Abstract
Rationale: Chitinases are enzymes that cleave chitin, which is present in fungal cells. Two types of human chitinases, chitotriosidase and acidic mammalian chitinase, and the chitinase-like protein, YKL-40, seem to play an important role in asthma. We hypothesized that exposure to environmental fungi may modulate the effect of chitinases in individuals with asthma.
Objectives: To explore whether interactions between high fungal exposure and common genetic variants in the two chitinases in humans, CHIT1 and CHIA, and the chitinase 3-like 1 gene, CHI3L1, are associated with severe asthma exacerbations and other asthma-related outcomes.
Methods: Forty-eight single nucleotide polymorphisms (SNPs) in CHIT1, CHIA, and CHI3L1 and one CHIT1 duplication were genotyped in 395 subjects and their parents as part of the Childhood Asthma Management Program. Household levels of mold (an index of fungal exposure) were determined on house dust samples. We conducted family-based association tests with gene–environment interactions. Our outcome was severe exacerbation, defined as emergency department visits and hospitalizations from asthma over a 4-year period, and our secondary outcomes included indices of lung function and allergy-related phenotypes.
Measurements and Main Results: Of the 395 subjects who had mold levels at randomization, 24% (95 subjects) had levels that were greater than 25,000 units per gram of house dust (high mold exposure). High mold exposure significantly modified the relation between three SNPs in CHIT1 (rs2486953, rs4950936, and rs1417149) and severe exacerbations (P for interaction 0.0010 for rs2486953, 0.0008 for rs4950936, and 0.0005 for rs1417149). High mold exposure did not significantly modify the relationship between any of the other variants and outcomes.
Conclusions: Environmental exposure to fungi, modifies the effect of CHIT1 SNPs on severe asthma exacerbations.
Keywords: chitinase, asthma, CHIA, CHIT1, CHI13L1
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Chitinases are enzymes that cleave chitin, which is present in fungal cells. Two types of human chitinases, chitotriosidase and acidic mammalian chitinase, and the chitinase-like protein, YKL-40, seem to play a role in asthma.
What This Study Adds to the Field
Fungal levels may modify the effect of variants in the chitinase gene, CHIT1, and chitinases may play an important role in asthma.
The pathogenesis of asthma, which affects up to 300 million people of all ages in the world and 15 million people in the United States (1), is mediated by immunologic responses that parallel those induced by parasitic infections. Furthermore, fungal exposure seems to be associated with severe asthma and admission to the intensive care unit for asthma (2, 3). Mammalian chitinases and a chitinase homolog that lacks chitinase activity may contribute to the pathogenesis of T-helper 2 (Th2) immune responses and thereby play an important role in asthma (4–6). Chitin is a polysaccharide that is present in fungal cells, crustaceans, insects, and parasitic nematodes, and chitinases cleave chitin (7–9). Chitin is present in the cell walls of fungi and provides architectural reinforcement to the cell walls; thus, exposure to fungi correlates with exposure to chitin (10). Chitinases seem to play an important role in Th2-mediated inflammation and allergic diseases, such as asthma. A study in rats found that intratracheal infection with a fungus, Cryptococcus neoformans, induces generalized chitinase activity in bronchoalveolar lavage fluid and lung, suggesting a link between respiratory fungal infection and asthma through the induction of chitinase (11).
Although chitin does not exist in humans, two chitinases, acidic mammalian chitinase (CHIA) and chitotriosidase (CHIT1), have been described in humans (7). A third protein, chitinase-like protein YKL-40 (also known as “human cartilage glycoprotein 39” and “chitinase 3-like 1”) also seems to play an important role in asthma (6). These two chitinases perform important defensive functions against chitin-containing pathogenic organisms in both the gastrointestinal tract and lungs. CHIT1 is primarily expressed in the lung, whereas CHIA is highly expressed in the lungs of patients with asthma, but not in normal subjects. In studies of mice, CHIA seems to play an influence the development of bronchial asthma (4). Inhibition of CHIA in mice leads to an abrogated Th2 inflammation, reduced bronchial hyperreactivity, and lower eosinophil counts (4). Studies have also found that chitin may protect mice from asthma (12). Levels of YKL-40 are higher in patients with asthma than healthy persons, and patients with more severe asthma have higher levels of YKL-40 (6).
We have previously reported that variants in CHITI, CHIA, and CHI3L1 are not associated with asthma or asthma-related phenotypes (13). However, we hypothesized that the effects of these genes may only be increased in the appropriate environmental context, in this case exposure to high levels of house dust fungi.
