Abstract
In the title coordination polymer, [Hg2Cl4(C6H12N2)]n, each HgII center within the chain is four-coordinated by one terminal Cl atom, two bridging μ2-Cl atoms, and one N-atom donor from a μ2-1,4-diazabicyclo[2.2.2]octane (μ2-daco) ligand in a distorted tetrahedral geometry. The daco ligand acts as an end-to-end bridging ligand and bridges adjacent HgII centers, forming a chain running along [001]. Weak C—H⋯Cl hydrogen-bonding interactions link the chains into a three-dimensional network. Comparison of the structural differences with previous findings suggests that the space between the two N donors, as well as the skeletal rigidity in N-heterocyclic linear ligands, may play an important role in the construction of such supramolecular networks.
Related literature
For a related structure, see: Pickardt et al. (1995 ▶). For functional materials, see: Chen, Kang & Su (2006 ▶); Fang et al. (2009 ▶); Liu et al. (2007 ▶); Ma et al. (2009 ▶); Tranchemontagne et al. (2009 ▶); Uemura et al. (2009 ▶); Xue et al. (2008 ▶). For N-containing hetercyclic bridging ligands, see: Batten et al. (2002 ▶); Chen et al. (2007 ▶); Culp et al. (2008 ▶); Kaim (1983 ▶); Leininger et al. (2000 ▶); Richardson & Steel (2003 ▶); Steel (2005 ▶). For 4,4′-bipyridine and pyrazine extended assemblies, see: Arpi et al. (2006 ▶); Chen, Wang et al. (2006 ▶); Choi et al. (2009 ▶); Derossi et al. (2007 ▶); Du et al. (2007 ▶); Liu et al. (2006 ▶); Li et al.(2008 ▶); Ramírez et al. (2009 ▶); Qiu et al. (2008 ▶); Nockemann & Meyer (2004 ▶); Xie & Wu (2007 ▶). For daco complexes, see: Dybtsev et al. (2004 ▶); Li et al. (2006 ▶); Rao & Rao (2007 ▶); Steel (2005 ▶). For factors determining the crystal packing, see: Kitagawa et al. (2004 ▶). For Hg—N and Hg—Cl bond distances and bond angles about Hg, see: Orpen et al. (1989 ▶); Wang et al. (2007 ▶).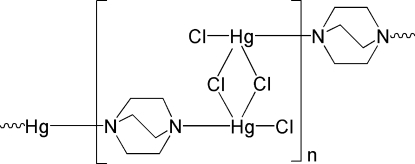
Experimental
Crystal data
[Hg2Cl4(C6H12N2)]
M r = 327.58
Orthorhombic,
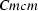
a = 9.2518 (9) Å
b = 8.8244 (9) Å
c = 14.7531 (8) Å
V = 1204.47 (18) Å3
Z = 8
Mo Kα radiation
μ = 26.31 mm−1
T = 293 K
0.05 × 0.04 × 0.02 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.353, T max = 0.621
1322 measured reflections
586 independent reflections
392 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.047
S = 0.96
586 reflections
39 parameters
6 restraints
H-atom parameters constrained
Δρmax = 1.41 e Å−3
Δρmin = −1.59 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809043839/su2148sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809043839/su2148Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2A⋯Cl2i | 0.97 | 2.77 | 3.708 (8) | 163 |
| C2—H2B⋯Cl2ii | 0.97 | 2.78 | 3.678 (8) | 154 |
Symmetry codes: (i) 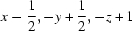 ; (ii)
; (ii) 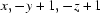 .
.
