Abstract
GeneSplicer is a new, flexible system for detecting splice sites in the genomic DNA of various eukaryotes. The system has been tested successfully using DNA from two reference organisms: the model plant Arabidopsis thaliana and human. It was compared to six programs representing the leading splice site detectors for each of these species: NetPlantGene, NetGene2, HSPL, NNSplice, GENIO and SpliceView. In each case GeneSplicer performed comparably to the best alternative, in terms of both accuracy and computational efficiency.
INTRODUCTION
Identification of protein coding genes in genomic DNA de novo requires that a program finds the locations of the start codons, all the exons and introns and the stop codon for each gene. The 5′ boundary or donor site of introns in most eukaryotes usually contains the dinucleotide GT (GU in pre-mRNA), while the 3′ boundary or acceptor site contains the dinucleotide AG. In addition to these dimers, a pyrimidine-rich region precedes the AG at the acceptor site, a shorter consensus follows the GT at the donor site, and a very weak consensus sequence appears at the branch point, ∼30 nt upstream from the acceptor site. These consensus sequences are recognized by a complex of proteins and small nuclear RNAs, known collectively as the spliceosome, which splices out the introns from pre-mRNA and produces the mature mRNA transcript. A number of computational methods have been developed to identify these splice sites, including both stand-alone splice site finders and gene finders, which identify splice sites as a subroutine. The performance of most gene finding systems is greatly influenced by their accuracy at determining splice sites. In theory, a program that could correctly identify all splice sites would do a nearly perfect job of ab initio gene finding, since it would identify all protein coding regions correctly (with the chance of a small error in the identification of the correct start site). Any reduction in the number of potential sites being considered by a gene finder will significantly reduce the number of alternative ways of parsing a DNA sequence into exons and introns, and therefore makes overall gene prediction easier.
Approximately 30% of the genes that are annotated in newly sequenced genomes such as Arabidopsis thaliana are, at present, purely the result of computational predictions. More accurate gene prediction is essential for future experimental work, which will attempt to validate and characterize these genes. As the genome sequences reach completion, the amount of training data increases too, making it possible to re-train gene and splice site predictors and improve their performance.
We have developed a new computational tool for detecting splice sites in eukaryotic mRNA by combining several techniques that have already proven successful in characterizing the patterns around the donor and acceptor sites. We use a decision tree method called maximal dependence decomposition (MDD), first introduced by Burge and Karlin (1), and enhance it with Markov models that capture additional dependencies among neighboring bases in a region around the splice site. This method considers only a small window around the splice junctions, which contains most of the information recognized by the spliceosome. Our algorithm also takes advantage of the fact that the coding and non-coding sequences switch at the splice junction, and this switch can sometimes be detected by considering sequence statistics in a larger window. In addition, by applying the local score optimality feature developed by Brendel and Kleffe (2), we increased the overall performance of the splice site detection system.
For the purpose of training and testing the new system, we considered two organisms for which extensive genomic sequence and confirmed genes are available: the model plant, A.thaliana, and human. We collected data sets containing 1323 genes for A.thaliana and 1115 genes for human to use in training and testing; these are described further below. The result of this study is a new system, called GeneSplicer, a statistical method that predicts splice sites by integrating multiple sources of evidence. (GeneSplicer is freely available; for details contact the corresponding author.)
Algorithm description
When performed in the cell, pre-mRNA splicing is not a purely deterministic process. Some transcripts are spliced into multiple alternative products; experimental evidence indicates that weak splice sites become active when mutations occur in nearby sites (3); and mis-splicing occurs at an unknown rate. Nonetheless, the cell is the best machinery we have for splicing, and therefore an algorithmic approach should first of all try to reproduce the biological mechanisms. Although the intermediates, products and reaction mechanism of splicing were characterized some years ago, pre-mRNA structural features that are important for this process have only just begun to be investigated (4), and signals such as exon splicing enhancers (short consensus sequences within exons) are still poorly understood. As a consequence, the best splice site algorithms available today employ a combination of simple biological modeling and more sophisticated statistical methods.
