Abstract
Introduction
As part of a clinical proteomics programme focused on diabetes and its complications, it was our goal to investigate the proteome of plasma in order to find improved candidate biomarkers to predict diabetic nephropathy.
Methods
Proteins derived from plasma from a cross-sectional cohort of 123 type 1 diabetic patients previously diagnosed as normoalbuminuric, microalbuminuric or macroalbuminuric were enriched with hexapeptide library beads and subsequently pooled within three groups. Proteins from the three groups were compared by online liquid chromatography and tandem mass spectrometry in three identical repetitions using isobaric mass tags (iTRAQ). The results were further analysed with ingenuity pathway analysis. Levels of apolipoprotein A1, A2, B, C3, E and J were analysed and validated by a multiplex immunoassay in 20 type 1 diabetic patients with macroalbuminuria and 10 with normoalbuminuria.
Results
A total of 112 proteins were identified in at least two out of three replicates. The global protein ratios were further evaluated by ingenuity pathway analysis, resulting in the recognition of apolipoprotein A2, B, C3, D and E as key nodes in the top-rated network. The multiplex immunoassay confirmed the overall protein expression patterns observed by the iTRAQ analysis.
Conclusion
The candidate biomarkers discovered in this cross-sectional cohort may turn out to be progression biomarkers and might have several clinical applications in the treatment and monitoring of diabetic nephropathy; however, they need to be confirmed in a longitudinal cohort.
Electronic supplementary material
The online version of this article (doi:10.1007/s12014-010-9053-0) contains supplementary material, which is available to authorized users.
Keywords: Quantitative proteomics, Diabetic nephropathy, iTRAQ, Protein networks, Pathway analysis, Multiplex immunoassay technology
Introduction
Approximately 8% of type 1 diabetic patients develop end-stage renal disease because of diabetic nephropathy 30 years after diagnosis [1]. As a consequence, diabetic nephropathy is the most common cause of renal failure in the developed world [2, 3]. Diabetic nephropathy advances through a number of recognisable steps from subclinical disease to the first measurable stage of microalbuminuria (MIC), defined as persistent urinary albumin excretion rates (UAER) of 30–300 mg/24 h, to macroalbuminuria/diabetic nephropathy (DMN) with UAER > 300 mg/24 h. DMN is characterised by declining renal function and ultimately end-stage renal disease. Although positive effects on the development and progression of diabetic nephropathy through strict control of blood glucose [4], blood pressure [5] and, in particular, blockade of the renin–angiotensin system (6, 7) have been reported, it still has not been enough to prevent the high incidence of end-stage kidney damage caused by diabetes. The urinary albumin excretion rate is used to identify patients at risk, but only a fraction of all patients with microalbuminuria progress to DMN. New biomarkers that can help in identifying normo- or microalbuminuric patients at risk for progression to DMN are needed as early intervention could further curb progression to DMN and end-stage renal disease.
Access to plasma samples from a cross-sectional study at the Steno Diabetes Center of type 1 diabetic patients with various degrees of diabetic nephropathy formed the basis for this study. The same cohort was previously investigated by fractionation coupled with surface-enhanced laser desorption/ionisation time-of-flight MS (SELDI-TOF-MS), which resulted in the discovery of differential levels of apolipoprotein C1 and apolipoprotein A1 along with transthyretin and cystatin C [8].
In this study, the cohort was further analysed using iTRAQ labelling (Applied Biosystems Inc.) and LC-MS/MS. In order to identify low abundant proteins in the plasma, we subjected the samples to hexapeptide library bead-based fractionation before proteolysis, iTRAQ labelling and LC-MS/MS. The hexapeptide library beads have been reported to have the capacity to reduce the most abundant proteins in plasma whilst simultaneously concentrating the most dilute species [9]. The library contains linear hexapeptides with millions of different ligands. The diversity and number of peptides should result in specific baits for a large number of proteins present in a complex proteome [10, 11]. Because the library has the same quantity of each ligand, hypothetically, the maximum amount of each protein bound is the same under overloading conditions [12, 13]. The iTRAQ labelling simplified the analysis and included protein identification information not present in previous studies. Evaluation of the iTRAQ results by ingenuity pathway analysis identified apolipoproteins A2, B, C3, D and E as central nodes in a network that was highly rated. For that reason, the apolipoprotein profile of type 1 diabetic patients were investigated further in a cohort of patient samples that were chosen based on the rate of loss of kidney function over a time period of 4 years. We compared the ten patients with the fastest rate of decline with those with the slowest rate of decline. Ten samples came from patients with DMN and a high loss of renal filtration based on the measurement of GFR (annual difference in ΔGFR > 5.2 ml min−1/1.73 m2, termed RD), and ten samples came from DMN patients with a relatively slow decline of renal filtration, ΔGFR < 3.3 (termed SlD). In addition, ten samples from normoalbuminuric patients were analysed. The samples were analysed by the cardiovascular disease (CVD) immunoplex assay from Merck (Whitehouse Station, NJ, USA) detecting apolipoprotein A1, A2, B, C3, E, H and J.
By exploring the plasma proteome using a different approach, we aimed to expand our list of candidate biomarkers for diabetic nephropathy. Biomarkers identifying MIC or DMN in this cross-sectional study can be evaluated as potential predictors of the development of diabetic nephropathy in longitudinal studies of diabetic patients.