Acknowledgments
This work was supported by the National Natural Science Funds of China (Nos. 20771095 and 20801049) and the start-up fund for PhDs in Natural Scientific Research of Zhengzhou University of Light Industry (Nos. 2007BSJJ001 and 2006BSJJ001 to CSL and SMF, respectively).
supplementary crystallographic information
Comment
The rational engineering and controlled preparation of novel coordination complexes are currently of great interest in coordination and supramolecular chemistry not only because of their intriguing structural diversities but also their potential applications as functional materials (Chen, Kang & Su, 2006; Fang et al., 2009; Liu et al., 2007; Ma et al., 2009; Tranchemontagne et al., 2009; Uemura et al., 2009; Xue et al., 2008). One of the most successful strategies for constructing such complexes has been the assembly reaction of different metal ions (as nodes) with well designed organic ligands (as building blocks), which, so far, has been at an evolutionary stage with the current focus mainly on understanding the factors to determine the crystal packing as well as exploring relevant potential properties (Kitagawa et al., 2004). N-containing heterocyclic ligands are extensively used as bridging ligands in coordination and metallosupramolecular chemistry (Batten et al., 2002; Chen et al., 2007; Culp et al., 2008; Kaim, 1983; Leininger et al., 2000; Richardson & Steel, 2003; Steel, 2005). For examples, the use of 4,4'-bipyridine (4bpy) and pyrazine (pyz) has resulted in a large number of extended assemblies including helical networks as well as diamondoid, honeycomb, square-grid, ribbon, grid, T-shaped, Ladder, brick wall, and octahedral frameworks (Arpi et al., 2006; Chen, Wang et al., 2006; Choi et al., 2009; Derossi et al., 2007; Du et al., 2007; Liu et al., 2006; Li et al., 2008; Ramírez et al., 2009; Qiu et al., 2008).
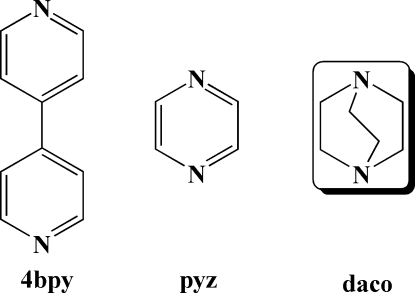
In comparison with 4bpy and pyz ligands (Fig. 3), however, 1,4-diazabicyclo[2.2.2]octane (daco) acts as a flexible N-containing heterocyclic bridging ligand with a skeletal separation of ca6 Å between two N donors, shorter than that of ca 7 and 11 Å for 4bpy and pyz, respectively (Dybtsev et al., 2004, Li et al., 2006; Rao & Rao, 2007; Steel, 2005). As part of a study on the effect of the space between two N donors or the skeletal rigidity in linear N-containing heterocyclic ligands on the self-assembly of coordination complexes, we have chosen to use 1,4-diazabicyclo[2.2.2]octane (daco) instead of 4bpy or pyz to construct the title compound, a new one-dimensional HgII coordination polymer.
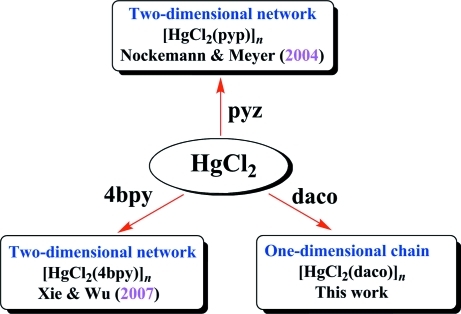
To explore the coordination possibility of a series of N-containing heterocyclic linear ligands, Xie & Wu (2007) as well as Nockemann & Meyer (2004) have selected 4,4'-bipyridine (4bpy) and pyrazine (pyz) to react with HgII atom, obtaining two different two-dimensional neutral networks, [HgCl2(4bpy)]n (A) and [HgCl2(pyz)]n (B), respectively (Fig. 4). In these two structures, the Hg atoms are both in octahedral coordination environments, coordinated by four µ2-Cl atoms and two µ2-4bpy for A as well as four µ2-Cl atoms and two µ2-pyz for B in trans positions. The relevant one-dimensional chain motifs bridged by µ2-4bpy or µ2-pyz in complexes A and B are further interlinked to form the two-dimensional layers via the µ2-Cl atoms. In this contribution, however, when we used HgCl2 to react with 1,4-diazabicyclo[2.2.2] octane (daco) under a conventional solution diffusion condition, the title one-dimensional coordination polymer, [Hg2Cl4(daco)]n, was produced.
