Abstract
Extracytoplasmic function (ECF) sigma factors are known to play an important role in the bacterial response to various environmental stresses and can significantly modulate their pathogenic potential. In the genome of Porphyromonas gingivalis W83, six putative ECF sigma factors were identified. To further evaluate their role in this organism, a PCR-based linear transformation method was used to inactivate five ECF sigma factor genes (PG0162, PG0214, PG0985, PG1660, and PG1827) by allelic exchange mutagenesis. All five isogenic mutants formed black-pigmented colonies on blood agar. Mutants defective in PG0985, PG1660 and PG1827 genes were more sensitive to 0.25 mM of hydrogen peroxide compared to the wild-type strain. Isogenic mutants of PG0162 and PG1660 showed a 50 percent decrease in gingipain activity. RT-PCR analysis showed that there was no alteration in the expression of rgpA, rgpB, and kgp gingipain genes in these mutants. Hemolytic and hemagglutination activities were decreased by more than 50 percent in the PG0162 mutant compared to the wild-type. Taken together, these findings suggest that ECF sigma factors can modulate important virulence factors in P. gingivalis. ECF sigma factors encoded by the PG0162 and PG1660 genes might also be involved in the post-transcriptional regulation of the gingipains.
Keywords: Porphyromonas gingivalis W83, ECF sigma factor, virulence
Introduction
The response and adaptation of bacteria to environmental stress is known to be mostly regulated at the level of transcription initiation. This regulation primarily involves alternative sigma factors which recruit RNA polymerase and facilitate specific promoter recognition and transcription initiation (Paget & Helmann, 2003). Extracytoplasmic function (ECF) sigma factor, the largest group of alternative sigma factors, plays a key role in adaption to environmental conditions (Staron et al., 2009). Furthermore, because bacteria-host interaction via surface structures is important in bacterial pathogenesis, ECF sigma factors also regulate virulence factors (Staron et al., 2009). This is well documented in Mycobacterium tuberculosis (Hahn et al., 2005), Staphylococcus aureus (Shaw et al., 2008), Pseudomonas aeruginosa (Wood & Ohman, 2009, Llamas et al., 2009), and Enterococcus faecalis (Le et al., 2010).
Porphyromonas gingivalis, an anaerobic gram negative bacterium, is an important etiological agent in adult chronic periodontitis. This organism possesses several cell surface associated virulence factors (e.g. hydrolytic enzymes, fimbriae, hemagglutinin, capsule, and lipopolysaccharide) that can directly or indirectly affect the periodontium (Yoshimura et al., 2009). In addition, to survive in the microenvironment of an advanced periodontal pocket it is necessary that the bacteria has the capacity to respond to environmental changes including temperature, pH, concentration of some nutrients, and oxygen tension. To date, little is known about the relationship between the regulation of adaptive mechanisms, virulence, and sigma factors in P. gingivalis.
The P. gingivalis W83 genome encodes 8 sigma factors, 6 of which belong to the extracytoplasmic function (ECF) sigma factor subfamily (PG0162, PG0214, PG0985, PG1318, PG1660, and PG1827) (Nelson et al., 2003). The PG1318 ECF sigma factor was recently shown to be involved in the regulation of mutation frequency in P. gingivalis (Kikuchi et al., 2009). In this study, we used a PCR-based linear transformation strategy to inactivate the remaining five putative ECF sigma factors, and analyzed the virulence related characteristics of these proteins in P. gingivalis W83. We now report that several of the ECF sigma factors may play a role in virulence regulation and adaptation to oxidative stress. ECF sigma factors encoded by the PG0162 and PG1660 genes are likely involved in the post-transcriptional regulation of the gingipains.
Materials and methods
Bacterial strains, plasmids, and culture conditions
Strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown in Brain Heart Infusion (BHI) broth supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, Mich.), hemin (5μg/ml), vitamin K (0.5μg/ml), and cysteine (0.1%) (Sigma-Aldrich, St. Louis, Mo). P. gingivalis strains were maintained in an anaerobic chamber (Coy Manufacturing, Ann Arbor, Mich.) in 10% H2, 10% CO2, and 80% N2 at 37°C. Growth rates for P. gingivalis strains were determined spectrophotometrically by measuring optical density at 600 nm [OD600]. Antibiotics were used at the following concentrations: erythromycin, 10μg/ml; tetracycline, 3μg/ml.
Table 1.
Strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | References |
|---|---|---|
| Strains | ||
| P. gingivalis | ||
| W83 | Wild-type | Henry et al. (2008) |
| FLL350 | PG0162-defective | This study |
| FLL351 | PG0214-defective | This study |
| FLL352 | PG0985-defective | This study |
| FLL354 | PG1660-defective | This study |
| FLL355 | PG1827-defective | This study |
| FLL350 C | FLL350 with pFLL350a | This study |
| Plasmids | ||
| pVA2198 | Spr, ermF-ermAM | Fletcher et al. (1995) |
| pT-COW | Apr, tetQ | Gardner et al. (1996) |
| pFLL350a | Apr, tetQ, Tc::PG0162 | This study |
Sensitivity to hydrogen peroxide
Sensitivity of P. gingivalis strains to hydrogen peroxide was tested as previously described (Henry et al., 2008). Briefly, P. gingivalis strains were grown to early log phase (OD600 ~0.2) in BHI broth. Hydrogen peroxide at a final concentration of 0.25 mM was then added to the cultures and further incubated at 37°C for 24 hours. The OD600 was measured at 3-hour intervals over a 24 hour period. Cell cultures without hydrogen peroxide were used as controls.
Construction of ECF sigma factor defective mutants
Long PCR-based fusion of several fragments was done as previously described (Shevchuk et al., 2004). Primers used in this study are listed in Table 2. One kilobase flanking fragments both upstream and downstream of the target genes were PCR amplified from chromosomal DNA of P. gingivalis W83. The ermF cassette was amplified from the pVA2198 (Fletcher et al., 1995) plasmid with oligonucleotide primers that contained overlapping nucleotides for the upstream and downstream fragments. These three fragments were fused together by using the forward primer of the upstream fragment and the reverse primer of the downstream fragment. The fusion PCR program consisted of 1 cycle of 5min at 94 °C, followed by 30 cycles of 30 sec at 94 °C, 30 sec at 55°C, and 4 min at 68°C, with a final extension of 5 min at 68 °C. This PCR-fused fragment was used to transform P. gingivalis W83 by electroporation as previously described (Abaibou et al., 2001). The cells were plated on BHI agar containing 10 μg/ml of erythromycin and incubated at 37 °C for 7 days. The correct gene replacement in the erythromycin resistant mutants was confirmed by colony PCR and DNA sequencing.
Table 2.
Primers used in this study
| Primers | Sequences (5’--3’) |
|---|---|
| Primers for PG0162 mutant construction | |
| PG0162_F1 | CGAAGAGGTGGAGGCCAACAATTACT |
| PG0162_R1 | TACCTTATTCCTCCTAGTTAGTCATGTGGAAACTGCTCATAGGAGAATC |
| PG0162_F3 | TTCGTAGTACCTGGAGGGAATAATCCATGTCGGCTTAGTCGATATTCCATA |
| PG0162_R3 | GTGCATCCTCTGCCAGCGTACATAT |
| Primers for PG0214 mutant construction | |
| PG0214_F1 | GCTGCTGCCACTATGCGCTTTTCGA |
| PG0214_R1 | TACCTTATTCCTCCTAGTTAGTCACTATGGGCGAATGCTTCTACTTCCAT |
| PG0214_F3 | TTCGTAGTACCTGGAGGGAATAATCGGCTCTCCAACTCAAACGATGATGA |
| PG0214_R3 | CAGGAGCGATACCAGATCCTCGGAA |
| Primers for PG0985 mutant construction | |
| PG0985_F1 | CTGACAGAAGAAGCTCGTAAGGAGT |
| PG0985_R1 | TACCTTATTCCTCCTAGTTAGTCAGAAAGCGATAGTATTCACGTCGTACT |
| PG0985_F3 | TTCGTAGTACCTGGAGGGAATAATCGAGGTGATGAAAGATGATGCGTG A |
| PG0985_R3 | CGTGAATGTCCCTTCCATGACAAGT |
| Primers for PG1660 mutant construction | |
| PG1660_F1 | CCCATGATAGAGACCATCTCCCCTT |
| PG1660_R1 | TACCTTATTCCTCCTAGTTAGTCATAGCTGTCGGATTGCTTCTCCATCT |
| PG1660_F3 | TTCGTAGTACCTGGAGGGAATAATCTGATGAGAAAATGAAACAGAATATA |
| PG1660_R3 | CTCGCGTAAGGAGATACAGCTTGAGA |
| Primers for PG1827 mutant construction | |
| PG1827_F1 | GACATAGGTGTTCGAACCGCCGATAT |
| PG1827_R1 | TACCTTATTCCTCCTAGTTAGTCAATAATAGACGGCATCAATGACATGAA |
| PG1827_F3 | TTCGTAGTACCTGGAGGGAATAATCAGGACATGAGATAACAGCCTGCTCA |
| PG1827_R3 | GATGCGTACAGGTGATCTTGGCACCA |
| Erm_F_r(3) | GATTATTCCCTCCAGGTACTACGAAGGATGAAATTTTTCA |