Materials and Methods
Patient Cohort
The cohort was collected as a part of the GENEX Study in 2004 [14, 15], and a total of 123 samples from Caucasian type 1 diabetic patients examined and diagnosed at Steno Diabetes Center had the adequate volume for the MS analysis. The patients were divided into three groups, normoalbuminuric (N, n = 42), MIC (n = 40) and DMN (n = 41), according to the UACR in at least two out of three consecutive urines collected in the outpatient clinic. The time between DMN diagnosis and sample collection for the GENEX cohort was variable. DMN patients were selected for the presence of DMN irrespective of its duration. All patients in the DMN group, except for three, were being treated to reduce their blood pressure, and consequently, their urinary albumin (U-albumin) at the time of the blood sample collection was not necessarily representative of the stage of the disease. At the time of plasma sampling, 26 out of 41 DMN patients still had high U-albumin levels, whilst 13 had intermediate levels and two had levels low enough to place them in the N group. Of the 15 patients whose levels were lower than at the point of diagnosis, five were put on a short break from their antihypertensive medication and three of them returned to DMN levels before continuing their medication. The two other patients had significant level increases of DMN values and support the original diagnosis. In a follow-up period 4 years after sampling, there were only three patients (two from MIC and one from N) who had progressed to diabetic nephropathy. The low progression rate may be explained by the stable disease state of this cohort at baseline without diabetic nephropathy despite long-standing diabetes.
The three diabetic groups were matched at group level by gender, age and duration of diabetes. Investigations and blood sampling were performed in the morning after an overnight fast and has been thoroughly described previously [8]. Comparisons of clinical data between the three groups were made with Prism 5 software (GraphPad Software, CA, USA). Values of p were two-sided, and a p value <0.05 was considered statistically significant. The Kruskal–Wallis (KW) test was used for ordinal data (gender). The distributions of continuous data were tested for normality by the D’Agostino–Pearson omnibus normality test. If distributions were normal, data were compared with one-way ANOVA, and if distributions were not normal, data were compared with a KW test.
The study was approved by the local ethics committee and all patients gave their informed consent.
Protein Purification and iTRAQ Sample Labelling
Patient plasma was fractioned with ProteoMiner beads as described previously [8]. Plasma proteins within each group were pooled and 100 μg protein was aliquoted three times for triplicate studies. Each replicate was precipitated with six times the volume of cold acetone separately, yielding a total of nine individual samples. The samples were reduced and blocked as suggested by the manufacturers. Proteins were digested overnight with Trypsin (2%, w/w), and the resulting peptides were subsequently labelled using iTRAQ 4-plex. The three samples from the DMN group were labelled with iTRAQ 114, the MIC group samples iTRAQ 115 and the N group samples iTRAQ 116, and dried in a vacuum centrifuge. The samples were reconstituted in 500 μL SCX loading buffer (Applied Biosystems) and mixed 1:1:1 to give a final of three mixed samples (each containing one iTRAQ 114-labelled DMN sample, one 115 iTRAQ-labelled MIC sample and so forth). The pH of the samples was adjusted to 2.5–3 with HCl and fractioned on a SCX column with 50, 100, 150, 200, 250, 300, 350, 400, 500 mM and 1 M KCl. The pH values of the fractions were adjusted to 2–3 and cleaned on C18 Stagetips (Proxeon, Odense, Denmark) using 50 μL 100% ACN as wetting solution, 50 μL 0.1% TFA for equilibrating the tips and washing the sample after loading, and, finally, elution of the peptides with 50 μL 0.1% TFA and 50% ACN. Afterwards, the samples were completely dried.
Nano-liquid Chromatography Tandem Mass Spectrometry
Peptides from the SCX fractions were dissolved in 0.3 μL 100% formic acid and diluted to 5 μL in 0.05% TFA. The peptides were loaded onto an EASY-nano LC system (Proxeon). Peptides were loaded directly onto a 20-cm, 100-μm inner diameter and 360-μm outer diameter, ReproSil-Pur C18 AQ 3 μm (Dr. Maisch, Ammerbuch-Entringen, Germany) reversed phase capillary column. The peptides were eluted using a gradient from 100% phase A (0.1% formic acid) to 40% phase B (0.1% formic acid, 80% acetonitrile) over 100 min at 200 nL/min directly into a LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA, USA). The LTQ-Orbitrap XL was operated in a data-independent mode automatically switching between MS, MS/MS and higher-energy collision dissociation (HCD) using a threshold of 30,000 for ion selection. For each MS scan, the two most abundant precursor ions were selected for fragmentation using MS/MS (normalised collision energy, 35; activation time, 30 ms) to facilitate peptide identification and then subsequently HCD (normalised collision energy, 55) to obtain reliable iTRAQ quantitative data [16]. Raw data were viewed in Xcalibur v2.1 (Thermo Scientific).
