was performed on human Ref-8v3 microarrays (illumina) as recommended by the manufacture. RNAs were isolated 48 h after the transfection of NCsiRNA or COP1-1siRNA to Huh7, HepG2 and Hep3B cells. Detailed procedures and pathway analysis are described in Supplementary Materials and Methods. The complete microarray data have been submitted to Gene Expression Omnibus database with accession number GSE21955 (http://www.ncbi.nlm.nih.gov/geo).
Results and Discussion
Silencing of COP1 inhibits proliferation and induces apoptosis of human HCC cells
To examine the biological effects of COP1 knockdown, two HCC cell lines with wild type (wt) p53 (HepG2) and mutant (mt) p53 (Huh7: Y220C) were treated with three COP1-specific (COP1-1, COP1-2 and COP1-3) siRNA for 4 days and analyzed for growth inhibition. This screen identified COP1-1 as the most potent siRNA. COP1 knockdown caused a strong reduction in growth rate in both cell lines which ranged between 84–88% and was paralleled by a similar degree of target mRNA silencing (Fig. 1A, Supplementary Fig. 1A). The Western blot experiments confirmed that the protein levels of COP1 were also reduced in COP1siRNA-treated HCC cell lines (Supplementary Fig. 1B and C)
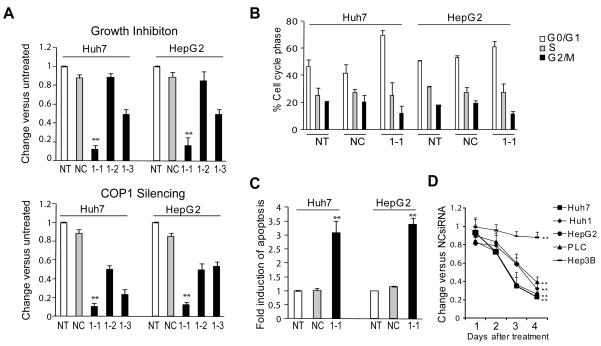
Figure 1.
siRNA knockdown of COP1 inhibits growth of HCC cells in vitro. A, Huh7 and HepG2 cells were transfected with COP1-specific siRNAs and examined at 4 days by MTT assay (top) and at 2 days by real-time RT-PCR (bottom). B, Cell cycle analysis 2 days after transfection. C, Detection of apoptosis 3 days after transfection. D, Effect of COP1-1siRNA knockdown on survival of HCC cells. The data are calculated relative to the negative control siRNA (NC) and presented as the mean ± SD of triplicate experiments. Statistical analysis was performed using Bootstrap t-test. NT, No treatment, COP1-1, COP1-2 and COP1-3 specific siRNA are shown as 1-1, 1-2, and 1-3. **, P < 0.01.
Analysis of cell cycle progression by FACS showed that COP1-1siRNA increased the G0/G1 population while decreasing the fraction of cells in G2/M phase in both Huh7 and HepG2 cells, consistent with a cell cycle arrest in G1-phase (Fig. 1B). Furthermore, COP1 treatment caused a strong induction of apoptotic cell death (Fig. 1C). Significantly, COP1 depletion was similarly effective in suppressing the growth of two additional HCC cell lines, Huh1 and PLC/PRF/5, expressing wt and mt p53 (R249S), respectively, whereas p53-null Hep3B cells were significantly more resistant (Fig. 1D).
Microarray analysis of global gene expression changes in COP1 siRNA-treated HCC cell lines
To understand the mechanism of action of COP1 in HCC cells, we performed expression profile analysis. For this purpose, three HCC cell lines with different genetic status of p53 were treated with either NCsiRNA or COP1-1siRNA for 48 hours and subjected to illumina microarray analysis. The number of differentially expressed genes which displayed more than a 2-fold change was 522 (179 up- and 343 down-regulated genes) and 462 (167 up- and 295 down-regulated genes) in COP1siRNA-treated HepG2 and Huh7 cells, respectively. Consistent with COP1 function as a negative regulator of p53 protein (11), several genes affected by COP1 inactivation were known/putative targets of p53. As expected, p53 was among the predominant pathways affected by differentially regulated genes by COP1 knockdown in HepG2 and Huh7 cells (Fig. 2A and B). In HepG2 cells, COP1 silencing increased expression of apoptosis-related (NOS2A and BIK) and anti-proliferative (BTG2, GLIPR1 and FHL2) genes which was paralleled by down-regulation of key molecules involved in a wide range of cellular responses to hypoxia (HIF1α ), growth (IGF1R, ABL1, POLK) and differentiation (HDAC5). Consistent with phenotypic changes, COP1-depleted Huh7 cells also displayed changes in p53-associated group of genes functionally involved in regulation of apoptosis, growth and differentiation including CASP6, GLIPR1, FHL2, GADD45A, ABL1, BCL6, and GDF15 genes. However, COP1 inactivation increased the p53 protein levels only in HepG2 cells with wt p53 and did not affect the p53 abundance in Huh7 cells which carry Y220C mutation increasing p53 protein stability (Supplementary Fig. 1B and C) (15). At present, the knowledge on the molecular basis for mutant p53 gain of function is limited (16), and we cannot exclude that COP1 inactivation does not activate classical p53 pathway in Huh7 cells or has an indirect effect on p53 pathway through intermediate molecules.