The objectives of this study were to assess whether fungal exposure modulates the effect of variants in CHIT1, CHIA, and CHI3L1 and one CHIT1 duplication on emergency department visits and hospitalizations from asthma. We hypothesized that exposure to fungi, as a source of environmental chitin exposure, would influence the association of single nucleotide polymorphisms (SNPs) in the genes of chitinases and chitinase-like protein because these genes have been found to be associated with asthma severity, fungal exposure is known to be associated with asthma symptoms, and higher fungal levels suggest higher chitin levels. Our secondary objectives were to assess whether fungal exposure modulates the association of these SNPs and changes in lung physiology that are associated with asthma or allergy-related phenotypes.
METHODS
Study Population
We used data from the Childhood Asthma Management Program (CAMP), a multicenter trial that enrolled children between the ages of 5 and 12 years with mild to moderate persistent asthma between 1993 and 1995 (14). We included data from 395 white subjects and their parents. Subjects were randomly assigned to receive budesonide, nedocromil, or placebo and were followed every 2 to 4 months for 4 years to study the long-term use of the medications. Details of this study have been previously published (14). The institutional review board at each of the eight participating institutions approved the study and parents or guardians of the subjects gave informed consent (14).
Measures
Mold measures (an index of fungal exposure) in the home environment were taken by research assistants. Details have been described previously (14, 15). Briefly, during a study visit, a CAMP-certified technician used a vacuum cleaner (Douglas Redivac model 6735; Scott-Fetzer, Walnut Ridge, AZ) fitted with a disposable filter to collect a dust sample 2 m2 from the upper part of the patient's mattress cover, 1 m2 of bedroom floor or carpet, 1 m2 of living room–family room floor or carpet, 1 m2 of the kitchen floor, and a major item of upholstered furniture to which the child was exposed. Each area was vacuumed for 2 minutes. The specimens were shipped frozen to the Dermatology Allergy Clinical Immunology Laboratory at Johns Hopkins University, Baltimore, MD, where they were analyzed. An agar plate was streaked for enumeration of mold colonies, and the colonies were not further identified. The mold colonies were reported in units per gram of house dust. We chose to stratify the mold levels by greater than or less than or equal to 25,000 units per gram as used in a previous CAMP study (15).
At each study visit, which occurred every 4 months, subjects reported on the number of hospitalizations and emergency department visits they had experienced since the last study visit (16). Research assistants obtained spirometry measurements on the subjects both before and after a bronchodilator at each study visit. Bronchodilator response was calculated at each visit as FEV1 ([postbronchodilator FEV1 – prebronchodilator FEV1]/prebronchodilator FEV1). Each year, the subjects' airway responsiveness to methacholine was measured by calculating the concentration of methacholine that caused a 20% decrease in the FEV1. The concentration that provoked a 20% decrease from postdiluent FEV 1 was obtained by linear interpolation of logarithmic dose–response curve expressed as PC 20 . Additional demographic information was obtained at an initial prerandomization study visit.
SNP Genotyping
SNPs in CHIT1, CHIA, and CHI3L1 were genotyped, in addition to a CHIT1 duplication, which has been previously studied (17). SNPs in CAMP were genotyped using the Infinium HumanHap550 genotyping at Illumina (San Diego, CA). Genotyping quality was evaluated using the program PLINK (V1.01). SNPs with low Illumina gencall scores, poor completion rates, or four or more parent–offspring genotyped inconsistencies were dropped. Using the Basic Local Alignment Search Tool, SNPs were further limited to those whose flanking sequences were reliably mapped to unique autosomal locations in the hg17 reference genome sequence. Mitochondrial and sex-linked markers were not included. The CHIT1 fragment analysis was performed using the Applied Biosystems 3100 Genetic Analyzer platform. Primers CHIT1_A1FGTCTGGATGAGGGGGTATCG-FAM and CHIT1 A1RGTTTCTTCCCTGCACAGGTCAGCTATC were used to polymerase chain reaction amplify the region containing the 24-bp duplication, and peaks were analyzed with Applied Biosystems GeneMapper software (Carlsbad, CA). Genotyping completion rate was 94%.
Family-Based Association Test Generalized Estimating Equations
Family-based association test generalized estimating equations (FBAT-GEE) is a method that has the ability to use genetic data from family members to assess potential associations between a disease phenotype and a gene allele (18, 19). This methodology has been robust in identifying the associations between SNPs with complex diseases, particularly in genome-wide association studies. We performed association analyses for each SNP and each phenotype using the FBAT-GEE approach, which has been described previously (20). We used Vansteelandt and coworkers' method (21), which uses causal inference to derive estimating equations that generate an estimate of the main genetic effect, β1, and the gene-by-environment interaction, β2, after accounting for the main genetic effect. The general principle behind FBAT interaction is that after removing the overall main genetic effect, the phenotype should not depend on the genotypes conditional on the environmental exposure under the null hypothesis (22).