Data Processing and Protein Sequence Database Searching
The data were processed using Proteome Discoverer, version 1.1.0.228. The files were subsequently submitted to an in-house MASCOT server (version 2.2.05, Matrix Science Ltd., London, UK) for database searching through the Proteome Discoverer programme. The data were searched against the human sequence database (NCBInr 20100602 (11186807 sequences; 3815639892 residues); taxonomy: Homo sapiens (human, 232199 sequences)). The search was performed choosing trypsin as specific enzyme. A maximum of one missed cleavage was allowed. Oxidation (M), iTRAQ 4plex (K) and iTRAQ 4plex (N-term) were chosen as variable modifications. The data obtained on the LTQ-orbitrap XL were searched with a peptide mass tolerance of 10 ppm and a fragment mass tolerance of 0.8 Da. Protein ratios were normalised using the overall median ratio for all the peptides in the sample for each separate ratio in every individual experiment. The ratio for a given protein was calculated by taking the average of all the peptides ratios that identified the protein. The final list of protein ratios was an average of the protein ratios of the three experiments and consisted only of proteins discovered in at least two of the three experiments.
Ingenuity Pathway Analysis
The iTRAQ results were evaluated using ingenuity pathway analysis (IPA; Ingenuity Systems, Mountain View, CA; www.ingenuity.com). This software analyses protein expression data in the context of known biological response and regulatory networks as well as other higher-order response pathways. IPA constructs hypothetical protein interaction clusters on the foundation of a regularly updated Ingenuity Pathways Knowledge Base, which is a very large database that consists of millions of individual relationships between proteins gathered from the biologic literature [17]. The data set that contained the differentially expressed proteins identified in the iTRAQ experiment was converted by IPA to ‘fold change’ and uploaded into IPA. No expression value cutoff was selected, and we were interested in looking at both up- and downregulated proteins. Hypothetical networks were generated from these proteins and other proteins from the database that were needed to fill out a protein cluster. Network generation was optimised for the inclusion of as many proteins from the input expression profile as possible and aimed for highly connected networks.
IPA computes a score for each possible network according to the fit of that network to the input proteins. The score is calculated as the negative base-10 logarithm of the p value that indicates the probability of the input proteins in a given network being found together as a result of random chance. Consequently, scores of 2 or higher have at least a 99% confidence of not being generated by random chance alone [17].
Bioplex Assay
Concentrations of Apo A1, A2, B, C3, E, H and J were simultaneously evaluated using a multiplex bead-based sandwich immunoassay kits (Novagen WideScreen ™ Human CVD Panel 1, Merck Chemicals Ltd. Nottingham, UK). Assays were performed following the manufacturer’s instructions. Briefly, seven distinct sets of fluorescently dyed beads loaded with capture monoclonal antibodies specific for each apolipoprotein to be tested were used. Plasma samples diluted 1:2,500 with dilution buffer (30 μL/well) or standards (30 μL/well) along with 10 μL of biotinylated antigens in blocking buffer were incubated with 10 μl of pre-mixed bead sets into the wells of a pre-wet 96-well microtitre plate. After incubation and washing, 20 μL of fluorescent detection antibody mixture (Streptavidin PE) was added for 30 min and the samples were washed and resuspended in assay buffer. The formation of different sandwich immunocomplexes on distinct bead sets was measured and quantified using the Bio-Plex Protein Array System (Bio-Rad Laboratories). A 50 μL volume was sampled from each well and the fluorescent signal of a minimum of 100 beads per region (chemokine/apolipoprotein) was evaluated and recorded. Values presenting a coefficient of variation beyond 15% were discarded before the final data analysis.
Data Analysis and Statistics
The clinical data were evaluated by the Prism software. Bio-plex data were analysed using the Bio-Plex Manager software version 3.0 (Bio-Rad Laboratories). Standard levels between 80% and 120% of the expected values were considered to be accurate and were used. Values were expressed as nanograms per millilitre and presented as mean (SD).
Results
Clinical Data
The clinical data of the GENEX cohort are summarised in Table 1. Patient groups in the cross-sectional cohort were matched with respect to gender, duration of diabetes and body mass index (BMI), but differed slightly by age (p = 0.01). The DMN group had significantly lower estimated glomerular filtration rates (eGFR) compared to the other groups (p < 0.0001), whilst equivalent eGFR values were observed in the N and MIC groups. There were no significant differences in systolic blood pressure or diastolic blood pressure for comparisons of all groups or for levels of serum cholesterol between the groups. Haemoglobin A1c (HbA1c) differed significantly for an all group comparison (p = 0.02); this was caused by a significant difference between the N and DMN groups.
Table 1.
