Abstract
In the title compound, C25H23N3O2, the dihedral angles formed by the pyrazolone ring with the three aromatic rings are 14.59 (7), 79.35 (5) and 87.10 (6)°. Three intramolecular C—H⋯O, C—H⋯N and N—H⋯O hydrogen-bond interactions are present. The crystal structure is stabilized by two weak intermolecular C—H⋯O and C—H⋯N hydrogen-bond interactions.
Related literature
For the biological activity of 1-phenyl-3-methyl-4-benzoylpyrazolon-5-one and its metal complexes, see: Li et al. (1997 ▶); Liu et al. (1980 ▶); Zhou et al. (1999 ▶). For a related structure, see: Wang et al. (2003 ▶).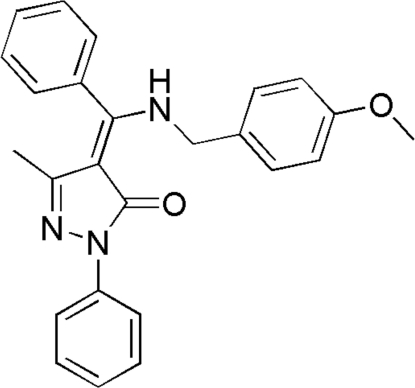
Experimental
Crystal data
C25H23N3O2
M r = 397.46
Orthorhombic,

a = 17.685 (4) Å
b = 11.613 (2) Å
c = 20.568 (4) Å
V = 4224.1 (15) Å3
Z = 8
Mo Kα radiation
μ = 0.08 mm−1
T = 113 K
0.20 × 0.18 × 0.12 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2005 ▶) T min = 0.984, T max = 0.990
32552 measured reflections
4852 independent reflections
4332 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.056
wR(F 2) = 0.135
S = 1.12
4852 reflections
278 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.23 e Å−3
Δρmin = −0.28 e Å−3
Data collection: CrystalClear (Rigaku, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809040458/fl2271sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809040458/fl2271Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3A⋯O1 | 0.96 (2) | 1.86 (2) | 2.6751 (17) | 141.3 (18) |
| C2—H2⋯O1 | 0.95 | 2.28 | 2.8956 (19) | 122 |
| C6—H6⋯N1 | 0.95 | 2.49 | 2.812 (2) | 100 |
| C25—H25C⋯O1i | 0.98 | 2.57 | 3.538 (2) | 169 |
| C24—H24⋯N1ii | 0.95 | 2.61 | 3.551 (2) | 174 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors gratefully acknowledge the financial support of the National Natural Science Foundation of China (grant No. 20772066).
supplementary crystallographic information
Comment
1-Phenyl-3-methyl-4-benzoylpyrazolon-5-one (HPMBP), an effective β-diketonate, is widely used and well known for its extractive ability. In recent years, HPMBP and its metal complexes have also been found to have good antibacterial and biological properties. Its metal complexes have analgesic activity (Liu et al., 1980; Li et al., 1997; Zhou et al., 1999). In order to develop new medicines, we have synthesized the title compound, (I), and its structure is reported here.
The structure of (I) is shown in Fig. 1. The dihedral angles formed by the pyrazolone ring with the three benzene rings C1···C6, C12···C17 and C19···C24 are 14.59 (7), 79.35 (5) and 87.10 (6)°, respectively. The O atom of the 3-methyl-1-phenylpyrazol-5-one moiety and the N atom of the amino group are available for coordination with metals. The pyrazole ring is planar and atoms O1, C7, C8, C11 and N3 are almost coplanar fwith an rmsd value of 0.0093 and the largest deviation is 0.0144 (10) Å for atom C7. The dihedral angle between this mean plane and the pyrazoline ring of PMBP is 3.53 (7)°, close to the value of 3.56 (3)° found in 4-{[3,4-dihydro-5- methyl-3-oxo-2-phenyl-2H-pyrazol-4-ylidene(phenyl)methylamino}- 1,5-dimethyl-2-phenyl-1H-pyrazol-3(2H)-one(Wang et al., 2003). The bond lengths within this part of the molecule lie between classical single- and double-bond lengths, indicating extensive conjugation. A strong intramolecular N3—H3A···O1 hydrogen bond (Table 1) is observed, leading to a keto-enamine form. The molecule is further stabilized by a C—H···O weak intramolecular hydrogen bond (Table 1). The crystal structure also involves two weak intermolecular(C—H···O and C—H···N) hydrogen-bond interactions(Fig. 2).