| Erm_F_P_5' | TGACTAACTAGGAGGAATAAGGTATTTGCAACATCATAGAAA |
| Primers for FLL350 complementation | |
| PG0162_Com_F | ATGGATCCGCATTAGTCGGGTTGATAAGCAGT |
| PG0162_Com_R | TTGGATCCCTAAGCCGACATGCCCATCATTTT |
| RT-PCR primers | |
| PG0161_RT_F | CCTTTTGGCGAATATGCTTATGACAGA |
| PG0161_RT_R | GAGCAACGTGAACGGGCCATAAGTTT |
| PG0162-RT_F | TGATCCATACGCTTCGTCGCGGA |
| PG0162-RT_R | GCAGATTGATCAGCGCATATCGCATT |
| PG0214-RT_F | GACCACTGCTCGGCGAATAGCCT |
| PG0214-RT_R | GGAGAGCCAGTGCTGTACGCAGT |
| PG0215-RT_F | GAAGAGGATCGCCTGCGTCGCTAT |
| PG0215-RT_R | CGAAAAGCTCCTGTGCCAACTCCT |
| PG0985-RT_F | CGACGACGAGGAGGCTGAGGAT |
| PG0985-RT_R | CACATTGCCCTCCGTCAGTCCCAA |
| PG0986-RT_F | CAAGAGGAGTTGGAGCTGTATTGCTT |
| PG0986-RT_R | CGAGACAATACTCCAATAGCTCCAT |
| PG1659-RT_F | GACTCAAACAACGCTTCGACGTGAT |
| PG1659-RT_R | CGATGCTTCGTGTTGCGATATAGTT |
| PG1660-RT_F | GTGGAGCGATACAGCGACATGCT |
| PG1660-RT_R | GTGGAGCTTGACCTTCACGTTGGA |
| PG1827-RT_F | CGCTGTTTATTAGGGTTTCTCACCTA |
| PG1827-RT_R | GTACATGGAGAATGGCTGCTTGAGA |
| PG1828-RT_F | GCTGTATCATTCGCTTCTTGCAACT |
| PG1828-RT_R | CTTGTTCAGTAGCTTCTTCAGCCTT |
| 16S_rRNA_F | AGGCAGCTTGCCATACTGCG |
| 16S_rRNA_R | ACTGTTAGCAACTACCGATGT |
| HagA_RT_F | GTGGTAGAGGCCGGTAAGACTTATCA |
| HagA_RT_R | CGTCACCTGTATGAGTAGCAGTGTT |
Complementation of the PG0162-defective mutant (FLL350)
A DNA fragment containing the PG0162 ORF with an upstream regulatory region was amplified from chromosomal DNA of P. gingivalis W83 using primer sets PG0162_Com_F and PG_0162_Com_R (Table 2). A BamHI restriction site was designed at the 5’ end of both primers to facilitate the subcloning of the PCR fragment. Both pT-COW (Gardner et al., 1996) and the BamHI-digested PCR fragment were ligated together and used to transform E. coli DH5α. The purified recombined plasmid designated pFLL350a was used to transform P. gingivalis FLL350 (PG0162::ermF) by electroporation. The transformants were selected on BHI agar plates with erythromycin and tetracycline.
Hemolytic activity assay
Hemolytic activity was determined as previously reported (Vanterpool et al., 2004). Briefly, bacterial cells from 24 hour cultures were harvested by centrifugation (10,000 g for 10 min), washed three times with phosphate buffered saline (PBS, 0.147 M NaCl, 0.01 M sodium phosphate, pH 7.4), and then resuspended to a final OD600 of 1.5. Sheep erythrocytes (Hemostat Laboratories, Dixon, CA) were harvested by centrifugation (4,400 g for 20 min) and washed with PBS until the supernatant was visibly free of hemoglobin pigment. The washed erythrocytes were suspended to a final concentration of 1% in PBS. Hemolytic activity was determined by mixing an equal volume of bacterial cells with 1% erythrocytes in PBS. This mixture was then incubated at 37°C for 4 hours. Samples (500 μl) were withdrawn and further spun (1,300 g for 5 min) in an Eppendorf 5403 centrifuge at room temperature. The OD405 of supernatant was determined by spectrophotometry. As a negative control, erythrocytes were used alone.
Hemagglutination studies
Hemagglutinin activity was determined as previously reported (Vanterpool et al., 2005a). Twenty-four hour cultures of P. gingivalis W83 and mutants were harvested by centrifugation (10,000 g for 10 min). Cells were washed twice in PBS buffer and resuspended to a final OD600 of 1.5. Sheep erythrocytes were washed twice with 1×PBS and resuspended in 1×PBS to a final concentration of 1%. An aliquot (100-μl volume) of the bacterial suspension was serially diluted two-fold with PBS in wells of a round-bottom 96-well microtiter plate. An equal volume (100 μl) of 1% sheep erythrocytes was mixed with each dilution and incubated at 4°C for 3 hours. Hemagglutination was visually assessed and the hemagglutination titer was determined as the last dilution that showed complete hemagglutination.