Clinical data of the GENEX cohort differentiated according to the level of albuminuria
| N | MIC | DMN | p value | |
|---|---|---|---|---|
| n, total | 42 | 40 | 41 | |
| Gender, male/female | 19/23 | 18/22 | 22/19 | 0.25 |
| Age (years) | 56 (11) | 55 (11) | 49 (10)a | 0.01b |
| DM duration (years) | 36 (11) | 36 (11) | 34 (10) | 0.58 |
| BMI (kg/m²) | 24.5 (2.8) | 25.2 (3.7) | 25.3 (4.5) | 0.62 |
| HbA1c (%) | 8.2 (1) | 8.8 (1.2) | 8.9 (1.1)a | 0.02* |
| Creatinine (μmol/l)b | 91 (82–96) | 90 (81–102) | 124 (99–172)a,c | <0.0001* |
| U-albumin (mg/g)b,d | 6 (4–8) | 23 (9–59) | 453 (195–936)a,c | <0.0001* |
| eGFR (ml min−1/1.73 m²) | 70.1 (10.7) | 68.6 (11.4) | 49.5 (18.5)a,c | <0.0001* |
| Cholesterol (mmol/l) | 4.8 (0.8) | 5 (1) | 4.9 (1) | 0.55 |
| Systolic BP (mmHg) | 138 (23) | 140 (23) | 144 (19) | 0.42 |
| Diastolic BP (mmHg) | 74 (10) | 73 (12) | 78 (10) | 0.09 |
Numbers are presented as mean (SD)
n numbers, DM diabetes mellitus, HbA1c haemoglobin A1c, U-albumin urinary albumin levels normalised to creatinine levels, eGFR estimated glomerular filtration rate, BP blood pressure
*p < 0.05
aIndicates that means differed significantly between DMN group and N group
bNumbers are presented as median (IQR 25%–75%)
cIndicates that means differed significantly between DMN group and MIC group
dSome patients had U-albumin levels reduced by antihypertensive medication which was not stopped when spot urine samples were collected for the study
Quantitative Comparison of the Three Patient Samples
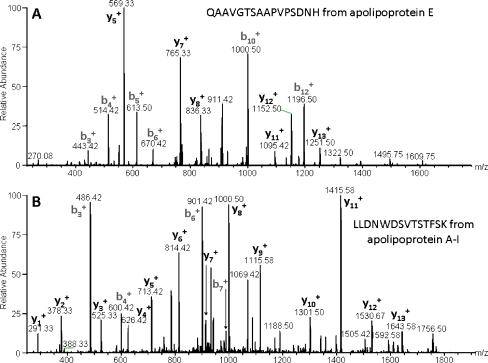
Protein identifications were initially accepted when based on peptide identifications with Mascot scores of ≥25 and a false discovery rate of <1%. This resulted in the identification of 3,365, 3,667 and 3,495 peptides, respectively, from the iTRAQ triplicate experiments. In total, 130, 133 and 122 proteins were identified in experiments 1, 2 and 3, respectively. The relative quantitation was measured as two pairwise ratios: the MIC group against the DMN group (iTRAQ115/iTRAQ114) and the N group against the DMN group (iTRAQ116/iTRAQ114). A total of 112 proteins were identified in at least two out of three iTRAQ runs. The molecular identity and abundance ratios of the 112 proteins are listed in Electronic Supplementary Materials. Examples of MS/MS spectra of peptides from apo E and A1 are presented in Fig. 1.
Fig. 1.
a MS/MS spectrum of the C-terminal peptide from apolipoprotein E (sequence QAAVGTSAAPVPSDNH). The assignment of the peptide is illustrated by the y- and b-ions in the spectrum. b MS/MS spectrum of the peptide with the sequence LLDNWDSVTSTFSK from apolipoprotein A1
Ingenuity Pathway Analysis
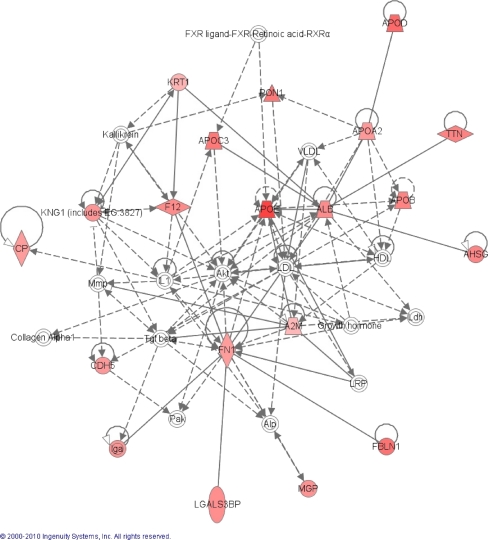
Figures 2 and 3a shows one of the largest protein clusters that were generated by the pathway analysis of the protein ratios between N/DMN. This network consists of a cluster of 35 proteins, of which 15 were not included in our list. The network was suggested to be involved in cardiovascular disease, genetic disorders and metabolic diseases, and had a score of 36. Apo A2, B, C3, D and E were all included in this protein network, and the results inspired us to look further into these apolipoproteins in plasma.
Fig. 2.
Top-rated network generated by ingenuity pathway analysis on the N/DMN ratio. Red nodes indicate that the protein is upregulated in the DMN group compared to the N group. Stronger red colours indicate higher regulation of the complex
Fig. 3.
a Network shapes. Figure legend for the IPA network. https://analysis.ingenuity.com/pa/info/help/help.htm#legend.htm. b Relationships. Figure legend for the molecular relationships in the IPA network. https://analysis.ingenuity.com/pa/info/help/help.htm#legend.htm
Multiplex Immunoassay Results
For further analysis of the aforementioned apolipoproteins and validation of the iTRAQ results, 30 plasma samples were selected for quantitative measurements of apolipoproteins by a multiplex immunoassay. The patient samples were chosen based on the rate of loss of kidney function over a time period of 4 years. Ten samples came from patients with DMN and a high loss of renal filtration based on the measurement of GFR (annual difference in ΔGFR > 5.2, termed RD), and ten samples came from DMN patients with a relatively slow decline of renal filtration, ΔGFR < 3.3 (termed SlD). In addition, ten samples from normoalbuminuric patients were analysed.