In the analysis for this study, the additive genetic model was used and a minimum of 20 informative families were required. We used an FBAT approach with GEE (23) for our outcomes. The main outcome was severe exacerbations defined as one or more hospitalizations or emergency department visits experienced during the 4 years of the study.
RESULTS
Descriptive Statistics
Our study population included 395 white subjects who had available genotype information and mold levels. The mean age was 8.7 years (SD 2.1). Table 1 provides the baseline demographic characteristics measured in our study population. The mean age of the subjects with mold levels of greater than 25,000 units per gram was 8.7 years, and 8.8 years for subjects with mold level of less than or equal to 25,000 units per gram (P = 0.68). Of the subjects with mold level greater than 25,000 units per gram, 33% were in the budesonide group, whereas 26% of subjects with mold level less than or equal to 25,000 units per gram were in the budesonide group; this difference was not statistically significant. A slightly higher percentage of subjects in the group with mold level greater than 25,000 units per gram were male (65% vs. 61%; P = 0.49). There were no differences between the groups of subjects with mold level greater than 25,000 units per gram and less than or equal to 25,000 units per gram with respect to the total number of hospitalization and emergency department visits over the 4-year period of the trial, pre-FEV1, bronchodilator response, FEV1 percent predicted, ln PC20, log10 IgE, log10 eosinophil, parental history of asthma or atopy.
TABLE 1.
DEMOGRAPHICS
| N = 395 Mean [SD] or Percent (n) | Mold Level >25,000 Units Per Gram (n = 95) | Mold Level ≤25,000 Units Per Gram (n = 300) | P Value |
|---|---|---|---|
| Age in years [SD] (range) | 8.70 [1.95] | 8.80 [2.16] | 0.68 |
| Treatment group | 0.23 | ||
| Budesonide | 31 (33%) | 78 (26%) | |
| Nedocromil | 22 (33%) | 95 (32%) | |
| Placebo | 42 (44%) | 127 (42%) | |
| Sex | 0.49 | ||
| Male | 62 (65%) | 184 (61%) | |
| Female | 33 (35%) | 116 (39%) | |
| Weight at baseline, kg | 30.76 [9.36] | 32.30 [11.07] | 0.22 |
| Height at baseline, cm | 131.08 [12.75] | 133.09 [14.04] | 0.22 |
| Total number of hospitalization and emergency department visits over 4-yr period | 0.64 | ||
| 0 | 67 | 209 | |
| 1 | 14 | 41 | |
| 2 | 7 | 19 | |
| ≥3 | 7 | 31 | |
| Baseline pre-FEV1 | 1.59 [0.43] | 1.64 [0.48] | 0.33 |
| Baseline bronchodilator response | 0.105 [0.093] | 0.11 [−0.11] | 0.79 |
| Baseline FEV1 percent predicted | 93.60 [13.79] | 93.3 [13.94] | 0.88 |
| Baseline 1n PC20 | −0.095 [1.24] | 0.069 [1.11] | 0.23 |
| Baseline log10 IgE | 2.61 [0.65] | 2.61 [0.66] | 0.98 |
| Baseline log10 eosinophil count | 2.56 [0.48] | 2.57 [0.48] | 0.91 |
| Paternal history of asthma | 0.27 | ||
| Present | 24 (26%) | 61 (21%) | |
| Absent | 67 (74%) | 231 (79%) | |
| Paternal history of atopy | 0.29 | ||