Experimental
Compound (I) was synthesized by refluxing a mixture of 1-phenyl-3- methyl-4-benzoylpyrazol-5-one (10 mmol) and 4-methoxybenzylamine (10 mmol) in ethanol (80 ml) over a steam bath for about 4 h. Excess solvent was removed by evaporation and the solution was cooled to room temperature. After 2 days a yellow solid was obtained and this was dried in air. The product was recrystallized from ethanol, to afford yellow crystals of (I) suitable for X-ray analysis.
Refinement
C-bonded H atoms were positioned geometrically, with C—H = 0.95–0.99 Å and amine H atoms (H3) were found in a difference map. Amine H atoms were refined freely, while C-bonded H atoms were included in the final cycles of refinement using a riding model, with Uiso(H) = 1.2Ueq(CH2 and CH) or 1.5Ueq(CH3).
Figures
Fig. 1.
View of the title compound, with displacement ellipsoids drawn at the 30% probability level.
Fig. 2.
Intermolecular hydrogen bonds (dashed line) in the structure of (I).
Crystal data
| C25H23N3O2 | F(000) = 1680 |
| Mr = 397.46 | Dx = 1.250 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 10389 reflections |
| a = 17.685 (4) Å | θ = 2.3–27.5° |
| b = 11.613 (2) Å | µ = 0.08 mm−1 |
| c = 20.568 (4) Å | T = 113 K |
| V = 4224.1 (15) Å3 | Block, yellow |
| Z = 8 | 0.20 × 0.18 × 0.12 mm |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 4852 independent reflections |
| Radiation source: rotating anode | 4332 reflections with I > 2σ(I) |
| confocal | Rint = 0.043 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.5°, θmin = 2.3° |
| ω and φ scans | h = −22→20 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2005) | k = −15→14 |
| Tmin = 0.984, Tmax = 0.990 | l = −20→26 |
| 32552 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.056 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.135 | w = 1/[σ2(Fo2) + (0.0569P)2 + 1.4565P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max = 0.001 |
| 4852 reflections | Δρmax = 0.23 e Å−3 |
| 278 parameters | Δρmin = −0.28 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0196 (15) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.03774 (6) | 0.05679 (9) | 0.39451 (5) | 0.0342 (3) | |
| O2 | 0.22937 (8) | −0.44370 (11) | 0.33811 (7) | 0.0532 (4) | |
| N1 | 0.04301 (7) | 0.34722 (11) | 0.44256 (6) | 0.0315 (3) | |
| N2 | 0.02452 (7) | 0.25809 (10) | 0.39929 (6) | 0.0290 (3) | |