The clinical data for the 30 patients chosen for the multiplex immunoassay is listed in Table 2.
Table 2.
Clinical data for patients chosen for the multiplex immunoassay
| RD | SlD | N | p value | |
|---|---|---|---|---|
| n, total | 10 | 10 | 10 | |
| GFR 2004a (mL/min) | 60.76 (11.3) | 54.7 (7.9) | – | 0.1051 |
| ΔGFRa (mL/min) over 4 years | 6.61 (1) | 2.17 (1.6) | – | 0.0002* |
| Age (years) | 42.20 (8.3) | 52.50 (9.2) | 57.90(11.7) | 0.0062* |
| Gender, %male | 50 | 50 | 20 | 0.2987 |
| Diabetes duration (years)a | 27.90 (11.5) | 37.10 (6) | 41.40 (10.3) | 0.017* |
| Systolic BP (mmHg) | 137.4 (18.8) | 133.0 (26.3) | 138.1 (18.9) | 0.9757 |
| Diastolic BP (mmHg) | 76.70 (9.5) | 69.40 (16) | 76.30 (11.4) | 0.3859 |
| BMI (kg/m2) | 25.86 (4.2) | 26.31 (6.5) | 24.01 (1.7) | 0.5544 |
| HbA1C (%) | 8.680 (1) | 8.480 (1) | 7.770 (0.8) | 0.0822 |
| Cholesterol (mmol/L) | 4.660 (0.6) | 4.920 (0.8) | 5.030 (5) | 0.5188 |
| Triglycerides (mmol/L) | 1.132 (0.4) | 1.067 (0.3) | 0.9870 (0.5) | 0.5569 |
Numbers are presented as mean (SD)
n numbers, eGFR estimated glomerular filtration rate, BP blood pressure, HbA1c haemoglobin A1c
*p < 0.05
aNon-parametric data were evaluated by a Kruskal–Wallis F test or a Mann–Whitney test when only two values were compared
There was no significant difference in GFR measured in 2004 between the RD and SlD groups. The distribution of gender, systolic and diastolic blood pressure, BMI, HbA1c, cholesterol and triglyceride values were not significant different across all three groups. The RD and SlD groups varied in the decline of renal function (ΔGFR) over a subsequent 4-year period (p = 0.0002). At follow-up, the three groups varied slightly in age and duration of diabetes. This was caused by one young patient (aged 27) in the RD group and one patient with only 2 years of diabetes also in the RD group.
Plasma concentrations of apo A1, A2, B, C3, E, H and J were detected in all the patients. Table 3 shows the concentrations of the measured apolipoproteins in the N, SlD and RD groups. The concentrations of the apolipoproteins did not differ between the three groups. Apo A1 concentrations correlated with systolic blood pressure (Spearman ρ coefficient = −0.45, p = 0.014), Apo A2 with triglycerides (Spearman ρ coefficient = 0.41, p = 0.0245), Apo B with age (Spearman ρ coefficient = 0.3642, p = 0.0478) and Apo C3 with cholesterol (Spearman ρ coefficient = 0.4361, p = 0.0160). There was no correlation between ΔGFR and concentrations of apolipoproteins in the SlD and RD groups, nor any between the GFR in 2004 and apolipoproteins in all three groups.
Table 3.
Concentrations of Apo A1, A2, B, C3, E, H and J in the plasma of the diabetic patients divided by GFR
| RD | SlD | N | p value | |
|---|---|---|---|---|
| Apo A1 (ng/mL) | 4.289e+006 (1.752e+006) | 4.653e+006 (1.715e+006) | 4.925e+006 (2.021e+006) | 0.7427 |
| Apo A2 (ng/mL) | 184,582 (47,323) | 237,641 (177,992) | 167,090 (66,080) | 0.7145 |
| Apo B (ng/mL) | 1.528e+006 (137,227) | 1.486e+006 (104,370) | 1.537e+006 (100,729) | 0.5790 |
| Apo C3 (ng/mL) | 66,449 (20,720) | 73,448 (48,535) | 56,741 (29,542) | 0.6600 |
| Apo E (ng/mL) | 32,008 (14,590) | 38,629 (9,400) | 37,414 (7,629) | 0.3682 |
| Apo H (ng/mL) | 164,944 (32,147) | 182,433 (95,217) | 129,301 (46,866) | 0.1048 |
| Apo J (ng/mL) | 137,605 (17,021) | 139,465 (29,007) | 136,150 (22,101) | 0.9502 |
Numbers are presented as mean (SD)
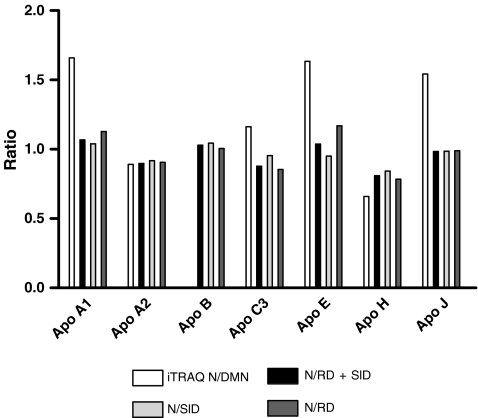
In order to compare the concentrations of the apolipoproteins from the multiplex immunoassay with the ratios of the apolipoproteins from the iTRAQ experiment, the average of the specified protein from the N group was divided by the average of the two macroalbuminuric (N/SlD + RD) groups and the SlD (N/SlD) and RD (N/RD) groups separately (Fig. 4). The ratios of the apolipoprotein in the multiplex immunoassay had, overall, the same pattern as first revealed by the iTRAQ analysis, except for apo C3 and apo J where the reversed relationship was seen between iTRAQ and the multiplex immunoassay.