Gingipain activity assays
The presence of Arg-X- and Lys-X-specific cysteine protease (Rgp and Kgp) activity was determined with a microplate reader (Bio-Rad, Hercules, CA) as previously reported (Vanterpool et al., 2005b). In brief, activity of arginine gingipains was measured with 1mM BAPNA (Nα-benzoyl-DL-arginine-p-nitroanilide) in activated protease buffer (0.2 M Tris-HCl, 0.1 M NaCl, 5 mM CaCl2, 10 mM L-cysteine, pH 7.6). Lysine gingipain activity was measured with ALNA (Ac-Lys-p-nitroanilide HCl). After incubating the substrate and culture, the reaction was stopped by the addition of 50 μl of glacial acetic acid. The O.D. at 405 nm was then measured against a blank sample containing no bacteria.
Reverse transcription (RT)-PCR analysis
Total RNA from P. gingivalis strains was extracted by using the SV Total RNA Isolation System (Promega Corp. Madison, WI) according to the manufacturer’s instructions. Complementary DNA was synthesized by using a Transcriptor High Fidelity cDNA Synthesis Kit (Roche Corp., Indianapolis, IN). The primers used as listed in Table 2. The PCR program consisted of 1 cycle of 5 min at 94 °C, followed by 30 cycles of 30 sec at 94 °C, 30 sec at 54°C, and 1 min at 68°C, with a final extension of 5 min at 68 °C.
Results and discussion
Construction of ECF sigma factor mutants in P. gingivalis W83
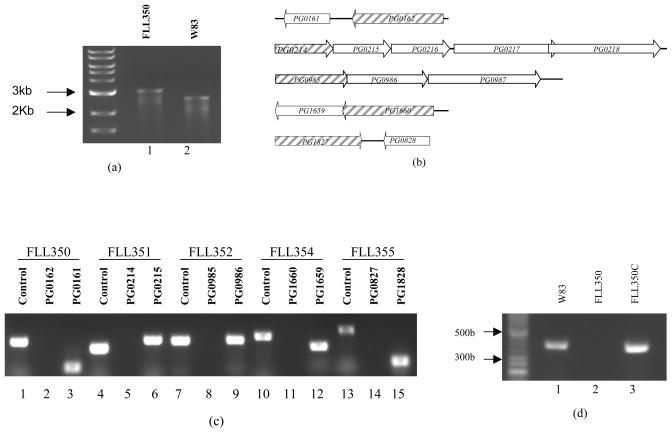
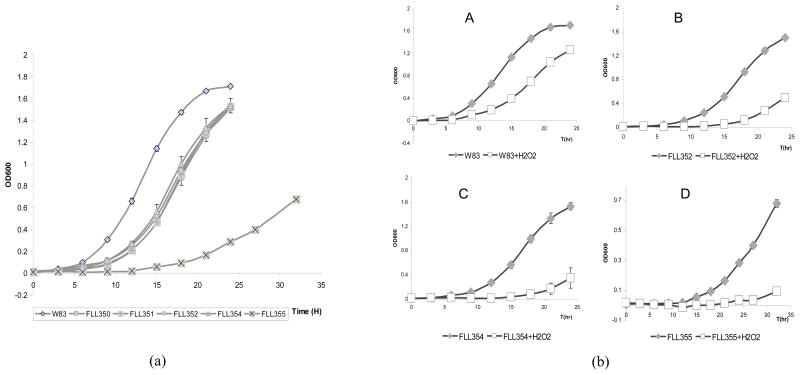
To construct ECF sigma factor isogenic mutants, PCR was used to fuse the upstream and downstream fragments of the target gene to ermF, generating a 3 kb-length fragment, which was then electrotransformed into P. gingivalis W83. To the promoter region upstream of the ATG start codon of ermF, we added a 20 base oligonucleotide 5’-TGACTAACTAGGAGGAATAA-3’ containing three stop codons separated by one nucleotide. An 18 base oligonucleotide 5-TACCTGGAGGGAATAATC-3’ containing a ribosome binding site was added downstream of the TAG stop codon of ermF, to eliminate the influence on expression of the other genes downstream of the target gene. Though W83 lacks a TraP, which was previously shown to be required for plasmid transfer in P. gingivalis (Tribble et al., 2007), PCR-based transformation worked with high efficiency in W83. We were able to construct 5 ECF sigma factor deletion mutants (PG0162, PG0214, PG0985, PG1660, and PG1827). These mutants were confirmed by colony PCR (Fig. 1a) and sequencing (data not shown). To rule out polar mutations arising from the inactivation of these genes, RT-PCR was used to amplify the sigma factor encoding genes and the downstream genes (Fig. 1b). As shown in Figure 1c, inactivation of the ECF genes had no effects on the expression of the downstream genes. FLL355 (PG1827::ermF) showed a slower growth rate compared to the other ECF mutants which were similar to the wild-type strain (Figure 2a). However, similar to the wild-type strain, all five ECF isogenic deletion mutants were black-pigmented on blood agar plates (data not shown).