| N3 | 0.11039 (8) | −0.01861 (11) | 0.50035 (6) | 0.0336 (3) | |
| C1 | −0.01072 (8) | 0.28454 (13) | 0.33938 (7) | 0.0295 (3) | |
| C2 | −0.04472 (9) | 0.19813 (14) | 0.30272 (7) | 0.0355 (4) | |
| H2 | −0.0454 | 0.1211 | 0.3182 | 0.043* | |
| C3 | −0.07760 (10) | 0.22525 (16) | 0.24341 (8) | 0.0411 (4) | |
| H3 | −0.1002 | 0.1662 | 0.2181 | 0.049* | |
| C4 | −0.07778 (9) | 0.33747 (17) | 0.22084 (8) | 0.0429 (4) | |
| H4 | −0.0998 | 0.3554 | 0.1799 | 0.051* | |
| C5 | −0.04572 (9) | 0.42288 (16) | 0.25824 (9) | 0.0438 (4) | |
| H5 | −0.0468 | 0.5002 | 0.2432 | 0.053* | |
| C6 | −0.01187 (9) | 0.39786 (14) | 0.31747 (8) | 0.0370 (4) | |
| H6 | 0.0103 | 0.4574 | 0.3428 | 0.044* | |
| C7 | 0.04567 (8) | 0.15137 (13) | 0.42300 (7) | 0.0283 (3) | |
| C8 | 0.07883 (8) | 0.17516 (12) | 0.48542 (7) | 0.0277 (3) | |
| C9 | 0.07448 (8) | 0.29768 (13) | 0.49309 (7) | 0.0299 (3) | |
| C10 | 0.09843 (10) | 0.37075 (15) | 0.54929 (8) | 0.0404 (4) | |
| H10A | 0.0878 | 0.4518 | 0.5396 | 0.061* | |
| H10B | 0.1528 | 0.3608 | 0.5568 | 0.061* | |
| H10C | 0.0704 | 0.3475 | 0.5883 | 0.061* | |
| C11 | 0.11209 (8) | 0.08831 (13) | 0.52273 (7) | 0.0282 (3) | |
| C12 | 0.15154 (8) | 0.11208 (13) | 0.58498 (7) | 0.0294 (3) | |
| C13 | 0.11171 (9) | 0.12018 (16) | 0.64250 (8) | 0.0404 (4) | |
| H13 | 0.0585 | 0.1095 | 0.6428 | 0.048* | |
| C14 | 0.14969 (11) | 0.1440 (2) | 0.69986 (9) | 0.0541 (5) | |
| H14 | 0.1223 | 0.1496 | 0.7395 | 0.065* | |
| C15 | 0.22707 (11) | 0.15964 (19) | 0.69975 (9) | 0.0521 (5) | |
| H15 | 0.2527 | 0.1767 | 0.7392 | 0.063* | |
| C16 | 0.26706 (10) | 0.15051 (18) | 0.64253 (9) | 0.0482 (5) | |
| H16 | 0.3203 | 0.1609 | 0.6425 | 0.058* | |
| C17 | 0.22957 (9) | 0.12621 (15) | 0.58515 (8) | 0.0391 (4) | |
| H17 | 0.2572 | 0.1192 | 0.5458 | 0.047* | |
| C18 | 0.14345 (10) | −0.12099 (14) | 0.53081 (8) | 0.0359 (4) | |
| H18A | 0.1058 | −0.1578 | 0.5597 | 0.043* | |
| H18B | 0.1877 | −0.0986 | 0.5574 | 0.043* | |
| C19 | 0.16749 (8) | −0.20464 (13) | 0.47878 (7) | 0.0306 (3) | |
| C20 | 0.22998 (9) | −0.18247 (13) | 0.44036 (8) | 0.0339 (3) | |
| H20 | 0.2575 | −0.1130 | 0.4464 | 0.041* | |
| C21 | 0.25327 (9) | −0.26025 (15) | 0.39292 (8) | 0.0372 (4) | |
| H21 | 0.2965 | −0.2448 | 0.3670 | 0.045* | |
| C22 | 0.21200 (10) | −0.36117 (14) | 0.38418 (8) | 0.0385 (4) | |
| C23 | 0.14925 (10) | −0.38372 (14) | 0.42166 (9) | 0.0407 (4) | |