Fig. 4.
Ratios of the lipoproteins identified by iTRAQ- and Luminex-based assays. The iTRAQ ratio is between the N/DMN and the Luminex ratio is the N/SlD, N/RD and N/SlD + RD ratios
Discussion
Identification of biomarkers in the blood of patients with high risk of developing diabetic nephropathy may have several clinical applications, including more aggressive control of glucose or arterial blood pressure, earlier onset of renoprotective treatment and a more tailored management of the disease.
Although several studies of urine, plasma and serum from T2D patients have been conducted [18–21], the plasma of T1D patients has not been fully examined. In this report, we present data to show that plasma obtained from three different stages of diabetic nephropathy in T1D patients contains candidate biomarkers for diabetic nephropathy.
Plasma samples from a cross-sectional study conducted at Steno Diabetes Center in 2004 formed the foundation of the investigations presented here. Previously, the plasma samples were fractioned with ProteoMiner beads and analysed on SELDI-TOF-MS [8]. The SELDI-TOF-MS approach relied on a side-by-side comparison of spectra generated from each patient and revealed a number of protein peaks significantly different between patients divided into groups according to albuminuria. The disadvantage of this methodology was the time-consuming job of identifying the various protein peaks in the spectra by immunoprecipitation, sequencing and Western blots. The combination of isobaric labelling using iTRAQ and LC-MS/MS offered a way to overcome this situation. Peptides are identified in the MS/MS mode, whilst quantitation is obtained from the iTRAQ reporter ion using higher-energy collision dissociation (HCD) and collision-induced dissociation (CID). A total of 112 proteins were identified in at least two out of three repetitions of the experiments. The protein number is relatively low compared to other published studies, such as those reported for a head and neck cancer biomarker discovery study and protein expression profiling of plasma from patients with placental diseases where 811 and 166 proteins were identified, respectively [22, 23]. An explanation could be the initial treatment with ProteoMiner beads; previously, Pernemalm et al. [24] evaluated the ProteoMiner technique on crude plasma and identified only 150 unique proteins by direct LC-MS/MS and GeLC-MS/MS with a start plasma volume comparable to ours. Additionally, a recent study demonstrated that iTRAQ isobaric tags significantly increased the average ion charge state of peptides, resulting in a considerable reduction in the number of peptides identified from a given sample when using CID and HCD peptide fragmentation [25].
Some of the more interesting proteins identified in this study are the lipoproteins where the majority are present in higher levels in MIC and N compared to DMN. Evaluation of the iTRAQ data by ingenuity pathway analysis identified apo A1, A2, B, C3, E and J (Clusterin) to contribute in the top-rated network (Fig. 2). Diabetes has been shown to be associated with the amounts of different classes of apolipoproteins, and accordingly, the apolipoproteins might contribute to the development of diabetic nephropathy and cardiovascular disease [26]. Apo B, Apo C1 and C3 are according to Hirano et al. [27] increased in patients with diabetic nephropathy compared to healthy individuals. We observed the same tendency in our results for apo C3 where higher levels of this protein were found in the groups with macroalbumninuria. Several reports have investigated different isoforms of apo E caused by three different alleles of the gene coding for apo E [28–30], and it has been suggested that the apo E4 isoform protects against diabetic nephropathy whilst the apo E2 isoform promotes the disease [31, 32]. We did not see any differences in the overall level of apo E in the different patient groups, suggesting that diagnostic potential of the protein potentially should be found in the different isoforms in contrast to the generally protein level. Apo A1 levels were lower in patients with macroalbuminuria in our study, although not statistically significant. Tolonen et al. [33] reported that low apo A1 levels predicted the progression of patients with macroalbuminuria to end-stage renal disease, and it might be reasonable to speculate that low levels of this protein in some way is associated to the development and progression of diabetic nephropathy. Whether the abnormal levels of the apolipoproteins are causing the development of diabetic nephropathy and therefore could be used as potential predictive markers or are just a consequence of the disease is difficult to say based on these cross-sectional studies presented here. The issue would ideally be addressed through further investigation in a longitudinal study.
To verify our results obtained with the iTRAQ investigation and ingenuity pathway analysis, we examined apo A1, A2, C3, E, H and J on an orthogonal platform in a series of patients with macroalbuminuria that had either a fast decline in kidney function (GFR, the RD group) and patients with macroalbuminuria and a slower decline in GFR (the SlD group) over 4 years. The ratios obtained in the iTRAQ analysis and the multiplex immuno-analysis appear quite similar. Differences could be explained by the initial treatment with ProteoMiner beads of the samples for the iTRAQ experiment, whereas the multiplex immunoassay was conducted on untreated plasma. Another explanation could be the difference in the number of plasma samples that constitutes the pool, i.e. the fact that the iTRAQ analysis is conducted in pools of plasma samples from a larger group of patients and therefore individuals with very high or low levels of protein would be diluted. In the multiplex immunoassay experiment, one patient had very high levels of apo A1, A2, C3 and H, and this resulted in a general higher average of these proteins. Another explanation could be potentially different isoforms of the apolipoproteins, with differences in which isoforms can be detected by the method employed. A recent paper by Borges et al. [34] reviews the microheterogenity of the human proteome. Evidently, polymorphisms, transcript variants and posttranslational modifications give rise to structurally similar but functionally distinct proteins whose detection is platform-dependent.