Fig. 1.
(a) Colony PCR confirmation of the mutant FLL350. The primers used were PG_0162_F1 and PG_0162_ R3. Lane 1, FLL350 PCR product (3 kb). Lane 2, W83 PCR product (2.5 kb). (b) Diagram of chromosomal structure of PG0162, PG0214, PG0985, PG1660 and PG1827. Hatched arrows indicate the ECF sigma factor encoding genes and white arrows indicate genes located downstream of ECF sigma factor genes. Arrow indicates the transcription orientation. (c) RT-PCR confirmation showing the non-polar effects of the inactivated ECF sigma factor genes is isogenic defective mutants. Lane 1, PG0162 product using W83 cDNA as template (379 bp). Lane 2, PG0162 product using cDNA of FLL350 as template. Lane 3, PG0161 product using cDNA of FLL350 as template (150 bp). Lane 4, PG0214 product using W83 cDNA as template (308 bp). Lane 5, PG0214 product using cDNA of FLL351 as template. Lane 6, PG0215 product using cDNA of FLL351 as template (382 bp). Lane 7, PG0985 product using W83 cDNA as template (367 bp). Lane 8, PG0985 product using cDNA of FLL352 as template. Lane 9, PG0986 product using cDNA of FLL352 as template (376 bp). Lane 10, PG1660 product using W83 cDNA as template (414 bp). Lane 11, PG1660 product using cDNA of FLL354 as template. Lane 12, PG1659 product using cDNA of FLL354 as template (310 bp). Lane 13, PG1827 product using W83 cDNA as template (491 bp). Lane 14, PG1827 product using cDNA of FLL355 as template. Lane 15, PG1828 product using cDNA of FLL355 as template (165 bp). (d) Transcriptional analyses of FLL350C by RT-PCR. Primers used were PG0162_RT_F and PG0162_RT_R as listed in Table 2. Lane 1, W83 cDNA. Lane 2, FLL350 cDNA. Lane 3, FLL350C cDNA.
Fig. 2.
Growth and H2O2 sensitivity of ECF sigma factor isogenic mutants. Strains grown to early log phase were treated with 0.25 mM H2O2 and further incubated for 24 hr. The cultures without H2O2 were used as controls. The results shown were representative of at least three independent replicates. Panel (a), Growth of P. gingivalis W83 and ECF sigma factor isogenic mutants in the absence of H2O2. Panel (b), Growth of P. gingivalis W83 and ECF sigma factor isogenic mutants in the presence of H2O2. A, W83. B, FLL352. C, FLL354. D, FLL355. Error bars represent the standard deviation.
Growth in the presence of hydrogen peroxide
The sensitivity to several environmental stresses including oxidative stress and involvement in pathogenesis of ECF sigma factor mutants have been described for several bacteria (Kallifidas et al., 2010, Staron et al., 2009, White et al., 2010). In the human oral cavity, P. gingivalis encounters oxidative stress from exposure to air and reactive oxidative species (ROS) generated by neutrophils or from other oral bacteria. ROS can cause damage to cell membranes, nucleic acids, and proteins (Imlay, 2003). While several organisms have evolved various mechanisms to protect themselves against oxidative stress, little is known about ROS sensing and adaptation/protection in anaerobic bacteria.
In order to evaluate the relationship between the sensitivity of P. gingivalis to hydrogen peroxide and ECF sigma factors, isogenic mutants defective in these factors were exposed to hydrogen peroxide. As shown in Figure 2, the growth of P. gingivalis isogenic mutants defective in PG0985 (FLL352), PG1660 (FLL354), and PG1827 (FLL355) was more retarded in the presence of hydrogen peroxide compared to the wild-type. PG0162 (FLL350) and PG0214 (FLL351) isogenic mutants and the wild-type showed a similar sensitivity to hydrogen peroxide (data not shown). This suggests that ECFs PG0985, PG1660, and PG1827 may play a role in H2O2-induced oxidative stress resistance in P. gingivalis.