| H23 | 0.1213 | −0.4527 | 0.4152 | 0.049* | |
| C24 | 0.12701 (9) | −0.30591 (14) | 0.46861 (8) | 0.0368 (4) | |
| H24 | 0.0836 | −0.3216 | 0.4943 | 0.044* | |
| C25 | 0.29322 (13) | −0.42679 (19) | 0.30302 (11) | 0.0608 (6) | |
| H25A | 0.2890 | −0.3545 | 0.2787 | 0.091* | |
| H25B | 0.3000 | −0.4908 | 0.2726 | 0.091* | |
| H25C | 0.3368 | −0.4228 | 0.3324 | 0.091* | |
| H3A | 0.0837 (12) | −0.0281 (18) | 0.4599 (11) | 0.057 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0433 (6) | 0.0283 (5) | 0.0310 (6) | 0.0064 (4) | −0.0066 (5) | −0.0041 (4) |
| O2 | 0.0675 (9) | 0.0387 (7) | 0.0535 (8) | 0.0109 (6) | −0.0160 (7) | −0.0121 (6) |
| N1 | 0.0359 (7) | 0.0267 (6) | 0.0319 (7) | 0.0000 (5) | −0.0018 (5) | −0.0022 (5) |
| N2 | 0.0344 (6) | 0.0255 (6) | 0.0269 (6) | 0.0027 (5) | −0.0032 (5) | −0.0009 (5) |
| N3 | 0.0400 (7) | 0.0312 (7) | 0.0296 (7) | 0.0079 (5) | −0.0056 (6) | 0.0008 (5) |
| C1 | 0.0267 (7) | 0.0347 (8) | 0.0272 (7) | 0.0056 (6) | 0.0025 (6) | 0.0042 (6) |
| C2 | 0.0405 (9) | 0.0354 (8) | 0.0307 (8) | 0.0088 (7) | −0.0035 (7) | −0.0005 (6) |
| C3 | 0.0411 (9) | 0.0521 (10) | 0.0302 (8) | 0.0103 (8) | −0.0047 (7) | −0.0026 (7) |
| C4 | 0.0336 (8) | 0.0629 (12) | 0.0320 (8) | 0.0079 (8) | −0.0004 (7) | 0.0131 (8) |
| C5 | 0.0352 (9) | 0.0487 (10) | 0.0477 (10) | −0.0009 (7) | −0.0018 (8) | 0.0218 (8) |
| C6 | 0.0329 (8) | 0.0357 (8) | 0.0425 (9) | −0.0016 (6) | −0.0005 (7) | 0.0108 (7) |
| C7 | 0.0299 (7) | 0.0273 (7) | 0.0276 (7) | 0.0031 (6) | 0.0002 (6) | −0.0004 (5) |
| C8 | 0.0280 (7) | 0.0285 (7) | 0.0267 (7) | 0.0015 (5) | −0.0001 (6) | −0.0012 (6) |
| C9 | 0.0299 (7) | 0.0297 (7) | 0.0303 (7) | −0.0006 (6) | 0.0018 (6) | −0.0013 (6) |
| C10 | 0.0497 (10) | 0.0336 (8) | 0.0378 (9) | −0.0015 (7) | −0.0051 (7) | −0.0052 (7) |
| C11 | 0.0254 (7) | 0.0323 (8) | 0.0269 (7) | 0.0018 (6) | 0.0022 (6) | 0.0015 (6) |
| C12 | 0.0286 (7) | 0.0318 (8) | 0.0279 (7) | −0.0016 (6) | −0.0013 (6) | 0.0031 (6) |
| C13 | 0.0307 (8) | 0.0582 (11) | 0.0323 (8) | −0.0071 (7) | 0.0015 (7) | −0.0060 (7) |
| C14 | 0.0499 (11) | 0.0811 (15) | 0.0314 (9) | −0.0092 (10) | 0.0008 (8) | −0.0084 (9) |
| C15 | 0.0494 (11) | 0.0669 (13) | 0.0401 (10) | −0.0066 (9) | −0.0157 (8) | −0.0040 (9) |
| C16 | 0.0314 (9) | 0.0588 (12) | 0.0544 (11) | −0.0081 (8) | −0.0117 (8) | 0.0021 (9) |