Originally, we hypothesised that apolipoprotein levels would be different between patients with macroalbuminuria that had a fast decline in kidney function (GFR, the RD group) and patients with macroalbuminuria and a lower fall in GFR (the SlD group). This was not observed in this study, possibly because of the relatively low number of samples in each group.
The candidate biomarkers discovered in this cross-sectional cohort may turn out to be progression biomarkers, but they need to be confirmed in a longitudinal cohort. The patients in the cohort all represent a stable disease state due to their long duration, and the proteins discovered in this study are indeed indicative of their current state, but temporal information about the changes needs to be investigated. The nature of the current cohort with a spread in U-albumin in the DMN and MIC group brings robustness and persistence to the actually obtained candidate biomarkers and reflects everyday clinical setting.
In this study, we investigated the plasma protein profiles from T1D patients with various degrees of albuminuria using hexapeptide library fractionation, iTRAQ labelling and LC-MS/MS, with the result of increasing our list of candidate biomarkers for diabetic nephropathy. Future research is aimed at analysing larger groups of samples. After establishing a model based on proteomic patterns for the separation of the three groups, the model will be validated in longitudinal sample sets to determine the diagnostic and predictive value of the obtained protein profiles. It is our goal to discover biomarkers able to predict, detect and monitor the progression of DMN and the effect of reno-protective intervention in diabetic patients.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
(DOC 182 kb)
Acknowledgements
This work was supported by funding from the Juvenile Diabetes Research Foundation, Paul and Erna Sehested Hansen Foundation, Steno Diabetes Center and Novo Nordisk A/S. We thank Peter Iversen for technical assistance with the Luminex immunoassays.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- MIC
Microalbuminuria
- UAER
Urinary albumin excretion rate
- DMN
Diabetic nephropathy
- SELDI-TOF-MS
Surface-enhanced laser desorption/ionisation time-of-flight mass spectrometry
- CVD
Cardiovascular disease
- Apo
Apolipoprotein
- N
Normoalbuminuric
- UACR
Urinary albumin creatinine ratio
- U-albumin
Urinary albumin
- GFR
Glomerular filtration rate
- KW
Kruskal–Wallis
- IPA
Ingenuity pathway analysis
- RD
Rapid decline
- SlD
Slow decline
- BMI
Body mass index
- HbA1c
Haemoglobin A1c
- HCD
Higher-energy collision dissociation
- CID
Collision-induced dissociation
References
- 1.Finne P, Reunanen A, Stenman S, Groop P-H, Gronhagen-Riska C. Incidence of end-stage renal disease in patients with type 1 diabetes. JAMA. 2005;294:1782–87. [DOI] [PubMed]
- 2.Daneman D. Type 1 diabetes. Lancet. 2006;367:847–58. [DOI] [PubMed]
- 3.Cameron JS. The discovery of diabetic nephropathy: from small print to centre stage. J Nephrol. 2006;19:75–87. [PubMed]
- 4.Kilpatrick ES, Rigby AS, Atkin SL. HbA1c variability and the risk of microvascular complications in type 1 diabetes: data from the DCCT. Diabetes Care. 2008;31:2198–202. [DOI] [PMC free article] [PubMed]
- 5.Mogensen CE. Long-term antihypertensive treatment inhibiting progression of diabetic nephropathy. Brit Med J. 1982;285:685–88. [DOI] [PMC free article] [PubMed]
- 6.Ahmad J. Renin-angiotensin system blockade in diabetic nephropathy. Diab Metab Synd: Clin Res Rev. 2008;2:135–58.