| Present | 42 (45%) | 112 (38% | |
| Absent | 52 (55%) | 179 (62%) | |
| Maternal history of asthma | 0.40 | ||
| Present | 22 (23%) | 82 (28%) | |
| Absent | 72 (77%) | 213 (72%) | |
| Maternal history of atopy | 0.86 | ||
| Present | 43 (46%) | 50 (47%) | |
| Absent | 50 (54%) | 156 (53%) |
FBAT Analysis
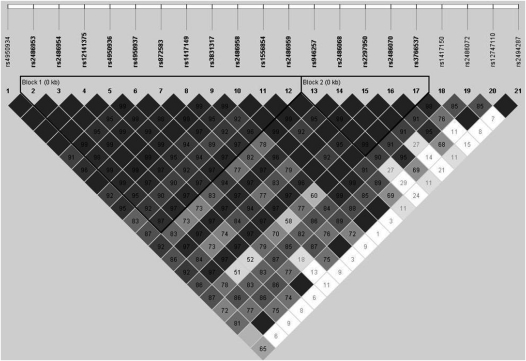
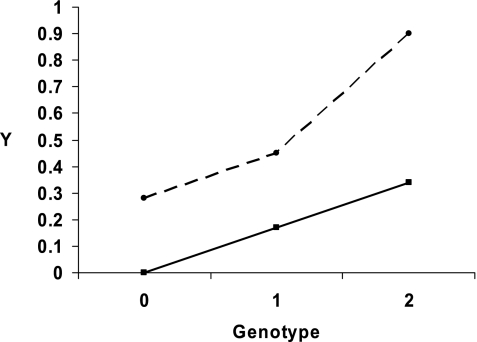
We studied 395 subjects and their parents, and there was one affected offspring within each family. Table 2 shows the findings for the SNP-by-mold level interaction on hospitalizations and emergency department visits. We present the number of informative families for each SNP and the uncorrected FBAT–interaction P values. Ten SNPs had significant FBAT-GEE P values (P < 0.05) for the interaction. After adjusting for multiple comparisons, the significant SNPs are bolded and the interaction estimate, β2, is given. Mold exposure significantly modified the relation between three SNPs in CHIT1 (rs2486953, rs4950936, and rs1417149) and one or more emergency department visit or hospitalization from asthma. We found that a mold level of greater than 25,000 units per gram modified the relationship of rs2486953 with one or more emergency department visits or hospitalizations (FBAT–interaction P value 0.0010); a mold level of greater than 25,000 units per gram modified the relationship of rs4950936 (FBAT–interaction P value 0.0008) and mold level modified rs1417149 (FBAT–interaction P value 0.0005). These three SNPs in CHIT1 are in linkage disequilibrium (Figure 1), and are located within an intron of CHIT1. Figure 2 depicts the increased number of emergency department visits and hospitalizations when exposed to a mold level of greater than 25,000 units per gram when possessing two copies of the genotype compared with one copy for rs2486953.
TABLE 2.
ASSOCIATION OF THE SINGLE NUCLEOTIDE POLYMORPHISMS IN CHIT1, CHIA, AND CHI3LI WITH ONE OR MORE HOSPITALIZATIONS OR EMERGENCY DEPARTMENT VISITS WITH GENE–ENVIRONMENT INTERACTION
| Gene | Marker | Allele | Minor Allele Frequency | Number of Informative Families | Beta Estimate for Interaction | FBAT-GEE P Value for Interaction | Beta Estimate for Main Effect | FBAT-GEE P Value for Main Effect |
|---|---|---|---|---|---|---|---|---|
| CHIT1 | rs4950934 | 1 | 0.11 | 160 | 0.0652 | 0.572 | 0.442 | −0.0462 |
| rs2486953 | 2 | 0.47 | 340 | 0.278 | 0.0010 | 0.173 | −0.0562 | |
| rs2486954 | 3 | 0.20 | 250 | 0.0871 | 0.283 | 0.794 | −0.0115 | |
| rs12141375 | 1 | 0.20 | 251 | 0.0871 | 0.283 | 0.794 | −0.0115 | |
| rs4950936 | 3 | 0.47 | 340 | 0.284 | 0.0008 | 0.138 | −0.0609 | |
| rs4950937 | 1 | 0.28 | 275 | 0.297 | 0.006 | 0.283 | −0.0561 | |
| rs872583 | 2 | 0.19 | 252 | 0.0688 | 0.399 | 0.810 | −0.0108 | |
| rs1417149 | 2 | 0.47 | 339 | 0.309 | 0.0005 | 0.148 | −0.0607 | |
| rs3831317* | 2 | 0.17 | 272 | 0.0050 | 0.961 | 0.525 | −0.0334 | |