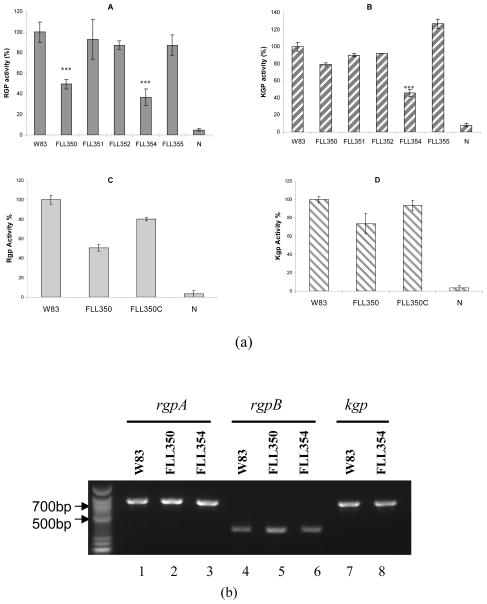
FLL350 and FLL354 showed decreased gingipain protease activity
Several reports have documented the multiple effects of gingipains, a major virulence factor of P. gingivalis (Sheets et al., 2006, Sheets et al., 2008). These gingipains, which are both extracellular and cell membrane-associated, are essential for growth and can also play a role in oxidative stress resistance (Sheets et al., 2008). In order to identify whether the sigma factors were involved in gingipain regulation, gingipain activity was measured in ECF sigma factor mutants. In comparison to the wild-type, Rgp gingipain activity was decreased by 50% and 60% in FLL350 (PG0162::ermF) and FLL354 (PG1660::ermF) respectively (Fig. 3a). Kgp activity was decreased by 20% in FLL350 and 50% in FLL354 compared to the wild-type strain (Figure 3a). Because gingipain activity can be regulated at the transcriptional and post-transcriptional levels (Tokuda et al., 1998), oligonucleotide primers as previously described (Vanterpool et al., 2005a) were used in RT-PCR analysis to determine whether these two sigma factors were involved in the transcriptional regulation of gingipain encoding genes. As shown in Figure 3b, the inactivation of PG0162 and PG1660 had no effect on the expression of rgpA, rgpB, or kgp at the transcriptional level. In FLL355 (PG1827::ermF), the Kgp activity showed a 25% increase over the wild-type. No change was observed in the transcription of the kgp gene in FLL355 (data not shown). Taken together, these results suggest that ECF sigma factors may be involved in the post-transcriptional regulation of gingipains. Post-transcriptional regulation of the gingipains in P. gingivalis is associated with its maturation pathway that is linked to the biosynthesis of surface carbohydrates (Paramonov et al., 2005, Shoji et al., 2002) and several other proteins including the PorR (Shoji et al., 2002), PorT (Nguyen et al., 2009, Sato et al., 2005), Sov (Saiki & Konishi, 2010) and VimA (Vanterpool et al., 2006). It is unclear how these factors are modulated by the ECF sigma factors and is an active area of further exploration in the laboratory.
Fig. 3.
(a) Proteolytic activity of ECF sigma factor mutants. P. gingivalis strains were grown to stationary phase in BHI media supplemented with hemin and vitamin K. Activities against Rgp or Kgp were tested in whole cell culture. The gingipain activities were normalized to W83 being 100% and the mutants reported as a percentage thereof. Asterisks indicate results significantly different from those of W83 (P<0.005). Error bars represent the standard deviation. A, Rgp activity. B, Kgp activity. C, Rgp activity of FLL350 complemented with PG0162. D, Kgp activity of FLL350 complemented with PG0162. (b) RT-PCR analysis of gingipain encoding genes in W83, FLL350 and FLL354.
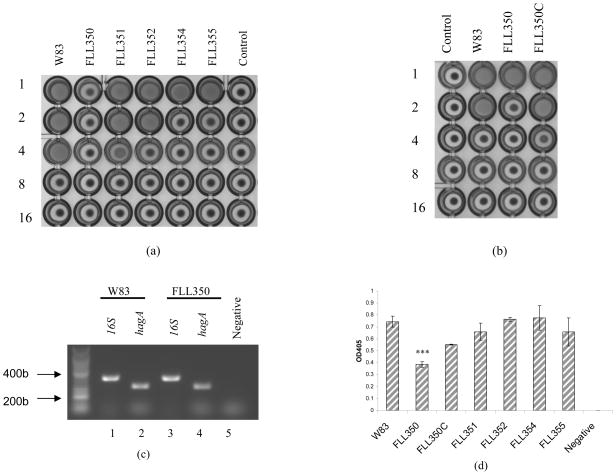
Hemagglutination activity is decreased in the ECF sigma factor-defective mutants
The correlation between gingipain activity and hemagglutination in P. gingivalis (Lewis et al., 1999, Shi et al., 1999, Vanterpool et al., 2005a) is related to the similar adhesion domains encoded by the hagA, rgpA, and kgp genes (Chen & Duncan, 2004). The hemagglutination potential of ECF sigma factor defective mutants was assessed. In comparison to the wild-type strain, there was a decrease in the hemagglutination activity in all the mutants. In FLL350, the level of hemagglutination activity was comparable to the negative control. This is in contrast to FLL354, which had the greatest reduction in gingipain activity but a higher hemagglutination activity. RT-PCR using hagA specific primers indicate no change in the expression of that gene in FLL350 (Fig. 4c).