When designing GeneSplicer, we tried to make use of the moderately successful techniques that were used to characterize the sequences around the splice sites in our previous gene finders (5,6). To improve the splice site detection, we combined the Markov modeling techniques described by Salzberg et al. (5,6) with the MDD described by Burge and Karlin (1) using the following algorithm.
The method begins with a set D of N aligned DNA sequences of length k, extracted from a set of donor (respectively acceptor) sites. For each of the k positions, let the most frequent base at that position be the consensus base. The variable Ci will be 1 if the nucleotide at position i matches the consensus at position i, 0 otherwise.
Next, compute the χ2 statistics between the variables Ci and Xj (which identifies the nucleotide at position j), for each i,j pair with i ≠ j. If strong dependencies are detected (defined as a χ2 value of at least 16.3, corresponding to a cutoff level of P = 0.001 with 3 degrees of freedom) between non-adjacent positions, then proceed as described by Burge and Karlin (1). (i) Compute the sum
![]()
for each position i. (ii) Choose i1 such that Si1 is maximal, and partition D into two subsets, Di1 containing all sequences with the consensus nucleotide at position i1, and Di2 = D – Di1 containing the remaining sequences. (iii) Recursively repeat steps 1 and 2 on each of the subsets Di1 and Di2 (thus, building a binary decision tree), until either: the k-1 level of the tree is reached; no significant dependencies are detected; or the number of the sequences in the subtree is too small for reliable Markov models to be constructed for them.
Each leaf of the tree thus constructed now contains a subset of the donor (or acceptor) sites used for training. We then construct first-order Markov chain models using a 16 base region around the donor sites and a 29 base region around the acceptor sites. (Higher-order Markov chains are always preferable when sufficient data is available; the decision to use a first-order Markov chain was made based on the amount of training data.) A set of ‘false’ splice sites were created from a large number of randomly chosen false sites, defined as GT and AG dinucleotides from the training data that did not correspond to true sites.
Finally, the score of a potential splice site is computed as the difference between the log-odds score returned for that sequence by the true Markov model and the score computed by the false Markov model. Details on how to compute the score of a Markov chain model for splice sites have been explained elsewhere (5,6).
In order to improve further the splice site detection mechanism we added another technique to the system. Based on the observation that a splice site is always surrounded by a coding region and a non-coding region, we constructed two second-order Markov models, one to model a coding region and another one to model a non-coding region near the splice site. We collected sequences of 80 bp on either side of the true splice sites, grouped them into coding or non-coding sets and then used these data to build the models. (Note that for exons and introns <80 bp, this procedure will include sequences from both coding and non-coding regions on the same side of the splice site. This event is relatively rare, and only slightly alters the Markov probabilities; the average exon and intron lengths in the A.thaliana data set are ∼216 and 157 bp, respectively.) The score of a splice site located at position k in the DNA sequence was then computed according to the following formulae:
S(k) = Scomb(k,16) + [Scod(k – 80) – Snoncod(k – 80)] + [Snoncod(k + 1) – Scod(k + 1)]
where k is the position of a donor site and:
S(k) = Scomb(k,29) + [Snoncod(k – 80) – Scod(k – 80)] + [Scod(k + 1) – Snoncod(k + 1)]
where k is the position of an acceptor site.
Here, Scomb(k,i) is the the score computed with our combined algorithm using MDD with Markov models at the leaf nodes of the tree, Scod(j) is the score of the coding Markov model computed on an 80 base substring starting at position j, and Snoncod(j) is the score of the non-coding Markov model computed on an 80 base substring starting at position j.
We used the training data to set a threshold for the splice site score computed by these formulae. We further eliminated a significant number of false positives by keeping only the splice sites whose score was maximal within a 60 bp DNA window, similarly to the locally optimal splice sites used by Brendel and Kleffe (2).
RESULTS AND DISCUSSION
To evaluate GeneSplicer, we needed databases of confirmed genes in which the splicing patterns were accurately annotated. We collected data and tested the system on DNA sequences from the A.thaliana and human genomes. These results are described below.