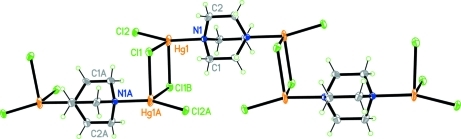
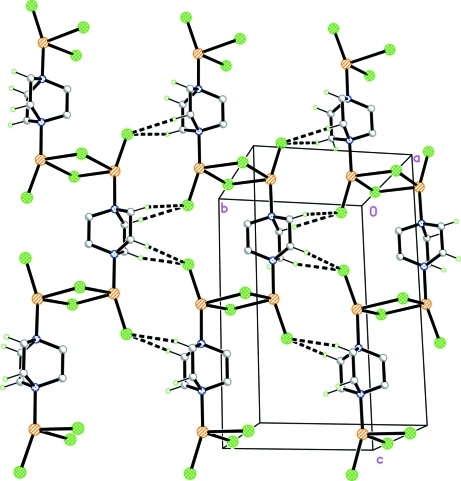
The crystal structure of the title compound consists of one-dimensional (1D) neutral [Hg2Cl4(daco)]n chains (Fig. 1). The HgII center is four-coordinated, by three Cl atoms (one terminal Cl and two µ2-Cl atoms) and one N-atom donor from one daco ligand, with a distorted tetrahedral geometry. Two µ2-Cl atoms bridge adjacent HgII centers to form one planar four-membered rings composed of Hg1–Cl1–Hg1i–Cl1ii with a non-bonding Hg(1)···Hg(1i) separation of 4.067 (1) Å [symmetry codes (i) = x, –y, 1–z; (ii) = 1–x, –y, 1–z]. Each daco ligand takes a µ2-bridging coordination mode to connect the adjacent HgIIcenters, generating a one-dimensional chain running along the [001] direction. The Hg–N and Hg–Cl bond distances as well as the bond angles around each HgII center are within the expected range for such complexes (Orpen et al., 1989; Wang et al., 2007). Moreover, adjacent 1D [Hg2Cl4(daco)]n chains are arranged into two-dimensional layers, running parallel to the (100) plane, by interchain C–H···Cl hydrogen-bonding interactions between the daco ligands and the terminal Cl atoms (Fig. 2 and Table 1).
Thus, in comparison with the previous findings, the present work reveals that the space between two N donors, or the skeletal rigidity of N-containing heterocyclic linear ligands, could play an important role in the final structure of the relate coordination complexes. This fact may offer the means to construct new coordination architectures with potentially useful properties by varying of the space between the two N donors, or the rigidity of skeleton of N-containing heterocyclic ligands. It is noteworthy that mercury(II) metal halide-daco materials are relatively rare, and to the best of our knowledge, only one halide-daco complex has been reported previously, viz. HgI2-daco (Pickardt et al., 1995). This complex, [HgI2(daco)]n, also exhibits a zigzag one-dimensional structure, but here the I-atoms are all terminally coordinated to the metal centers, while in the title complex one of the Cl atoms act as a bridging atom.
Experimental
A solution of 1,4-Diazabicyclo[2.2.2]octane (daco) (0.05 mmol) in CH3OH (10 ml) was carefully layered on top of an aqueous solution (15 ml) of HgCl2 (0.1 mmol) in a test tube. Colorless single crystals suitable for X-ray analysis appeared at the tube wall after ca one month at room temperature (yield ~30% based on daco). Elemental analysis calculated for (C3H6Cl2HgN): H 1.85, C 11.00, N 4.28%; found: H 1.78, C 10.87, N 4.24%.
