Abstract
Synthetic biology is a nascent technical discipline that seeks to enable the design and construction of novel biological systems to meet pressing societal needs. However, engineering biology still requires much trial and error because we lack effective approaches for connecting basic “parts” into higher-order networks that behave as predicted. Developing strategies for improving the performance and sophistication of our designs is informed by two overarching perspectives: “bottom-up” and “top-down” considerations. Using this framework, we describe a conceptual model for developing novel biological systems that function and interact with existing biological components in a predictable fashion. We discuss this model in the context of three topical areas: biochemical transformations, cellular devices and therapeutics, and approaches that expand the chemistry of life. Ten years after the construction of synthetic biology's first devices, the drive to look beyond what does exist to what can exist is ushering in an era of biology by design.
1. Introduction
Our understanding of physical laws and knowledge of material properties allow us to engineer bridges that do not collapse and car engines that convert energy into mechanical motion. Engineering biology, however, is different. Even the simplest bacterium comprises a system whose complexity is humbling [1, 2]. In recognition of such challenges, the central goal of synthetic biology is to transform biology into a system that can be engineered just as we engineer bridges and mechanical systems today (reviewed in [3–8]). This approach promises to provide an improved understanding of the living world and will enable us, in the coming years, to harness the diverse repertoire of biology for compelling applications. These include next-generation biofuels, renewable “green” chemicals and industrial feedstocks, programmable and personalizable biological therapies, materials with novel properties, cheap and deployable diagnostics and therapeutics to promote global health, and technologies enabling environmental stewardship and remediation. The guiding question is this: how can we make biology engineerable? The new paradigm of biology by design can be summarized as follows:
conceive a desired biological function,
design an engineered biological system to perform this function,
build the system,
the system performs as predicted.
Achieving this ambitious goal will require an improved understanding of the mechanisms by which biological parts function and by which they interact with one another and their environment.
The first wave of synthetic biology has focused on the development of simple biological modules, involving anywhere between two and ten well-characterized biological “parts” (e.g., genes) that are connected to perform defined functions (reviewed in [7]). In principle, these “parts” should still operate in a predictable manner when transplanted into different biological contexts. This conceptual framework connects synthetic biology with other engineering disciplines. Whether the parts are electrical, chemical, physical, or biological, engineers depend on the predictable behavior of connected components to design and construct complex systems. For example, the automotive industry must develop engines that work within the context of an entire automobile. Construction of an engine is possible due to an understanding of how the individual components (e.g., pistons, crankshafts, and spark plugs) interact with one another, how these individual functions can be assembled to achieve a desired task (e.g., conversion of energy released by combusting fuel into mechanical motion), and how this assembled system interacts with the broader context of the vehicle (e.g., drawing fuel from the fuel lines and powering the transmission to transmit energy to the wheels). When synthetic biologists design simple biological systems from individual components in order to perform novel functions, these systems often do not behave as expected, particularly when transported from one biological context to another. In effect, a synthetic biologist today must endeavor to design an engine from imperfectly characterized parts and without knowing how the engine will eventually connect to the rest of the vehicle.
To overcome the limitations of our incomplete knowledge of biology, synthetic biologists approach the construction of user-defined functions through an iterative design cycle that incorporates both bottom-up and top-down design perspectives (Figure 1).
Figure 1.
General conceptual framework for incorporating top-down and bottom-up perspectives in the synthetic biology design process. Due to our incomplete knowledge of biology, the design of biological systems through synthetic biology is currently an iterative process that incorporates both top-down and bottom-up design considerations. First, a design objective is identified. Next, a suitable synthetic biological system is designed given the known properties of well-characterized components (bottom-up). The synthetic system is then constructed and inserted into a larger biological context with which the synthetic system may interact (top-down), and performance of the combined system is assessed. If the system fails to meet performance requirements, this new information can be used to refine the design and repeat the cycle. Our ever-improving understanding of biology should reduce the number of iterations necessary to achieve a specific design objective.
-
Bottom-up considerations include the following: What is the desired function of the synthetic system? What parts could be used to construct this system (e.g., promoters, ribosome binding sites, etc.)? Do the parts exist and do they require additional characterization? How should the parts be configured? What is the predicted behavior of the synthetic module? Conceptually, this design perspective focuses on the characteristics of individual modules (which may be parts or subnetworks of parts) and their assembly into novel configurations in isolation from the endogenous cellular context into which they will eventually be placed.
-
Top-down considerations include the following: How might the engineered module be decoupled from or insulated from aspects of the broader biological context (e.g., metabolic state, cell cycle progression, and epigenetic modifications) that may complicate the function of the engineered network? What interactions are necessary to connect a synthetic controller to the endogenous system it will control? How can the synthetic system take advantage of the existing biological infrastructure? The top-down perspective focuses on potential interactions—both desirable and undesirable—between the engineered subsystem and its biological context, and on development of strategies for harnessing or compensating for these influences.