Data collection
The Arabidopsis database for evaluating GeneSplicer was constructed by searching all the genes from chromosome (chr) II as of late 1999 (7) against a non-redundant protein database and an EST database. We retained in the data only those genes confirmed by homology across their full length, and we carefully checked the borders of the genes for non-consensus splice sites or other evidence of error. This process resulted in a set of 1131 genes. We then added to this set the 474 genes collected from GenBank and used to train an Arabidopsis version of Genscan (C.Burge, personal communication). These 474 genes span all five Arabidopsis chromosomes, and 23 were already included in the chr II data. After eliminating these duplicates, we were left with 1582 genes. This number was further reduced by performing pairwise alignments between all genes to remove homologous sequences. This resulted in a non-redundant set of 1323 genes that was used for training and testing the algorithm. This database contains 5490 splice sites of each type (acceptor or donor). We also collected all the ‘false’ splice sites in the data, defined as sequences containing the consensus GT or AG dinucleotide that were not annotated as splice sites. The numbers of true and false splice sites present in the data are shown in Table 1. All the results in this paper refer to standard GT/AG splice sites, leaving detection of the much less frequent non-consensus splice sites as a feature to be implemented in the future. Therefore some of the results below indicate a difference between the number of donor and acceptor sites.
Table 1. Number of genes, true and false splice sites in the data sets for A.thaliana and human.
| Database |
Genes |
Donor sites |
Acceptor sites |
False donors |
False acceptors |
| Arabidopsis | 1323 | 5440 | 5488 | 351 615 | 417 939 |
| Arabidopsis, chr II only | 928 | 3354 | 3391 | 181 659 | 221 491 |
| Human | 1115 | 5733 | 5733 | 478 983 | 650 099 |
For our evaluation of GeneSplicer on human DNA, we used the Exon–Intron Database (8) to collect a confirmed gene set. We extracted only fully annotated human genes with experimental supporting evidence. After removing genes with unknown introns, we had 2532 genes, which were further screened for homologous sequences using the same technique as the one we used for A.thaliana. This process resulted in a non-redundant data set of 1115 genes. (Note that because EID is built automatically by parsing GenBank records, there are likely to be incorrectly annotated genes included in this data set.) The total number of true and false splice sites for this data set is shown in Table 1.
We used a 5-fold cross-validation in all our experiments to estimate the splice site detection accuracy, as follows: (i) the data set was randomly divided into five equal-sized disjoint partitions. (ii) For each partition, we used all data outside the partition to train GeneSplicer. We then tested the program on the data in the partition. (iii) The reported accuracy represents the average of the accuracies computed on all five partitions.
GeneSplicer on A.thaliana
The accuracy of GeneSplicer computed on all five partitions used in the cross-validation experiment for several false negative rates of the splice sites is shown in Table 2, which shows the relationship between false negative and false positive rates. In order to evaluate GeneSplicer in context, we compared its results with those of NetPlantGene, a well-known splice site detection system for A.thaliana (9). We also analyzed the results of NetGene2 (9,10; http://genome.cbs.dtu.dk/services/NetGene2/), a newer system which was designed to replace NetPlantGene. Since the latter system was not strictly an improvement when tested on our data set, we report here results from both systems. All genes in the A.thaliana chr II subset of our non-redundant data set (Table 1) were submitted to be analyzed by the Web servers at http://genome.cbs.dtu.dk/services/NetGene2/ for NetGene2 and at http://genome.cbs.dtu.dk/services/NetPGene/ for NetPlantGene. We computed the number of false negatives and false positives returned by these servers for this data. Because all of the genes in our data set are publicly available from GenBank, and because we do not have the ability to re-train either of these systems, we cannot discriminate between those genes that were included in the training sets of NetGene2 or NetPlantGene. Therefore, we present GeneSplicer results for both the training and test data.