| C17 | 0.0301 (8) | 0.0485 (10) | 0.0386 (9) | −0.0048 (7) | 0.0014 (7) | 0.0069 (7) |
| C18 | 0.0413 (9) | 0.0319 (8) | 0.0345 (8) | 0.0082 (7) | −0.0011 (7) | 0.0058 (6) |
| C19 | 0.0329 (7) | 0.0262 (7) | 0.0328 (8) | 0.0039 (6) | −0.0060 (6) | 0.0046 (6) |
| C20 | 0.0355 (8) | 0.0295 (8) | 0.0368 (8) | −0.0004 (6) | −0.0045 (6) | 0.0012 (6) |
| C21 | 0.0370 (8) | 0.0384 (9) | 0.0361 (8) | 0.0049 (7) | −0.0006 (7) | 0.0013 (7) |
| C22 | 0.0477 (9) | 0.0313 (8) | 0.0363 (8) | 0.0100 (7) | −0.0142 (7) | −0.0058 (7) |
| C23 | 0.0437 (9) | 0.0297 (8) | 0.0486 (10) | −0.0033 (7) | −0.0167 (8) | 0.0020 (7) |
| C24 | 0.0338 (8) | 0.0338 (8) | 0.0427 (9) | −0.0014 (6) | −0.0095 (7) | 0.0070 (7) |
| C25 | 0.0666 (13) | 0.0546 (12) | 0.0614 (13) | 0.0193 (10) | −0.0103 (11) | −0.0208 (10) |
Geometric parameters (Å, °)
| O1—C7 | 1.2527 (18) | C11—C12 | 1.484 (2) |
| O2—C25 | 1.354 (3) | C12—C13 | 1.380 (2) |
| O2—C22 | 1.382 (2) | C12—C17 | 1.390 (2) |
| N1—C9 | 1.3118 (19) | C13—C14 | 1.386 (2) |
| N1—N2 | 1.4037 (17) | C13—H13 | 0.9500 |
| N2—C7 | 1.3834 (19) | C14—C15 | 1.381 (3) |
| N2—C1 | 1.4145 (19) | C14—H14 | 0.9500 |
| N3—C11 | 1.325 (2) | C15—C16 | 1.377 (3) |
| N3—C18 | 1.4656 (19) | C15—H15 | 0.9500 |
| N3—H3A | 0.96 (2) | C16—C17 | 1.383 (2) |
| C1—C6 | 1.391 (2) | C16—H16 | 0.9500 |
| C1—C2 | 1.392 (2) | C17—H17 | 0.9500 |
| C2—C3 | 1.388 (2) | C18—C19 | 1.507 (2) |
| C2—H2 | 0.9500 | C18—H18A | 0.9900 |
| C3—C4 | 1.383 (3) | C18—H18B | 0.9900 |
| C3—H3 | 0.9500 | C19—C20 | 1.383 (2) |
| C4—C5 | 1.377 (3) | C19—C24 | 1.393 (2) |
| C4—H4 | 0.9500 | C20—C21 | 1.392 (2) |
| C5—C6 | 1.388 (2) | C20—H20 | 0.9500 |
| C5—H5 | 0.9500 | C21—C22 | 1.392 (2) |
| C6—H6 | 0.9500 | C21—H21 | 0.9500 |
| C7—C8 | 1.438 (2) | C22—C23 | 1.376 (3) |
| C8—C11 | 1.397 (2) | C23—C24 | 1.380 (2) |
| C8—C9 | 1.434 (2) | C23—H23 | 0.9500 |
| C9—C10 | 1.495 (2) | C24—H24 | 0.9500 |
| C10—H10A | 0.9800 | C25—H25A | 0.9800 |
| C10—H10B | 0.9800 | C25—H25B | 0.9800 |
| C10—H10C | 0.9800 | C25—H25C | 0.9800 |
| C25—O2—C22 | 116.75 (16) | C17—C12—C11 | 119.41 (14) |
| C9—N1—N2 | 106.13 (12) | C12—C13—C14 | 119.75 (15) |
| C7—N2—N1 | 111.97 (11) | C12—C13—H13 | 120.1 |
| C7—N2—C1 | 128.37 (13) | C14—C13—H13 | 120.1 |
| N1—N2—C1 | 119.66 (12) | C15—C14—C13 | 120.36 (17) |
| C11—N3—C18 | 127.07 (13) | C15—C14—H14 | 119.8 |
| C11—N3—H3A | 114.7 (12) | C13—C14—H14 | 119.8 |