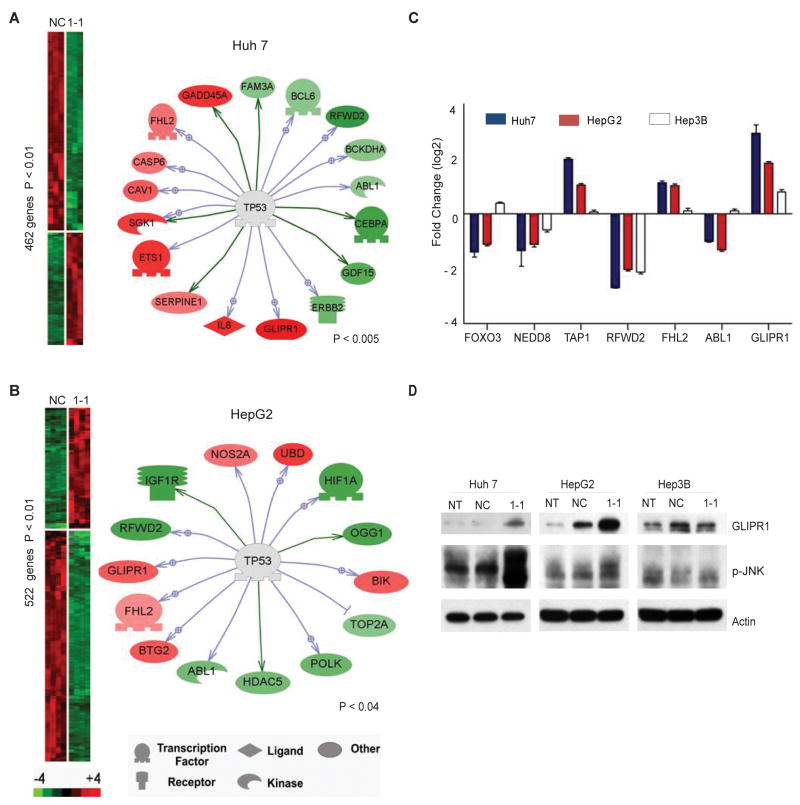
Figure 2.
Changes in gene expression following COP1 knockdown. A,B, Heat-map overview of genes up- and downregulated at 48 h after COP1 inactivation in Huh7 (A) and HepG2 (B) cells. The means of the intensity log ratios from COP1-1siRNA treated cells were calculated relative to the negative control siRNA-treated cells. P < 0.01 by Bootstrap t-test. Expression targets of p53 are shown to the right. C, Fold-changes of genes commonly dysregulated and functionally associated with p53. D, Western blot analysis of GLIPR1 and phosphorylated JNK in Huh7, HepG2, and Hep3B cells that were untreated or treated with the indicated siRNA for 48 h. Actin was included as a loading control.
To further explore the molecular mechanisms of COP1 response, we have generated a common COP1 knockdown signature consisting of 78 deregulated genes (Supplementary Table 2) Using the Ingenuity Pathway Analysis software, we then identified common statistically significant pathway networks (score >19) which were strongly associated with NF-κB, HNF4α , p53 and TNF, suggesting that molecular alterations in the diverse oncogenic pathways may cooperatively result in the growth inhibition of HCC cells in response to COP1 inactivation (Supplementary Fig. 2, Supplementary Table 3). Given the statistical and biological relevance to the study, we focused on the p53 network (#3). Our results showed that despite a limited gene to gene overlap, the expressions of seven genes, known to be associated with p53 pathway, such as FOXO3, NEDD8, TAP1, RFWD2 (COP1), FHL2, ABL1 and GLIPR1, was commonly deregulated (Fig. 2C). Knockdown of COP1 in p53-null Hep3B cells did not affect any of these genes, except for the RFWD2 (COP1) target gene (Fig. 2C). The p53-null cells were also significantly more resistant to growth inhibition caused by COP1 silencing, suggesting that COP1 knockdown phenotype is associated with p53 function. In particular, our microarray analysis identified a common upregulation of glioma pathogenesis-related protein 1 (GLIPR1) (Fig. 2C and D). GLIPR1 is a novel p53 target gene shown to exert tumor suppressor activities through upregulation of ROS-JNK pathway in p53+/+ and p53+/− genetic background (17). Indeed, increase in GLIPR1 protein and JNK phosporylation were found only in Hun7 and HepG2 but not in p53-null Hep3B cells, suggesting that activation of GLIPR1/JNK pathway might be a common mechanism of growth inhibition and apoptotic induction engaged by COP1 inactivation.