Table 2. False negative and false positive rates for acceptor and donor site detection on five equal-sized disjoint partitions of a 1323 gene A.thaliana data set.
| True sites missed (%) | False Positives (%) | ||||||
| |
|
Part. 1 |
Part. 2 |
Part. 3 |
Part. 4 |
Part. 5 |
Average |
| Acceptor site detection (5488 true sites) | 3 | 13.64 | 8.06 | 9.23 | 11.04 | 16.46 | 11.7 |
| 5 | 5.66 | 4.05 | 4.88 | 4.71 | 4.95 | 4.9 | |
| 7 | 4.03 | 2.87 | 3.31 | 3.08 | 3.38 | 3.3 | |
| 8 | 3.48 | 2.51 | 3.05 | 2.74 | 2.73 | 2.9 | |
| 10 | 2.93 | 2.04 | 2.38 | 2.39 | 2.38 | 2.4 | |
| 15 | 1.71 | 1.39 | 1.62 | 1.75 | 1.69 | 1.6 | |
| 20 | 1.12 | 0.99 | 1.06 | 1.23 | 1.26 | 1.1 | |
| 30 | 0.68 | 0.58 | 0.61 | 0.70 | 0.71 | 0.7 | |
| Donor site detection (5440 true sites) | 3 | 6.37 | 4.54 | 3.75 | 3.61 | 5.04 | 4.7 |
| 5 | 3.36 | 2.85 | 2.66 | 2.52 | 2.83 | 2.8 | |
| 7 | 1.73 | 2.32 | 1.88 | 1.80 | 1.87 | 1.9 | |
| 8 | 1.61 | 2.07 | 1.71 | 1.58 | 1.65 | 1.7 | |
| 10 | 1.36 | 1.50 | 1.23 | 1.28 | 1.38 | 1.4 | |
| 15 | 0.95 | 0.95 | 0.83 | 0.97 | 0.87 | 0.9 | |
| 20 | 0.66 | 0.60 | 0.61 | 0.69 | 0.62 | 0.6 | |
| 30 | 0.41 | 0.44 | 0.36 | 0.41 | 0.40 | 0.4 |
To make the comparison appropriate, we computed a complete ROC (receiver operating characteristics) plot for GeneSplicer. This plot contains all combinations of values of sensitivity and specificity. We can then compare GeneSplicer to each of the other systems by setting its false negative rate to be the same, and then comparing the differences in false positives. This is shown in Table 3.
Table 3. False negative and false positive rates for acceptor and donor site detection for GeneSplicer, NetPlantGene and NetGene2 on the A.thaliana data set.
| False negatives (%) | False positives (%) | ||||
| (true sites missed) | GeneSplicer | NetGene2 (chr II data) | NetPlantGene (chr II data) | ||
| |
|
Train |
Test |
|
|
| Acceptor site detection | 9.5 | 1.9 | 2.5 | - | 4.0 |
| 15.4 | 1.2 | 1.6 | 1.5 | - | |
| Donor site detection | 5.9 | 1.7 | 2.3 | 2.7 | - |
| 7.7 | 1.3 | 1.8 | - | 2.1 |
If we set the threshold in GeneSplicer to have the same sensitivity (missing the same number of true splice sites) as NetPlantGene, then GeneSplicer reports 38% fewer falsely predicted acceptor sites and 14% fewer false donor sites, considering GeneSplicer’s performance only on the test set. When compared to NetGene2, again at the same level of sensitivity, GeneSplicer introduced slightly more false positive acceptor sites (1.6 versus 1.5%) and 15% fewer false donor sites. One can also see from Table 3 that NetGene2 misses many more true acceptor sites than NetPlantGene (15.4 versus 9.5%), and somewhat fewer true donor sites (5.9 versus 7.7%). The false negative rate is a fully adjustable parameter in GeneSplicer; by default it is currently set so that it will miss appoximately the same number of donor sites as NetGene2 and the same number of acceptor sites as NetPlantGene.
GeneSplicer on human genes
Next, we trained GeneSplicer on our non-redundant human data set, and measured its accuracy using the 5-fold cross-validation method described above. The performance of GeneSplicer on this database is presented in Table 4.