| C18—N3—H3A | 118.2 (13) | C16—C15—C14 | 120.02 (17) |
| C6—C1—C2 | 120.02 (14) | C16—C15—H15 | 120.0 |
| C6—C1—N2 | 119.60 (14) | C14—C15—H15 | 120.0 |
| C2—C1—N2 | 120.38 (14) | C15—C16—C17 | 119.93 (16) |
| C3—C2—C1 | 119.58 (16) | C15—C16—H16 | 120.0 |
| C3—C2—H2 | 120.2 | C17—C16—H16 | 120.0 |
| C1—C2—H2 | 120.2 | C16—C17—C12 | 120.16 (16) |
| C4—C3—C2 | 120.65 (17) | C16—C17—H17 | 119.9 |
| C4—C3—H3 | 119.7 | C12—C17—H17 | 119.9 |
| C2—C3—H3 | 119.7 | N3—C18—C19 | 109.38 (12) |
| C5—C4—C3 | 119.34 (16) | N3—C18—H18A | 109.8 |
| C5—C4—H4 | 120.3 | C19—C18—H18A | 109.8 |
| C3—C4—H4 | 120.3 | N3—C18—H18B | 109.8 |
| C4—C5—C6 | 121.13 (16) | C19—C18—H18B | 109.8 |
| C4—C5—H5 | 119.4 | H18A—C18—H18B | 108.2 |
| C6—C5—H5 | 119.4 | C20—C19—C24 | 118.83 (15) |
| C5—C6—C1 | 119.25 (16) | C20—C19—C18 | 120.76 (14) |
| C5—C6—H6 | 120.4 | C24—C19—C18 | 120.40 (14) |
| C1—C6—H6 | 120.4 | C19—C20—C21 | 121.08 (15) |
| O1—C7—N2 | 126.17 (13) | C19—C20—H20 | 119.5 |
| O1—C7—C8 | 129.17 (14) | C21—C20—H20 | 119.5 |
| N2—C7—C8 | 104.65 (12) | C20—C21—C22 | 118.79 (16) |
| C11—C8—C9 | 132.73 (14) | C20—C21—H21 | 120.6 |
| C11—C8—C7 | 121.55 (13) | C22—C21—H21 | 120.6 |
| C9—C8—C7 | 105.49 (12) | C23—C22—O2 | 115.55 (16) |
| N1—C9—C8 | 111.77 (13) | C23—C22—C21 | 120.70 (15) |
| N1—C9—C10 | 118.93 (14) | O2—C22—C21 | 123.75 (17) |
| C8—C9—C10 | 129.28 (14) | C22—C23—C24 | 119.82 (15) |
| C9—C10—H10A | 109.5 | C22—C23—H23 | 120.1 |
| C9—C10—H10B | 109.5 | C24—C23—H23 | 120.1 |
| H10A—C10—H10B | 109.5 | C23—C24—C19 | 120.77 (16) |
| C9—C10—H10C | 109.5 | C23—C24—H24 | 119.6 |
| H10A—C10—H10C | 109.5 | C19—C24—H24 | 119.6 |
| H10B—C10—H10C | 109.5 | O2—C25—H25A | 109.5 |
| N3—C11—C8 | 118.43 (13) | O2—C25—H25B | 109.5 |
| N3—C11—C12 | 119.01 (13) | H25A—C25—H25B | 109.5 |
| C8—C11—C12 | 122.52 (13) | O2—C25—H25C | 109.5 |
| C13—C12—C17 | 119.77 (14) | H25A—C25—H25C | 109.5 |
| C13—C12—C11 | 120.82 (13) | H25B—C25—H25C | 109.5 |
| C9—N1—N2—C7 | −0.73 (16) | C7—C8—C11—N3 | −2.2 (2) |
| C9—N1—N2—C1 | 180.00 (12) | C9—C8—C11—C12 | 1.8 (2) |
| C7—N2—C1—C6 | −165.17 (15) | C7—C8—C11—C12 | 175.44 (13) |
| N1—N2—C1—C6 | 14.0 (2) | N3—C11—C12—C13 | −97.80 (19) |
| C7—N2—C1—C2 | 15.4 (2) | C8—C11—C12—C13 | 84.6 (2) |
| N1—N2—C1—C2 | −165.43 (13) | N3—C11—C12—C17 | 82.15 (19) |
| C6—C1—C2—C3 | 2.0 (2) | C8—C11—C12—C17 | −95.49 (18) |
| N2—C1—C2—C3 | −178.63 (14) | C17—C12—C13—C14 | 0.9 (3) |