Systemic delivery of COP1siRNA by SNALP suppresses liver tumor growth in vivo
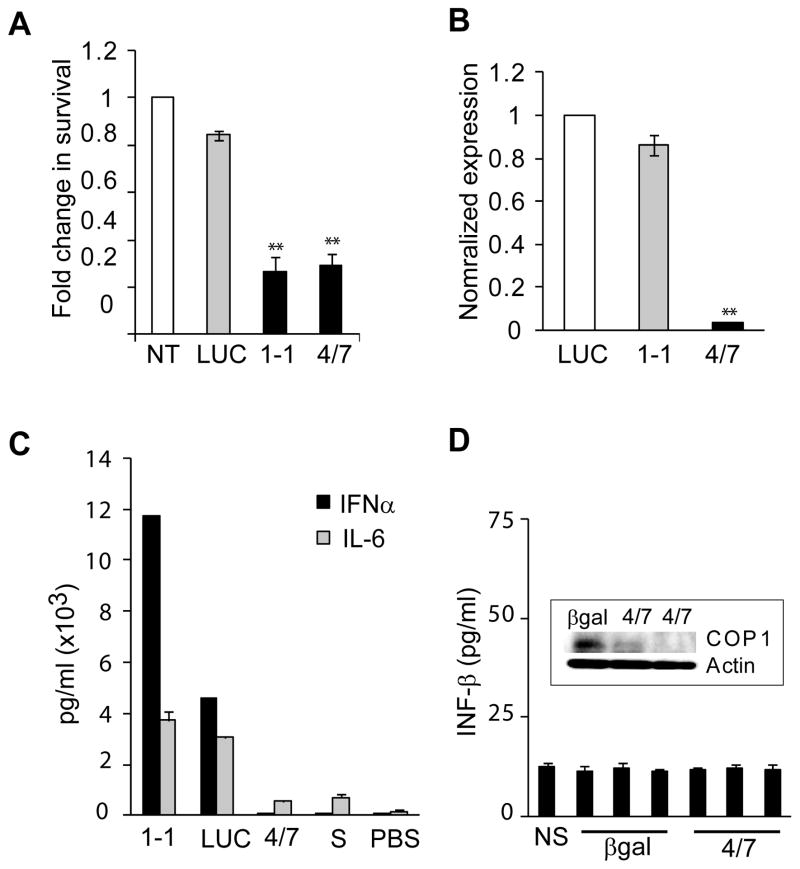
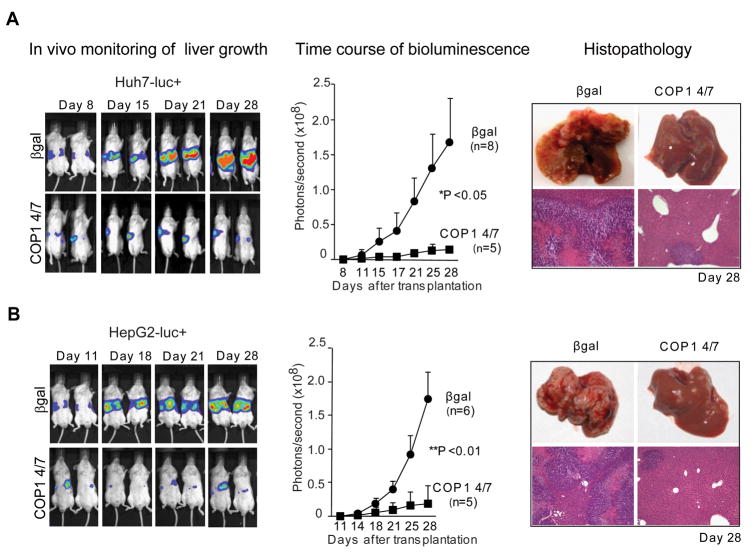
Ultimately, we confirmed the therapeutic potential of COP1 in vivo, using two human xenograft models. First, statistically significant inhibition of tumor growth was observed in a subcutaneous model of transplantation in nude/athymic mice (Supplementary Fig. 3). Direct injections of native COP1-1siRNA into the tumors established from Huh7 cells caused a dose-dependent reduction in tumor mass. As a final validation of antitumor efficacy of COP1 in vivo, we established an orthotopic xenograft model in SCID/Beige mice using luciferase-expressing HCC reporter cell lines and a SNALP formulation optimized for delivery of siRNA into liver. Recently, we have described the development of SNALP as an effective systemic delivery vehicle for targeting siRNA to murine and primate liver as well as solid tumors and have demonstrated robust therapeutic silencing of endogenous hepatocyte, tumor and viral gene transcripts in the absence of any measurable immune response (12–14). To prevent immune activation by the formulated siRNA, the native COP1-1 and non-targeting control β gal478 sequences were modified by selective incorporation of 2’-O-methyl (2’OMe) uridine or guanosine nucleosides into siRNA duplex (18). COP1-4/7 was selected as the most effective 2’OMe-modified siRNA for growth inhibition (>70%) and target mRNA silencing (>90%) (Fig.3A and B). The modified COP1 4/7 caused minimal activation of cytokines, such as IFN-α and IL-6 (Fig. 3C). Additionally, systemic injection of SNALP-COP1 4/7 did not increase the production of IFN-β in serum collected 48 h after delivery (Fig. 3D). Four intravenous injections of SNALP-COP1 4/7 significantly suppressed the growth of Huh7-luc+- or HepG2-luc+-derived tumors in liver as compared to a control group receiving SNALP-β gal478 based on bioluminescence imaging and microscopic examination (Fig. 4). In both cases, a dose of 2 mg/kg showed a potent and long lasting effect resulting in a more than 12- and 9-fold decrease in tumor growth, respectively, 10 days after the last treatment and thereby exceeding the NCI criteria for promising therapeutic compounds. In conclusion, this is the first demonstration that COP1 is an important regulator of HCC growth and survival, and may represent a promising molecular target for systemic therapy of a wide spectrum of human HCC.
Figure 3.
Selection of COP1 4/7siRNA for in vivo application based on the inhibition of tumor cell growth and minimal cytokine induction. A, Inhibition of Huh7-luc+ cell growth after transfection of SNALP-formulated COP1-1 (native) or COP1 4/7 siRNA (a modified variant). The siRNA transfectants were examined by MTT assay at 4 days after treatment. B, Real-Time RT-PCR analysis of COP1 gene expression in Huh7-luc+ cells treated with the indicated siRNA. **, P < 0.01 (n=3) by Bootstrap t-test. C, Quantification of cytokines after luciferase (LUC) or COP1 targeting. Culture supernatants of Flt3L-derived dendrocytes were assayed for IFN-α and IL-6 using ELISA at 24 h after siRNA treatment. Data are shown are the means ± SD of triplicate experiments. D, Serum levels of IFN-β and downregulation of COP1 protein levels in Huh7-derived tumors (inset) 48 h after a single i.v. administration of encapsulated siRNA (2 mg/kg) targeting β-galactosidase (β gal) or COP1 into immunodeficient mice. Each bar represents the mean picogram of IFN-β ± SD of duplicate experiments. NS, normal serum, S, SNALP.