Table 4. False negative and false positive rates for acceptor and donor site detection on five equal-sized disjoint partitions of a 1115 gene human data set.
| True sites | False Positives (%) | ||||||
| |
missed (%) |
Part. 1 |
Part. 2 |
Part. 3 |
Part. 4 |
Part. 5 |
Average |
| Acceptor site detection (5773 true sites) | 3 | 7.18 | 10.57 | 13.22 | 7.17 | 8.34 | 9.3 |
| 5 | 5.31 | 6.90 | 5.64 | 4.94 | 5.97 | 5.8 | |
| 7 | 4.31 | 5.21 | 4.99 | 3.91 | 5.16 | 4.7 | |
| 8 | 3.76 | 4.78 | 4.43 | 3.64 | 4.71 | 4.3 | |
| 10 | 3.16 | 4.10 | 3.80 | 3.22 | 4.16 | 3.7 | |
| 15 | 2.34 | 3.12 | 2.55 | 2.13 | 2.89 | 2.6 | |
| 20 | 1.60 | 2.48 | 2.07 | 1.46 | 2.17 | 1.9 | |
| 40 | 0.50 | 1.40 | 0.73 | 0.56 | 0.92 | 0.8 | |
| Donor site detection (5773 true sites) | 3 | 16.63 | 12.00 | 21.39 | 9.10 | 14.16 | 14.7 |
| 5 | 5.98 | 6.66 | 5.91 | 5.21 | 8.02 | 6.4 | |
| 7 | 4.46 | 5.45 | 3.78 | 4.04 | 6.48 | 4.8 | |
| 8 | 3.96 | 4.34 | 3.45 | 3.39 | 5.58 | 4.1 | |
| 10 | 3.34 | 3.73 | 2.99 | 2.93 | 4.36 | 3.5 | |
| 15 | 2.41 | 2.65 | 2.02 | 1.99 | 3.38 | 2.5 | |
| 20 | 1.85 | 1.87 | 1.44 | 1.49 | 2.41 | 1.8 | |
| 40 | 0.75 | 0.52 | 0.51 | 0.66 | 0.99 | 0.7 | |
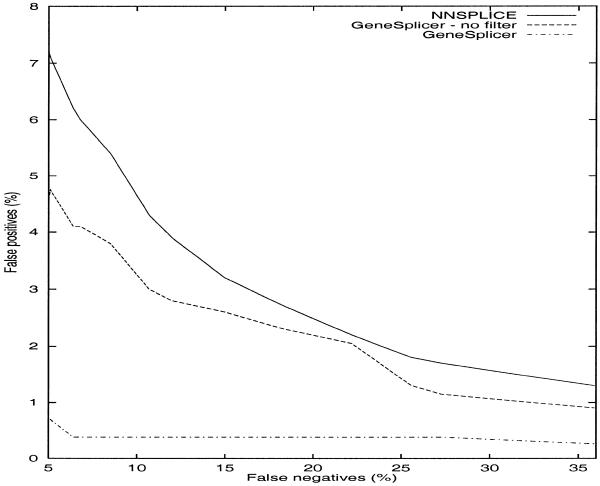
The splice site algorithms for plant genomes have not all been trained on human data, and conversely the algorithms for human have not been trained on plants. Therefore, in order to conduct a comparison on the human data, we had to consider a different set of programs from those used above. First we compared it to NNSplice, which is a splice site predictor based on neural networks (11; http://www.fruitfly.org/seq_tools/splice.html). For this comparison, we used the collection of data for human splice sites used in the GENIE system (http://www.fruitfly.org/sequence/human-datasets.html). GeneSplicer was trained and tested using the same data as NNSplice (11), using a training set of 1116 genes and a test set of 208 genes. The results of GeneSplicer on the test data were compared with those reported at the NNSplice Web site (http://www.fruitfly.org/seq_tools/splice.html). This comparison is shown in Figure 1 for acceptor sites and Figure 2 for donors. We also included, for comparison, the results of GeneSplicer before filtering the splice sites whose score was not locally maximal.
Figure 1.
False positive versus false negative rates on human acceptor sites for GeneSplicer both with and without the local maximal score filter, and for NNSplice.