- 7.Lewis EJ, Hunsicker LG, Bain RP, Rohde RD, and The Collaborative Study, G. The effect of angiotensin-converting-enzyme inhibition on diabetic nephropathy. N Engl J Med. 1993;329:1456–62. [DOI] [PubMed]
- 8.Overgaard A, Hansen H, Lajer M, Pedersen L, Tarnow L, Rossing P, et al. Plasma proteome analysis of patients with type 1 diabetes with diabetic nephropathy. Proteome Science. 2010;8:4. [DOI] [PMC free article] [PubMed]
- 9.Boschetti E, Righetti PG. The ProteoMiner in the proteomic arena: a non-depleting tool for discovering low-abundance species. J Proteomics. 2008;71:255–64. [DOI] [PubMed]
- 10.Lam KS, Lebl M, Krchnak V. The ‘one-bead-one-compound’ combinatorial library method. Chem Rev. 1997;97:411–48. [DOI] [PubMed]
- 11.Furka Á, Sebestyén F, Asgedom M, Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991;37:487–93. [DOI] [PubMed]
- 12.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein equalizer technology: the quest for a democratic proteome. Proteomics. 2006;6:3980–92. [DOI] [PubMed]
- 13.Righetti PG, Castagna A, Antonioli P, Boschetti E. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis. 2005;26:297–319. [DOI] [PubMed]
- 14.Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, et al. Urinary proteomics in diabetes and CKD. J Am Soc Nephrol. 2008;19:1283–90. [DOI] [PMC free article] [PubMed]
- 15.Nielsen SE, Sugaya T, Tarnow L, Lajer M, Schjoedt KJ, Astrup AS, et al. Tubular and glomerular injury in diabetes and the impact of ACE inhibition. Diabetes Care. 2009;32:1684–88. [DOI] [PMC free article] [PubMed]
- 16.Rewitz KF, Larsen MR, Lobner-Olesen A, Rybczynski R, O’Connor MB, Gilbert LI. A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem Mol Biol. 2009;39:475–83. [DOI] [PubMed]
- 17.Hoorn EJ, Hoffert JD, Knepper MA. Combined proteomics and pathways analysis of collecting duct reveals a protein regulatory network activated in vasopressin escape. J Am Soc Nephrol. 2005;16:2852–63. [DOI] [PMC free article] [PubMed]
- 18.Apakkan Aksun S, Özmen D, Özmen B, Parildar Z, Mutaf I, Turgan N, et al. β2-Microglobulin and cystatin C in type 2 diabetes: assessment of diabetic nephropathy. Exp Clin Endocrinol Diabetes. 2003;112:195–200. [DOI] [PubMed]
- 19.Gu W, Zou LX, Shan PF, Chen YD. Analysis of urinary proteomic patterns for diabetic nephropathy by ProteinChip. Proteom Clin APPL. 2008;2:744–50. [DOI] [PubMed]
- 20.Hong CY, Chia KS. Markers of diabetic nephropathy. J Diabetes its Complicat. 1998;12:43–60. [DOI] [PubMed]
- 21.Hyun-Jung K, Hyun-Syuk Y, Chan-Wha K. Proteomics in diabetic nephropathy. Proteom Clin APPL. 2008;2:301–11. [DOI] [PubMed]
- 22.Ralhan R, DeSouza LV, Matta A, Chandra Tripathi S, Ghanny S, Datta Gupta S, et al. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:1162–73. [DOI] [PMC free article] [PubMed]
- 23.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, WalkerA, et al. Differential protein expression profiling by iTRAQ-2dlc-ms/ms of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res. 2006;5:1143–54. [DOI] [PubMed]
- 24.Pernemalm M, Orre LM, Lengqvist J, Wikstrom P, Lewensohn R, Lehtio J. Evaluation of three principally different intact protein prefractionation methods for plasma biomarker discovery. J Proteome Res. 2008;7:2712–22. [DOI] [PubMed]
- 25.Thingholm T, Palmisano G, Kjeldsen F, Larsen MR. Undesirable charge-enhancement of isobaric tagged phosphopeptides leads to reduced identification efficiency. J Proteome Res. 2010;9:4045–52. [DOI] [PubMed]
- 26.Rutledge JC, Ng KF, Aung HH, Wilson DW. Role of triglyceride-rich lipoproteins in diabetic nephropathy. Nat Rev Nephrol. 2010;6:361–70. [DOI] [PubMed]
- 27.Hirano T, Sakaue T, Misaki A, Murayama S, Takahashi T, Okada K, et al. Very low-density lipoprotein–apoprotein CI is increased in diabetic nephropathy: comparison with apoprotein CIII. Kidney Int. 2003;63:2171–77. [DOI] [PubMed]
- 28.Onuma T, Laffel LM, Angelico MC, Krolewski AS. Apolipoprotein E genotypes and risk of diabetic nephropathy. J Am Soc Nephrol. 1996;7:1075–78. [DOI] [PubMed]
- 29.Araki S, Moczulski DK, Hanna L, Scott LJ, Warram JH, Krolewski AS. APOE polymorphisms and the development of diabetic nephropathy in type 1 diabetes: results of case-control and family-based studies. Diabetes. 2000;44:2190–95. [DOI] [PubMed]
- 30.Kimura H, Suzuki Y, Gejyo F, Karasawa R, Miyazaki R, Suzuki S, et al. Apolipoprotein E4 reduces risk of diabetic nephropathy in patients with NIDDM. Am J Kidney Dis. 1998;31:666–73. [DOI] [PubMed]
- 31.Eto M, Horita K, Morikawa A, Nakata H, Okada M, Saito M, et al. Increased frequency of apolipoprotein ε2 allele in non-insulin dependent diabetic (NIDDM) patients with nephropathy. Clin Genet. 1995;48:288–92. [DOI] [PubMed]
- 32.Liberopoulos E, Siamopoulos K, Elisaf M. Apolipoprotein E and renal disease. Am J Kidney Dis. 2004;43:223–33. [DOI] [PubMed]
- 33.Tolonen N, Forsblom C, Thorn L, Wadén J, Rosengård-Bärlund M, Saraheimo M, et al. Lipid abnormalities predict progression of renal disease in patients with type 1 diabetes. Diabetologia. 2009;52:2522–30. [DOI] [PubMed]
- 34.Borges CR, Rehder DS, Jarvis JW, Schaab MR, Oran PE, Nelson RW. Full-length characterization of proteins in human populations. Clin Chem. 2010;56:202–11. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 182 kb)