Fig. 4.
(a) Hemagglutination activity of ECF sigma factor mutants. Hemagglutinin activities of P. gingivalis were assessed in cells serially diluted and incubated with sheep erythrocytes at 4°C for 3 h in a round-bottom microtiter plate. The titer was defined as the last dilution that showed full agglutination. Dilution folds were listed on the left. (b) Hemagglutination activity of ECF sigma factor mutants. Dilution folds were listed on the left. (c) RT-PCR analysis of hagA in FLL350. Lane 1, 16S rDNA, W83 cDNA as template (404b). Lane 2, hagA, W83 cDNA as template (312b). Lane 3, 16SrDNA, FLL350 cDNA as template. Lane 4, hagA, FLL350 cDNA as template. Lane 5, hagA, FLL350 RNA as template. (d) Hemolytic activity ECF sigma factor mutants. Erythrocytes were used as negative control. The hemolytic activity of W83 was normalized as 100% and the mutants reported here as percentage. Error bars represent the standard deviation. The data shown are an average of three or more replicates. Asterisks indicate results significantly different from that of W83 (P<0.005).
PG0162 can regulate hemolytic activity
While gingipains have been observed to have hemolytic activity (Lewis et al., 1999, Shah & Gharbia, 1989), hemolysin can be independent of their catalytic association (Deshpande & Khan, 1999). Several putative hemolysin genes have been identified in the P. gingivalis genome (Nelson et al., 2003) and cloned in E. coli (Karunakaran & Holt, 1993). The hemolysins produced by P. gingivalis provide the bacterium with heme-containing molecules that are required for their in vivo survival. Hemolytic activities of all the ECF-defective mutants in this study were similar to those of the wild-type except FLL350 (Figure 4d). FLL350 mutant showed a 50% reduction in those activities compared to the parent strain. Complementation of FLL350 with a wild-type copy of PG0162 restored some of the hemolytic activity of this mutant [FLL350c, (Figure 4)]. Gingipain and hemagglutination activities were also restored in FLL350c (Figure 3 and 4), thus confirming a role for PG0162 in virulence regulation in P. gingivalis W83. It is noteworthy that although FLL354 had the lowest gingipain activity, its hemolysin profile was similar to the wild-type strain. Taken together, this is consistent with previous observations suggesting that hemolysin and gingipain activities can be distinct from each other (Deshpande & Khan, 1999).
Collectively, our study showed that ECF sigma factors PG0162 and PG1660 play an important role in the regulation of gingipain, hemolytic, and hemagglutination activities, and could likely modulate the virulence potential of P. gingivalis. Because the exterior surface structures and factors of the infectious bacteria are the first to come in contact with the host and must respond and adapt to the host environment, our observations are consistent with the role of ECF sigma factors in the regulation of virulence associated genes [reviewed in (Brooks & Buchanan, 2008) ]. The activity of ECF sigma factors is most often negatively regulated by direct interaction with cognate anti-sigma factors, which prevent their association with the core RNA polymerase or facilitate holoenzyme dissociation [reviewed in (Staron et al., 2009)]. While PG1660 appear to have a putative cognate anti-sigmafactor PG1659 (http://www.oralgen.lanl.gov/, http://www.cbs.dtu.dk/services/TMHMM/ ), a similar putative component is missing for PG0162. It is noteworthy that regulation of PG0162 on virulence observed in this study is occurring at the post-transcriptional level. It is likely that PG0162 may be involved in a unique and complex regulatory mechanism and that requires further evaluation.