| rs2486958 | 2 | 0.49 | 339 | 0.223 | 0.0100 | 0.364 | −0.0399 | |
| rs1556854 | 2 | 0.49 | 340 | 0.247 | 0.007 | 0.371 | −0.0397 | |
| rs2486959 | 3 | 0.17 | 234 | 0.0397 | 0.649 | 0.935 | 0.0038 | |
| rs946257 | 3 | 0.31 | 289 | 0.270 | 0.0082 | 0.311 | −0.0567 | |
| rs2486068 | 3 | 0.17 | 233 | 0.0449 | 0.634 | 0.871 | −0.0080 | |
| rs2297950 | 4 | 0.31 | 288 | 0.259 | 0.0121 | 0.345 | −0.0539 | |
| rs2486070 | 1 | 0.17 | 232 | 0.0522 | 0.578 | 0.782 | −0.0136 | |
| rs3766537 | 4 | 0.19 | 241 | 0.154 | 0.204 | 0.347 | 0.0538 | |
| rs1417150 | 2 | 0.47 | 330 | 0.1200 | 0.0122 | 0.133 | −0.0646 | |
| rs2486072 | 3 | 0.35 | 322 | 0.0533 | 0.558 | 0.365 | 0.0408 | |
| rs12747110 | 4 | 0.01 | 28 | 0.901 | 0.0141 | 0.681 | 0.0597 | |
| rs2494287 | 4 | 0.13 | 177 | 0.202 | 0.156 | 0.9208 | −0.0060 | |
| CHIA | rs4240529 | 1 | 0.28 | 222 | 0.0783 | 0.456 | 0.1138 | −0.0693 |
| rs4272622 | 2 | 0.19 | 167 | −0.158 | 0.194 | 0.4753 | −0.0447 | |
| rs11102233 | 4 | 0.26 | 208 | −0.249 | 0.061 | 0.6353 | −0.0274 | |
| rs12401737 | 4 | 0.46 | 264 | −0.0252 | 0.775 | 0.5242 | −0.0299 | |
| rs10857871 | 2 | 0.21 | 188 | 0.191 | 0.089 | 0.0112 | −0.1237 | |
| rs3806448 | 1 | 0.48 | 261 | −0.0786 | 0.471 | 0.8169 | −0.0118 | |
| rs10494132 | 2 | 0.22 | 208 | 0.0817 | 0.597 | 0.0036 | 0.2003 | |
| rs3806446 | 2 | 0.45 | 270 | −0.0072 | 0.943 | 0.3928 | −0.0435 | |
| rs7411387 | 2 | 0.40 | 239 | −0.0299 | 0.763 | 0.0591 | −0.0904 | |
| rs11584291 | 4 | 0.31 | 243 | 0.0633 | 0.581 | 0.6792 | −0.0233 | |
| rs4240530 | 2 | 0.29 | 245 | −0.0876 | 0.414 | 0.3206 | 0.0506 | |
| rs12127313 | 1 | 0.14 | 149 | 0.118 | 0.425 | 0.3740 | 0.0545 | |
| rs10494133 | 2 | 0.14 | 159 | −0.0685 | 0.601 | 0.5605 | 0.0444 | |
| rs3818822 | 1 | 0.10 | 124 | −0.0623 | 0.672 | 0.5302 | 0.0387 | |
| rs12034576 | 3 | 0.33 | 242 | −0.0233 | 0.846 | 0.9829 | 0.0011 | |
| rs10494134 | 4 | 0.46 | 269 | 0.0301 | 0.789 | 0.6086 | 0.0236 | |
| rs2275253 | 1 | 0.29 | 236 | 0.0180 | 0.893 | 0.1084 | −0.0948 | |
| rs2275254 | 4 | 0.40 | 261 | −0.0062 | 0.964 | 0.3007 | −0.0573 | |
| rs2256721 | 4 | 0.29 | 225 | 0.0061 | 0.968 | 0.1387 | −0.0955 | |
| rs2820093 | 4 | 0.10 | 126 | −0.0605 | 0.685 | 0.5542 | 0.0378 | |
| rs2282290 | 3 | 0.47 | 267 | −0.0563 | 0.609 | 0.6820 | 0.0208 | |
| rs12034177 | 2 | 0.33 | 241 | −0.0204 | 0.864 | 0.9840 | −0.0010 | |
| rs10776724 | 2 | 0.45 | 262 | −0.0202 | 0.886 | 0.3560 | −0.0539 | |
| rs12137697 | 4 | 0.13 | 141 | 0.146 | 0.329 | 0.6063 | 0.0330 | |
| CHI3L1 | rs7542294 | 1 | 0.15 | 168 | −0.212 | 0.399 | 0.8317 | −0.0186 |
| rs880633 | 2 | 0.49 | 254 | 0.047 | 0.585 | 0.8918 | 0.0068 | |
| rs10399805 | 1 | 0.13 | 149 | −0.178 | 0.505 | 0.3133 | −0.0958 | |
| rs946261 | 2 | 0.40 | 252 | −0.105 | 0.342 | 0.8310 | −0.0115 |
Definition of abbreviation: FBAT-GEE = family-based association test generalized estimating equations.
rs3831317 is a 24-bp duplication in CHIT1.
Significant SNPs are bolded.
Figure 1.