Figure 2.
False positive versus false negative rates on human donor sites for GeneSplicer both with and without the local maximal score filter, and for NNSplice.
For the remaining systems, we did not have access to complete descriptions of their training and test data; therefore, we used our own test data to compare GeneSplicer with five of the best splice site recognition systems currently available: NetGene2 (trained on human data) (10; http://genome.cbs.dtu.dk/services/NetGene2/); HSPL (12,13; http://genomic.sanger.ac.uk/); NNSplice (11; http://www.fruitfly.org/seq_tools/splice.html); the GENIO splice site and exon predictor (14,15; http://genio.informatik.uni-stuttgart.de/GENIO/splice/); and SpliceView (16; http://l25.itba.mi.cnr.it/~webgene/wwwspliceview.html). Since all five of these splice site predictors offer a Web page where the DNA sequences may be submitted, we submitted all 1115 genes in our non-redundant human data set to each site. We used the default parameters for each of the Web-based predictors.
Note that, as with the Arabidopsis data, all our test data are publicly available. Thus, it is not possible, without access to source code for each of the other systems, to measure their accuracy on a data set that was not included in the training data. This restriction will tend to make the other systems look better, since in some cases the results will be mixing accuracy on training and test data, so we present GeneSplicer’s accuracy on the test data as well as on the entire human data set (including the training data set). The results are shown in Table 5.
Table 5. False negative and false positive rates of acceptor and donor site detection compared for GeneSplicer, NetGene2, HSPL, NNSplice, the GENIO splice site and exon predictor, and SpliceView on the human data set.
| True sites missed (%) | False Positives (%) | |||||||
| GeneSplicer | NetG2 | HSPL | NNSplice | GENIO | SpliceView | |||
| |
|
(test) |
(all data) |
(all data) |
(all data) |
(all data) |
(all data) |
(all data) |
| Acceptor sites (5733 true sites) | 5.6 | 5.4 | 4.1 | - | - | - | - | 16.3 |
| 6.4 | 4.9 | 3.7 | 4.6 | - | - | - | - | |
| 19.2 | 2.0 | 1.4 | - | - | - | 3.3 | - | |
| 20.4 | 1.9 | 1.3 | - | 1.5 | - | - | - | |
| 33.0 | 1.2 | 0.7 | - | - | 4.8 | - | - | |
| Donor sites (5733 true sites) | 6.0 | 5.4 | 4.3 | 2.5 | - | - | - | - |
| 7.0 | 4.9 | 3.8 | - | - | - | - | 11.9 | |
| 10.4 | 3.3 | 2.6 | - | 2.5 | - | - | - | |
| 11.2 | 3.1 | 2.5 | - | - | - | 4 | - | |
| 25.3 | 1.4 | 1.4 | - | - | 4.1 | - | - |
Table 5 shows that in comparison with three of the systems—NNSplice, GENIO and SpliceView—GeneSplicer has substantially lower error rates for both donor and acceptor sites. HSPL has a false positive rate on acceptor sites that falls in between GeneSplicer’s rate for the test set and the complete set. Its false positive rate for donor sites is slightly better than GeneSplicer’s rate on the complete set (2.5 versus 2.6%). NetGene2 has a false positive rate for acceptor sites (4.6%) that is in between GeneSplicer’s rate on the test set and the complete set (4.9 and 3.7%, respectively), while on donor sites it has the lowest false positive rate of all systems. Overall, NetGene2 appears to be the best for donor site prediction, while for acceptor sites either GeneSplicer, NetGene2 or HSPL perform comparably. One advantage of GeneSplicer for the latter task is that its thresholds can be adjusted by the user to vary the false negative and false positive rates.