Acknowledgments
This work was supported by Loma Linda University and Public Health Grant DE13664 and DE019730 from NIDCR (to H.M.F)
Reference List
- 1.Abaibou H, Chen Z, Olango GJ, Liu Y, Edwards J, Fletcher HM. VimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect Immun. 2001;69:325–335. doi: 10.1128/IAI.69.1.325-335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks BE, Buchanan SK. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim Biophys Acta. 2008;1778:1930–1945. doi: 10.1016/j.bbamem.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T, Duncan MJ. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb Pathog. 2004;36:205–209. doi: 10.1016/j.micpath.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Deshpande RG, Khan MB. Purification and characterization of hemolysin from Porphyromonas gingivalis A7436. FEMS Microbiol Lett. 1999;176:387–394. doi: 10.1111/j.1574-6968.1999.tb13688.x. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed â-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl Environ Microbiol. 1996;62:196–202. doi: 10.1128/aem.62.1.196-202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn MY, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry LG, Sandberg L, Zhang K, Fletcher HM. DNA repair of 8-oxo-7,8-dihydroguanine lesions in Porphyromonas gingivalis. J Bacteriol. 2008;190:7985–7993. doi: 10.1128/JB.00919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 10.Kallifidas D, Thomas D, Doughty P, Paget MS. The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology. 2010;156:1661–1672. doi: 10.1099/mic.0.037804-0. [DOI] [PubMed] [Google Scholar]
- 11.Karunakaran T, Holt SC. Cloning of two distinct hemolysin genes from Porphyromonas (Bacteroides) gingivalis in Escherichia coli. Microb Pathog. 1993;15:37–49. doi: 10.1006/mpat.1993.1055. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi Y, Ohara N, Ueda O, Hirai K, Shibata Y, Nakayama K, Fujimura S. Porphyromonas gingivalis mutant defective in a putative extracytoplasmic function sigma factor shows a mutator phenotype. Oral Microbiol Immunol. 2009;24:377–383. doi: 10.1111/j.1399-302X.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- 13.Le JA, Torelli R, Sanguinetti M, Giard JC, Hartke A, Auffray Y, Benachour A. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One. 2010;5:e9658. doi: 10.1371/journal.pone.0009658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llamas MA, van der SA, Chu BC, Sparrius M, Vogel HJ, Bitter W. A Novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 2009;5:e1000572. doi: 10.1371/journal.ppat.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen KA, Zylicz J, Szczesny P, Sroka A, Hunter N, Potempa J. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology. 2009;155:328–337. doi: 10.1099/mic.0.024323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramonov N, Rangarajan M, Hashim A, Gallagher A, duse-Opoku J, Slaney JM, Hounsell E, Curtis MA. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 20.Saiki K, Konishi K. The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis. FEMS Microbiol Lett. 2010;302:166–174. doi: 10.1111/j.1574-6968.2009.01848.x. [DOI] [PubMed] [Google Scholar]
- 21.Sato K, Sakai E, Veith PD, et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem. 2005;280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 22.Shah HN, Gharbia SE. Lysis of erythrocytes by the secreted cysteine proteinase of Porphyromonas gingivalis W83. FEMS Microbiol Lett. 1989;52:213–217. doi: 10.1016/0378-1097(89)90199-7. [DOI] [PubMed] [Google Scholar]
- 23.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. Identification and characterization of ós, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One. 2008;3:e3844. doi: 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheets SM, Potempa J, Travis J, Fletcher HM, Casiano CA. Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induce caspase-independent apoptosis. Infect Immun. 2006;74:5667–5678. doi: 10.1128/IAI.01140-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheets SM, Robles-Price AG, McKenzie RM, Casiano CA, Fletcher HM. Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci. 2008;13:3215–3238. doi: 10.2741/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis: Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 28.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 29.Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 30.Tokuda M, Chen W, Karunakaran T, Kuramitsu HK. Regulation of protease expression in Porphyromonas gingivalis. Infect Immun. 1998;66:5232–5237. doi: 10.1128/iai.66.11.5232-5237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tribble GD, Lamont GJ, Progulske-Fox A, Lamont RJ. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J Bacteriol. 2007;189:6382–6388. doi: 10.1128/JB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanterpool E, Roy F, Fletcher HM. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect Immun. 2005a;73:3971–3982. doi: 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanterpool E, Roy F, Fletcher HM. The vimE gene downstream of vimA is independently expressed and is involved in modulating proteolytic activity in Porphyromonas gingivalis W83. Infect Immun. 2004;72:5555–5564. doi: 10.1128/IAI.72.10.5555-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanterpool E, Roy F, Sandberg L, Fletcher HM. Altered gingipain maturation in vimA− and vimE− defective isogenic mutants of Porphyromonas gingivalis. Infect Immun. 2005b;73:1357–1366. doi: 10.1128/IAI.73.3.1357-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanterpool E, Roy F, Zhan W, Sheets SM, Sangberg L, Fletcher HM. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology. 2006;152:3383–3389. doi: 10.1099/mic.0.29146-0. [DOI] [PubMed] [Google Scholar]
- 36.White MJ, He H, Penoske RM, Twining SS, Zahrt TC. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J Bacteriol. 2010;192:1498–1510. doi: 10.1128/JB.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood LF, Ohman DE. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis. J Periodontal Res. 2009;44:1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]