Linkage disequilibrium plot for CHIT1 demonstrating that rs2486953, rs4950936, and rs1417149 are in linkage disequilibrium with each other. Linkage disequilibrium is measured as D′, with darker gray colors indicating higher values. The number in each box represents the r2 between the two corresponding SNPs.
Figure 2.
Depiction of the effect of mold level >25,000 units per gram and genotype on the outcome of hospitalizations and emergency department visits for rs2486953. E = 1 refers to exposure to a mold level of >25,000 units per gram and E = 0 refers to a mold level of ≤25,000 units per gram. G stands for genotype and there are three possibilities for the number of copies of rs2486953: 0 copies, 1 copy, or 2 copies. Y refers to the probability of having one or more hospitalizations and emergency department visits. This figure demonstrates that with exposure to a mold level of >25,000 units per gram, having two copies of the minor allele produces a much higher probability of experiencing hospitalizations and emergency department visits than with no exposure to this mold level.
Mold exposure did not modify the relationship between polymorphisms in CHIA or CHI3L1 with hospitalizations or emergency department visits. Furthermore, SNP-by-mold level interaction was not associated with the secondary asthma or allergy phenotypes. More specifically, mold exposure did not modify the relationship between polymorphisms of CHIT1, CHIA, or CHI3L1 and pre-FEV1, bronchodilator response, FEV1 percent predicted, ln PC20, log10 IgE, and log10 eosinophil.
DISCUSSION
Mold levels may modify the effect of variants in the chitinase gene, CHIT1, on emergency department visits and hospitalizations from asthma. We also found that mold levels do not modify the association between other variants in CHIT1, and variants in both CHIA and CHI3L1 and childhood asthma or asthma-related phenotypes. To our knowledge, this was the first study to examine the effect of mold levels on the association of SNPs in the genes of both chitinases and chitinase-like proteins with asthma and allergy-related phenotypes. Strengths of our study include the availability of mold levels in a well-defined clinical trial, the availability of outcomes over a 4-year time period, and a family-based design that avoids issues with population stratification.
Our results support increasing evidence that CHIT1, which is primarily expressed in the lung, plays an important role in the pathophysiology of asthma in the proper environmental context of exposure to chitin, which was approximated by mold levels (24). Our results are supported by a study of workers in the snow crab–processing industry who are exposed to high levels of chitin (found in the exoskeletons of crustaceans), which found that cumulative exposure to snow crab allergens is associated with prevalence of occupational asthma and allergy in a dose–response manner, even after adjusting for age, sex, and smoking (25). Furthermore, intranasal administration of chitin to mice induces the accumulation of IL-4 expressing innate immune cells, inducing eosinophils and basophils (26), further lending support to the notion that exposure to chitin induces an allergic phenotype. Chitinases seem to be able negatively to regulate the tissue infiltration of eosinophils and basophils (26). Thus, the level of enzymatic activity of chitinases may be protective against development of allergies or asthma by breaking down chitin. Additional evidence supporting our hypothesis is a pilot study that found that subjects with the CHIT1 genotype that correlates with decreased levels of chitotriosidase had increased susceptibility to filarial infection (27). These findings in the pathophysiology of asthma support our finding that CHIT1 may be associated with hospitalizations and emergency department visits from asthma in the setting of varying mold exposures.
Previous studies have found conflicting results on whether genes of chitinases and chitinase-like proteins are not associated with asthma-related phenotypes. Some studies have suggested that variants in CHIT1, CHIA, and CHI3L1 are not associated with asthma or other asthma phenotypes (13, 28, 29). On the other hand, previous studies found that polymorphisms in CHIA are associated with asthma and IgE levels (30, 31). Ober and coworkers (32) concluded that SNPs in CHI3L1 are associated with bronchial hyperresponsiveness in the Hutterite population. Because this is a relatively isolated population, with similar environmental exposure, this finding may avoid some confounding effects of genetic and environmental heterogeneity (32). One potential reason for the conflicting findings in the literature is environmental heterogeneity in exposure to sources of chitin between populations, and we show that accounting for environmental exposure to mold levels may help to clarify these genetic associations.
Despite the strengths of our study, a few caveats deserve mention. First, our sample size of 395 subjects was relatively small. Nevertheless, we did find that mold exposure significantly modified the relation between three SNPs in CHIT1 and one or more emergency department visits or hospitalizations from asthma. Second, our analysis was limited to one population and we did not have a replication population for study; thus, our results may not be generalizable to other populations. To our knowledge, no other longitudinal clinical trials of asthma have measured mold levels as an exposure; thus, we do not have other populations to replicate our findings. A recent review article on gene-by-environment interaction in asthma mentioned that the study of gene–environment interaction in relation to asthma is in its infancy (33). Thus, our findings provide support for future studies examining gene–environment interactions in asthma, and should encourage evaluation of environmental exposures in these studies. In addition, we only included mold measurement from the randomization visit. Although a second mold measurement was attempted at the 3-year visit, 23% of subjects did not have a mold measurement, and FBAT does not allow repeated measurements for the environment variable. Finally, it is likely that mold exposure is only one source of chitin exposure. A comprehensive evaluation of all sources of environmental chitin exposure was beyond the scope of this study.