CONCLUSIONS
The comparison of GeneSplicer to other splice site predictors indicates that GeneSplicer is comparable to the best predictors for both human and plant data, and considerably better than most systems. The comparison holds up even though our test data probably included some genes used to train most of the other systems in the comparison. Of the systems evaluated, only NetGene2 and GeneSplicer are available for both human and Arabidopsis sequences. In addition to its accuracy, an advantage of GeneSplicer over all the other systems is its computational efficiency. First, in terms of memory usage and input sequence length, GeneSplicer has no discernible limits, being able to process the entire 20 Mb sequence of chr II of A.thaliana. All the other systems limit submissions (through their Web sites) to a few kilobases, and locally installed versions run so slowly that large submissions are impractical. (The GeneSplicer code is freely available; contact the authors for download information.) Second, in terms of speed, GeneSplicer surpasses all of the systems we tested. Not all systems were available for downloading, and running through a Web server is obviously slower than running locally, but in one local comparison, GeneSplicer took 1 min to predict splice sites in a 1 Mb sequence extracted from A.thaliana, while NetGene2 processes the same sequence in 1 h (running both systems on a 450 MHz Pentium II Linux computer). While the accuracy of these systems is the most important feature, GeneSplicer’s computational efficiency makes it much easier to run on large DNA sequences, which are becoming increasingly common as genome sequencing progress accelerates.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Maria-Ines Benito for helpful discussions on Arabidopsis splicing, and to Chris Burge for providing his Arabidopsis training data. This work was supported in part by NSF grants KDI-9980088 and IIS-9902923 and by NIH grant R01-LM06845 to S.L.S.
References
- 1.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 2.Brendel V. and Kleffe,J. (1998) Prediction of locally optimal splice sites in plant pre-mRNA with applications to gene identification in Arabidopsis thaliana genomic DNA. Nucleic Acids Res., 26, 4748–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berget S. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 4.Moore M.J., Query,C.C. and Sharp,P.A. (1993) In Gesteland,R. and Atkins,J. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357.
- 5.Salzberg S., Delcher,A., Fasman,K. and Henderson,J. (1998) A decision tree system for finding genes in DNA. J. Comput. Biol., 5, 667–680. [DOI] [PubMed] [Google Scholar]
- 6.Salzberg S.L., Pertea,M., Delcher,A.L., Gardner,M.J. and Tettelin,H. (1999) Interpolated Markov models for eukaryotic gene finding. Genomics, 59, 24–31. [DOI] [PubMed] [Google Scholar]
- 7.Lin X., Kaul,S., Rounsley,S., Shea,T.P., Benito,M.-I., Town,C.D., Fujii,C.Y., Mason,T., Bowman,C.L., Barnstead,M. et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature, 402, 761–768. [DOI] [PubMed] [Google Scholar]
- 8.Saxonov S., Daizadeh,I., Fedorov,A. and Gilbert,W. (2000) An exhaustive database of protein-coding intron-containing genes. Nucleic Acids Res., 28, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebsgaard S.M., Korning,P.G., Tolstrup,N., Engelbrecht,J., Rouze,P. and Brunak,S. (1996) Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res., 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunak S., Engelbrecht,J. and Knudsen,S. (1991) Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol., 220, 49–65. [DOI] [PubMed] [Google Scholar]
- 11.Reese M.G., Eeckman,F.H., Kulp,D. and Haussler,D. (1997) Improved splice site detection in Genie. J. Comput. Biol., 4, 311–323. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard T., Birney,E., Bruskiewich,R., Clamp,M., Gilbert,J., King,A., Pocock,M. and Wilming,L. (1999) Abstracts of Papers Presented at the 1999 Meeting on Genome Sequencing and Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 114.
- 13.Solovyev V.V., Salamov,A.A. and Lawrence,C.B. (1994) Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res., 22, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mache N. and Levi,P. (1998) EST/STS Guided Identification of Genes in Human Genomic DNA. ISMB98 Poster, Montreal, Canada.
- 15.Mache N. and Levi,P. (1998) GENIO—A Non-Redundant Eukaryotic Gene Database of Annotated Sites and sequences. RECOMB-98 Poster, New York.
- 16.Rogozin I.B. and Milanesi,L. (1997) Analysis of donor splice signals in different organisms. J. Mol. Evol., 45, 50–59. [DOI] [PubMed] [Google Scholar]