Refinement
H atoms were included in calculated positions and treated as riding atoms: C—H = 0.97 Å, with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The one-dimensional molecular structure of the title complex, propagating in the [001] direction. Displacement ellipsoids are drawn at the 30% probability level. The atoms labeled with the suffixes A and B are generated by the symmetry operations (x, -y, -z + 1; -x + 1, -y, -z + 1), respectively.
Fig. 2.
The two-dimensional net, viewed along the a axis, formed by the interchain C–H···Cl hydrogen-bonds (dashed lines). For clarity, only H-atoms involved in hydrogen-bonding are shown.
Fig. 3.
Structures of 4bpy, pyz and daco.
Fig. 4.
Reaction scheme.
Crystal data
| [Hg2Cl4(C6H12N2)] | F(000) = 1160 |
| Mr = 327.58 | Dx = 3.613 Mg m−3 |
| Orthorhombic, Cmcm | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2c 2 | Cell parameters from 453 reflections |
| a = 9.2518 (9) Å | θ = 3.2–30.3° |
| b = 8.8244 (9) Å | µ = 26.31 mm−1 |
| c = 14.7531 (8) Å | T = 293 K |
| V = 1204.47 (18) Å3 | Block, colorless |
| Z = 8 | 0.05 × 0.04 × 0.02 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 586 independent reflections |
| Radiation source: fine-focus sealed tube | 392 reflections with I > 2σ(I) |
| graphite | Rint = 0.040 |
| φ and ω scans | θmax = 25.0°, θmin = 3.2° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −10→10 |
| Tmin = 0.353, Tmax = 0.621 | k = −10→8 |
| 1322 measured reflections | l = −17→9 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.047 | H-atom parameters constrained |
| S = 0.96 | w = 1/[σ2(Fo2) + (0.0085P)2] where P = (Fo2 + 2Fc2)/3 |
| 586 reflections | (Δ/σ)max < 0.001 |
| 39 parameters | Δρmax = 1.41 e Å−3 |
| 6 restraints | Δρmin = −1.59 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Hg1 | 0.5000 | 0.22915 (6) | 0.51455 (3) | 0.0533 (2) | |
| C1 | 0.5000 | 0.0735 (12) | 0.6983 (7) | 0.036 (3) | |
| H1A | 0.5849 | 0.0205 | 0.6763 | 0.043* | 0.50 |
| H1B | 0.4151 | 0.0205 | 0.6763 | 0.043* | 0.50 |
| C2 | 0.3708 (8) | 0.3114 (9) | 0.6975 (5) | 0.032 (2) | |
| H2A | 0.2843 | 0.2615 | 0.6754 | 0.039* | |
| H2B | 0.3704 | 0.4150 | 0.6754 | 0.039* | |
| Cl1 | 0.2906 (3) | 0.0000 | 0.5000 | 0.0344 (8) | |
| Cl2 | 0.5000 | 0.3087 (3) | 0.3653 (2) | 0.0349 (9) | |
| N1 | 0.5000 | 0.2318 (11) | 0.6631 (6) | 0.022 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Hg1 | 0.0692 (4) | 0.0785 (5) | 0.0122 (3) | 0.000 | 0.000 | 0.0076 (3) |
| C1 | 0.055 (9) | 0.017 (6) | 0.036 (8) | 0.000 | 0.000 | 0.006 (5) |
| C2 | 0.016 (5) | 0.050 (6) | 0.031 (5) | −0.003 (4) | −0.003 (4) | 0.008 (4) |