Both in vitro and in vivo studies have demonstrated that chitin and chitin derivatives have important immunologic effects and play an important role in pulmonary inflammation (34). The literature suggesting the importance of chitinases in the pathophysiology of asthma is strong (4–9, 12), and chitinases may play a role in future targets for asthma therapy (24). In future genetic studies of asthma, measurements of fungal levels could contribute important knowledge on the pathophysiology of asthma.
In conclusion, fungal levels may modulate the effect of variants in the chitinase gene, CHIT1, on emergency department visits and hospitalizations. This finding supports the important role that chitinases have in asthma.
Acknowledgments
The authors thank Brooke Schuemann, M.P.H., for her assistance with preparation of the phenotype data. The authors also thank all subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by the National Heart, Lung, and Blood Institute, for collection of CAMP Genetic Ancillary Study data. All work on data collected from the CAMP Genetic Ancillary Study was conducted at Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subject's protections. CAMP is supported by U01 HL076419, U01 HL65899, P01 HL083069, and T32 HL07427 from the National Heart, Lung, and Blood Institute, National Institutes of Health.
Supported by the Childhood Asthma Management Program through contracts NO1-HR-16044, 16045, 16046, 16047, 16048, 16049, 16050, 16051, and 16,052 with the National Heart, Lung, and Blood Institute and General Clinical Research Center grants M01RR00051, M01RR0099718–24, M01RR02719–14, and RR00036 from the National Center for Research Resources. This work was also supported by U01 HL65899. Dr. Litonjua is supported by R01 AI056230.
Originally Published in Press as DOI: 10.1164/rccm.201003-0322OC on June 10, 2010
Author Disclosure: A.C.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.L-S. received up to $1,000 from Golden Helix for providing statistical support. C.A.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J.K. holds a patent from the Massachusetts Institute of Technology (US Patent 6,703,228) for methods and products related to genotyping and DNA analysis, receiving $0 in the past 3 years. A.A.L. received $1,001–$5,000 from Up To Date in author royalties and more than $100,001 from the National Institutes of Health in sponsored grants.
References
- 1.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 2004;59:469–478. [DOI] [PubMed] [Google Scholar]
- 2.Black PN, Udy AA, Brodie SM. Sensitivity to fungal allergens is a risk factor for life-threatening asthma. Allergy 2000;55:501–504. [DOI] [PubMed] [Google Scholar]
- 3.Zureik M, Neukirch C, Leynaert B, Liard R, Bousquet J, Neukirch F. Sensitisation to airborne moulds and severity of asthma: cross sectional study from European Community respiratory health survey. BMJ 2002;325:411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 2004;304:1678–1682. [DOI] [PubMed] [Google Scholar]
- 5.Ray A, Cohn L. Th2 cells and GATA-3 in asthma: new insights into the regulation of airway inflammation. J Clin Invest 1999;104:985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, Dziura JD, Reed J, Coyle AJ, Kiener P, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med 2007;357:2016–2027. [DOI] [PubMed] [Google Scholar]
- 7.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem 2001;276:6770–6778. [DOI] [PubMed] [Google Scholar]
- 8.Shahabuddin M, Vinetz JM. Chitinases of human parasites and their implications as antiparasitic targets. EXS 1999;87:223–234. [DOI] [PubMed] [Google Scholar]
- 9.Shahabuddin M, Toyoshima T, Aikawa M, Kaslow DC. Transmission-blocking activity of a chitinase inhibitor and activation of malarial parasite chitinase by mosquito protease. Proc Natl Acad Sci USA 1993;90:4266–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickey BF. Exoskeletons and exhalation. N Engl J Med 2007;357:2082–2084. [DOI] [PubMed] [Google Scholar]
- 11.Vicencio AG, Narain S, Du Z, Zeng WY, Ritch J, Casadevall A, Goldman DL. Pulmonary cryptococcosis induces chitinase in the rat. Respir Res 2008;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibata Y, Foster LA, Bradfield JF, Myrvik QN. Oral administration of chitin down-regulates serum IgE levels and lung eosinophilia in the allergic mouse. J Immunol 2000;164:1314–1321. [DOI] [PubMed] [Google Scholar]