Figure 4.
Systemic delivery of COP1 4/7siRNA by SNALP suppresses human HCC growth in orthotopic xenograft model. SCID/beige mice received Huh7-luc+ (A) and HepG2-luc+ (B) cells through intrasplenic injection resulting in tumorous growth in the liver. Mice were randomly assigned either to control (SNALP-β gal478) or treatment (SNALP-COP1 4/7) group based on the intensity of bioluminescence before initiation of COP1 4/7siRNA therapy at day 8 for Huh7 and day 11 day for HepG2. Two mg/kg of SNALP-β gal478 and SNALP-COP1 4/7 were injected into tail vein at the time indicated. Representative in vivo bioluminescence imaging of Huh7- and HepG2-xenografts are shown on the left. Images were set at the same pseudocolor scale to show the relative bioluminescence changes over time. Quantification of bioluminescence (middle panels). The total flux is plotted as photon/second. *, P < 0.05 (n=8 vs. n=5) by Student’s t-test; **, P < 0.01, (n=8 vs. n=6) by Mann-Whitney U-test. Histopathological evaluation (right panels). Representative photos of gross liver morphology at 28 days after transplantation are shown. H&E staining, original magnification X50.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We thank Joseph A. Frank for helpful discussions and Jeonghoon Heo for assistance during in vivo studies.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–27. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–36. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 6.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 7.Feitelson MA, Sun B, Satiroglu Tufan NL, et al. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 8.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–25. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–76. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 10.Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–25. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 11.Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–7. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 14.Judge AD, Robbins M, Tavakoli I, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–73. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu IC, Tokiwa W, Bennett RA, et al. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- 16.Weisz L, Oren M, Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Fattah EA, Cao G, et al. Glioma pathogenesis-related protein 1 exerts tumor suppressor activities through proapoptotic reactive oxygen species-cJun-NH2 kinase signaling. Cancer Res. 2008;68:434–43. doi: 10.1158/0008-5472.CAN-07-2931. [DOI] [PubMed] [Google Scholar]
- 18.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2005;13:494–504. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.