| Cl1 | 0.0244 (17) | 0.0511 (18) | 0.028 (2) | 0.000 | 0.000 | −0.0067 (17) |
| Cl2 | 0.042 (2) | 0.043 (2) | 0.0198 (16) | 0.000 | 0.000 | 0.0094 (13) |
| N1 | 0.022 (2) | 0.022 (2) | 0.022 (2) | 0.000 | 0.000 | 0.0000 (10) |
Geometric parameters (Å, °)
| Hg1—N1 | 2.192 (8) | C1—H1B | 0.9700 |
| Hg1—Cl2 | 2.311 (3) | C2—N1 | 1.476 (9) |
| Hg1—Cl1i | 2.8083 (17) | C2—C2ii | 1.549 (15) |
| Hg1—Cl1 | 2.8083 (17) | C2—H2A | 0.9700 |
| C1—N1 | 1.490 (13) | C2—H2B | 0.9700 |
| C1—C1ii | 1.52 (2) | Cl1—Hg1i | 2.8083 (17) |
| C1—H1A | 0.9700 | N1—C2iii | 1.476 (9) |
| N1—Hg1—Cl2 | 161.7 (3) | N1—C2—H2A | 109.6 |
| N1—Hg1—Cl1i | 94.82 (18) | C2ii—C2—H2A | 109.6 |
| Cl2—Hg1—Cl1i | 98.39 (5) | N1—C2—H2B | 109.6 |
| N1—Hg1—Cl1 | 94.82 (18) | C2ii—C2—H2B | 109.6 |
| Cl2—Hg1—Cl1 | 98.39 (5) | H2A—C2—H2B | 108.2 |
| Cl1i—Hg1—Cl1 | 87.21 (7) | Hg1—Cl1—Hg1i | 92.79 (7) |
| N1—C1—C1ii | 110.4 (5) | C2iii—N1—C2 | 108.1 (8) |
| N1—C1—H1A | 109.6 | C2iii—N1—C1 | 109.1 (6) |
| C1ii—C1—H1A | 109.6 | C2—N1—C1 | 109.1 (6) |
| N1—C1—H1B | 109.6 | C2iii—N1—Hg1 | 110.4 (5) |
| C1ii—C1—H1B | 109.6 | C2—N1—Hg1 | 110.4 (5) |
| H1A—C1—H1B | 108.1 | C1—N1—Hg1 | 109.8 (6) |
| N1—C2—C2ii | 110.1 (4) |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) x, y, −z+3/2; (iii) −x+1, y, z.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2A···Cl2iv | 0.97 | 2.77 | 3.708 (8) | 163 |
| C2—H2B···Cl2v | 0.97 | 2.78 | 3.678 (8) | 154 |
Symmetry codes: (iv) x−1/2, −y+1/2, −z+1; (v) x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2148).
References
- Arpi, M., Gramlich, V., Rosair, G. M., Batten, S. R., Masuda, J. D., El Fallah, M. S., Ribas, J., Sutter, J.-P., Desplanches, C. & Mitra, S. (2006). Cryst. Growth Des.6, 2355–2368.
- Batten, S. R., Harris, A. R., Murray, K. S. & Smith, J. P. (2002). Cryst. Growth Des.2, 87–89.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, C.-L., Kang, B.-S. & Su, C.-Y. (2006). Aust. J. Chem.59, 3–18.
- Chen, B., Ma, S., Zapata, F., Fronczek, F. R., Lobkovsky, E. B. & Zhou, H.-C. (2007). Inorg. Chem.46, 1233–1236. [DOI] [PubMed]
- Chen, W.-T., Wang, M.-S., Liu, X., Guo, G.-C. & Huang, J.-S. (2006). Cryst. Growth Des.6, 2289–2300.
- Choi, E.-Y., Barron, P. M., Novotny, R. W., Son, H.-T., Hu, C. & Choe, W. (2009). Inorg. Chem.48, 426–428. [DOI] [PubMed]
- Culp, J. T., Smith, M. R., Bittner, E. & Bockrath, B. (2008). J. Am. Chem. Soc.130, 12427–12434. [DOI] [PubMed]
- Derossi, S., Casanova, M., Iengo, E., Zangrando, E., Stener, M. & Alessio, E. (2007). Inorg. Chem.46, 11243–11253. [DOI] [PubMed]
- Du, Z.-Y., Li, X.-L., Liu, Q.-Y. & Mao, J.-G. (2007). Cryst. Growth Des.7, 1501–1507.