- 13.Wu AC, Lasky-Su J, Rogers CA, Klanderman BJ, Litonjua A. Polymorphisms of chitinases and chitinase-like proteins are not associated with asthma or other asthma phenotypes. J Allergy Clin Immunol 2010;125:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N Engl J Med 2000;343:1054–1063. [DOI] [PubMed] [Google Scholar]
- 15.Nelson HS, Szefler SJ, Jacobs J, Huss K, Shapiro G, Sternberg AL. The relationships among environmental allergen sensitization, allergen exposure, pulmonary function, and bronchial hyperresponsiveness in the Childhood Asthma Management Program. J Allergy Clin Immunol 1999;104:775–785. [DOI] [PubMed] [Google Scholar]
- 16.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 17.Seibold MA, Donnelly S, Solon M, Innes A, Woodruff PG, Boot RG, Burchard EG, Fahy JV. Chitotriosidase is the primary active chitinase in the human lung and is modulated by genotype and smoking habit. J Allergy Clin Immunol 2008;122:944–950, e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange C, van Steen K, Andrew T, Lyon H, DeMeo DL, Raby B, Murphy A, Silverman EK, MacGregor A, Weiss ST, et al. A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat Appl Genet Mol Biol 2004;3:Article17. [DOI] [PubMed]
- 20.Sonuga-Barke EJ, Lasky-Su J, Neale BM, Oades R, Chen W, Franke B, Buitelaar J, Banaschewski T, Ebstein R, Gill M, et al. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet 2008;147B:1359–1368. [DOI] [PubMed] [Google Scholar]
- 21.Vansteelandt S, Demeo DL, Lasky-Su J, Smoller JW, Murphy AJ, McQueen M, Schneiter K, Celedon JC, Weiss ST, Silverman EK, et al. Testing and estimating gene-environment interactions in family-based association studies. Biometrics 2008;64:458–467. [DOI] [PubMed] [Google Scholar]
- 22.Lasky-Su J, Biederman J, Doyle AE, Wilens T, Monuteaux M, Smoller JW, Faraone S. Family based association analysis of statistically derived quantitative traits for drug use in ADHD and the dopamine transporter gene. Addict Behav 2006;31:1088–1099. [DOI] [PubMed] [Google Scholar]
- 23.Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics 2003;4:195–206. [DOI] [PubMed] [Google Scholar]
- 24.Donnelly LE, Barnes PJ. Acidic mammalian chitinase–a potential target for asthma therapy. Trends Pharmacol Sci 2004;25:509–511. [DOI] [PubMed] [Google Scholar]
- 25.Gautrin D, Cartier A, Howse D, Horth-Susin L, Jong M, Swanson M, Lehrer S, Fox G, Neis B. Occupational asthma and allergy in snow crab processing in Newfoundland and Labrador. Occup Environ Med 2010;67:17–23. [DOI] [PubMed] [Google Scholar]
- 26.Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 2007;447:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi EH, Zimmerman PA, Foster CB, Zhu S, Kumaraswami V, Nutman TB, Chanock SJ. Genetic polymorphisms in molecules of innate immunity and susceptibility to infection with Wuchereria bancrofti in South India. Genes Immun 2001;2:248–253. [DOI] [PubMed] [Google Scholar]
- 28.Bierbaum S, Superti-Furga A, Heinzmann A. Genetic polymorphisms of chitotriosidase in caucasian children with bronchial asthma. Int J Immunogenet 2006;33:201–204. [DOI] [PubMed] [Google Scholar]
- 29.Sohn MH, Lee JH, Kim KW, Kim SW, Lee SH, Kim KE, Kim KH, Lee CG, Elias JA, Lee MG. Genetic variation in the promoter region of chitinase 3-like 1 is associated with atopy. Am J Respir Crit Care Med 2009;179:449–456. [DOI] [PubMed] [Google Scholar]
- 30.Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med 2005;172:1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee R, Batra J, Das S, Sharma SK, Ghosh B. Genetic association of acidic mammalian chitinase with atopic asthma and serum total IgE levels. J Allergy Clin Immunol 2008;122:202–208, e201–207. [DOI] [PubMed] [Google Scholar]
- 32.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 2008;358:1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.London SJ, Romieu I. Gene by environment interaction in asthma. Annu Rev Public Health 2009;30:55–80. [DOI] [PubMed] [Google Scholar]
- 34.Lee CG, Da Silva CA, Lee JY, Hartl D, Elias JA. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol 2008;20:684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]