| C1—C2—C3—C4 | −0.9 (2) | C11—C12—C13—C14 | −179.12 (17) |
| C2—C3—C4—C5 | −0.8 (3) | C12—C13—C14—C15 | 0.0 (3) |
| C3—C4—C5—C6 | 1.4 (3) | C13—C14—C15—C16 | −0.6 (3) |
| C4—C5—C6—C1 | −0.3 (2) | C14—C15—C16—C17 | 0.3 (3) |
| C2—C1—C6—C5 | −1.4 (2) | C15—C16—C17—C12 | 0.6 (3) |
| N2—C1—C6—C5 | 179.19 (14) | C13—C12—C17—C16 | −1.2 (3) |
| N1—N2—C7—O1 | −178.48 (14) | C11—C12—C17—C16 | 178.80 (16) |
| C1—N2—C7—O1 | 0.7 (2) | C11—N3—C18—C19 | −148.95 (15) |
| N1—N2—C7—C8 | 0.62 (16) | N3—C18—C19—C20 | 72.31 (18) |
| C1—N2—C7—C8 | 179.82 (13) | N3—C18—C19—C24 | −107.74 (16) |
| O1—C7—C8—C11 | 3.6 (2) | C24—C19—C20—C21 | −1.0 (2) |
| N2—C7—C8—C11 | −175.44 (13) | C18—C19—C20—C21 | 178.92 (14) |
| O1—C7—C8—C9 | 178.78 (15) | C19—C20—C21—C22 | 0.6 (2) |
| N2—C7—C8—C9 | −0.28 (15) | C25—O2—C22—C23 | −176.45 (16) |
| N2—N1—C9—C8 | 0.52 (16) | C25—O2—C22—C21 | 4.4 (2) |
| N2—N1—C9—C10 | −178.04 (13) | C20—C21—C22—C23 | 0.1 (2) |
| C11—C8—C9—N1 | 174.23 (15) | C20—C21—C22—O2 | 179.21 (14) |
| C7—C8—C9—N1 | −0.16 (17) | O2—C22—C23—C24 | −179.45 (14) |
| C11—C8—C9—C10 | −7.4 (3) | C21—C22—C23—C24 | −0.3 (2) |
| C7—C8—C9—C10 | 178.22 (15) | C22—C23—C24—C19 | −0.2 (2) |
| C18—N3—C11—C8 | 178.28 (14) | C20—C19—C24—C23 | 0.9 (2) |
| C18—N3—C11—C12 | 0.5 (2) | C18—C19—C24—C23 | −179.10 (14) |
| C9—C8—C11—N3 | −175.86 (15) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3A···O1 | 0.96 (2) | 1.86 (2) | 2.6751 (17) | 141.3 (18) |
| C2—H2···O1 | 0.95 | 2.28 | 2.8956 (19) | 122 |
| C6—H6···N1 | 0.95 | 2.49 | 2.812 (2) | 100 |
| C25—H25C···O1i | 0.98 | 2.57 | 3.538 (2) | 169 |
| C24—H24···N1ii | 0.95 | 2.61 | 3.551 (2) | 174 |
Symmetry codes: (i) −x+1/2, y−1/2, z; (ii) −x, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FL2271).
References
- Li, J.-Z., Yu, W.-J. & Du, X.-Y. (1997). Chin. J. Appl. Chem.14, 98–100.
- Liu, J.-M., Yang, R.-D. & Ma, T.-R. (1980). Chem. J. Chin. Univ.1, 23–29.
- Rigaku (2005). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, J.-L., Yang, Y., Zhang, X. & Miao, F.-M. (2003). Acta Cryst. E59, o430–o432.
- Zhou, Y.-P., Yang, Zh.-Y., Yu, H.-J. & Yang, R.-D. (1999). Chin. J. Appl. Chem.16, 37–41.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809040458/fl2271sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809040458/fl2271Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