- Dybtsev, D. N., Chun, H. & Kim, K. (2004). Angew. Chem. Int. Ed.43, 5033–5036. [DOI] [PubMed]
- Fang, Z.-L., Yu, R.-M., He, J.-G., Zhang, Q.-S., Zhao, Z.-G. & Lu, C.-Z. (2009). Inorg. Chem.48, 7691–7697. [DOI] [PubMed]
- Kaim, W. (1983). Angew. Chem. Int. Ed.22, 171–190.
- Kitagawa, S., Kitaura, R. & Noro, S. (2004). Angew. Chem. Int. Ed.43, 2334–2375. [DOI] [PubMed]
- Leininger, S., Olenyuk, B. & Stang, P. J. (2000). Chem. Rev.100, 853–908. [DOI] [PubMed]
- Li, C.-J., Hu, S., Li, W., Lam, C.-K., Zheng, Y.-Z. & Tong, M.-L. (2006). Eur. J. Inorg. Chem. pp. 1931–1935.
- Li, Y., Xie, L., Liu, Y., Yang, R. & Li, X. (2008). Inorg. Chem.4, 10372–10377. [DOI] [PubMed]
- Liu, T., Chen, Y.-H., Zhang, Y.-J., Wang, Z.-M. & Gao, S. (2006). Inorg. Chem.45, 9148–9150. [DOI] [PubMed]
- Liu, C.-S., Wang, J.-J., Yan, L.-F., Chang, Z., Bu, X.-H., Sanũdo, E. C. & Ribas, J. (2007). Inorg. Chem.46, 6299–6310. [DOI] [PubMed]
- Ma, L.-F., Wang, Y.-Y., Liu, J.-Q., Yang, G.-P., Du, M. & Wang, L.-Y. (2009). CrystEngComm, 11, 1800–1802.
- Nockemann, P. & Meyer, G. (2004). Acta Cryst. E60, m744–m746.
- Orpen, A. G., Brammer, L., Aleen, F. H., Kennard, O., Watson, D. G. & Taylor, R. (1989). J. Chem. Soc. Dalton Trans. pp. S1–83.
- Pickardt, J., Shen, J. & Gong, G.-T. (1995). Z. Naturforsch Teil B, 50, 833–836.
- Qiu, Y., Liu, Z., Li, Y., Deng, H., Zeng, R. & Zeller, M. (2008). Inorg. Chem.47, 5122–5128. [DOI] [PubMed]
- Ramírez, J., Stadler, A.-M., Rogez, G., Drillon, M. & Lehn, J.-M. (2009). Inorg. Chem.48, 2456–2463. [DOI] [PubMed]
- Rao, K. P. & Rao, C. N. R. (2007). Inorg. Chem.46, 2511–2518. [DOI] [PubMed]
- Richardson, C. & Steel, P. J. (2003). Dalton Trans. pp. 992–1000.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst D65, 148–155. [DOI] [PMC free article] [PubMed]
- Steel, P. J. (2005). Acc. Chem. Res.38, 243–250. [DOI] [PubMed]
- Tranchemontagne, D. J., Mendoza-Cortés, J. L., O’Keeffe, M. & Yaghi, O. M. (2009). Chem. Soc. Rev.38, 1257–1283. [DOI] [PubMed]
- Uemura, T., Yanai, N. & Kitagawa, S. (2009). Chem. Soc. Rev.38, 1228–1236. [DOI] [PubMed]
- Wang, X.-F., Lv, Y., Okamura, T., Kawaguchi, H., Wu, G., Sun, W.-Y. & Ueyama, N. (2007). Cryst. Growth Des.7, 1125–1133.
- Xie, Y.-M. & Wu, J.-H. (2007). Acta Cryst. C63, m220–m221. [DOI] [PubMed]
- Xue, M., Zhu, G., Zhang, Y., Fang, Q., Hewitt, I. J. & Qiu, S. (2008). Cryst. Growth Des.8, 427–434.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809043839/su2148sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809043839/su2148Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report