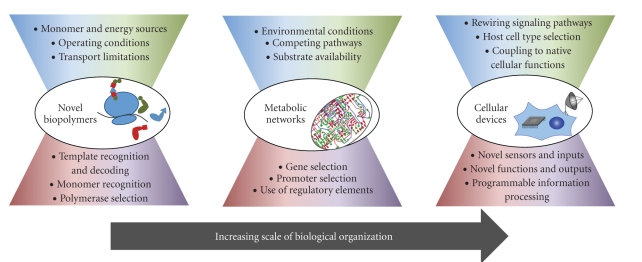
Top-down and bottom-up perspectives guide our development of improved strategies for engineering biology (Figure 1), even in the face of incomplete information. Here, we discuss these perspectives in the context of three rapidly progressing areas which exemplify the opportunities and challenges encountered at different scales of biological organization: biochemical transformations, cellular devices and therapeutics, and approaches that expand the chemistry of life (Figure 2). Each of these topics is leading to new applications and advancing frontiers in synthetic biology.
Figure 2.
Application of the general framework to specific design objectives. At each scale of biological organization, designing synthetic biological systems invokes unique instances of the top-down and bottom-up considerations described in Figure 1.
2. Applications and New Frontiers
2.1. Biochemical Transformations
In the nearly four decades since the molecular biology revolution, it has become clear that the biological world provides a rich and diverse repertoire that may help to address some of humanity's most significant challenges. For example, harnessing biology for production of small molecules has led to applications in biofuels, chemical feedstocks, and therapeutics [3, 9]. However, to achieve these advances, substantial investments of resources, time, and labor are currently necessary. For example, it has been estimated that engineering microbes for expression of the antimalarial drug artemisinin required 150-person years of work [10].
Biochemical transformations comprise a family of applications for which synthetic biology may complement and expand efforts in metabolic and genetic engineering, by providing a robust technological framework for addressing common challenges. These challenges include targeting substrate flux towards a specific product, avoiding the loss of intermediates to competing pathways, reducing the accumulation of toxic intermediates, and preventing saturation of pathway enzymes [11, 12]. Synthetic biology tools are used to remove rate-limiting steps and increase titers of target biochemicals through an iterative design cycle of analysis, design, and implementation (Figure 1). Each cycle invokes both bottom-up and top-down considerations (Figure 2). Bottom-up considerations may include gene selection, promoter selection, and use of regulatory elements. Top-down considerations may include selection of the host organism, potential interactions of the synthetic metabolic network with existing cellular components, and competition with native pathways that impact substrate or intermediate availability. Host selection must often include making a choice between transferring heterologous pathways to a well-characterized organism or improving the production capacity (or other properties) of organisms for which robust genetic tools are not available.
Despite the numerous design requirements and constraints, many biochemical transformation projects in biotechnology have been realized. Here, we highlight four types: (i) incorporating nonnative genes to extend natural metabolism, (ii) incorporating whole pathways to add function, (iii) creating new pathways that have never existed before in nature, and (iv) organizing pathways to enhance activity (e.g., through compartmentalization or scaffolds). It should be noted that significant engineering of host metabolism is often a necessity to ensure precursor availability and inhibition of competitive pathways [12]. Moreover, in many cases the scope of genetic modifications requires multiple approaches to achieve a desired user-defined objective [13–15].
To extend natural metabolism, network optimization and selection of nonnative enzymes that catalyze the formation of a desired biochemical is required. In one example of this approach, Liao and colleagues converted 2-keto acid metabolites into branched alcohols for use as biofuels [13]. This was accomplished in Escherichia coli by first engineering the amino acid biosynthetic pathways that supply precursors required for 2-keto acid metabolite production. Then, incorporation of 2-keto acid decarboxylases and alcohol dehydrogenases from a variety of organisms yielded the desired alcohols. In later work, Atsumi et al. improved production titers for isobutanol through the use of alcohol dehydrogenases from different organisms, Saccharomyces cerevisiae and Lactococcus lactis, rather than the native alcohol dehydrogenase of E. coli [16]. In another example, an E. coli strain was engineered to convert simple sugar to biodiesel through modified fatty-acid biosynthesis pathways [15]. Here, native host metabolism was reprogrammed by modulating more than 8 enzymes (such as thioesterase, acyl-ligase, and wax ester synthase) to overproduce fatty acids, deregulate fatty acids, and produce fatty acid ethyl and methyl esters (biodiesel). Then, incorporation of nonnative hemicellulase genes enabled direct conversion of biomass to biodiesel in a single E. coli strain. Other poignant examples of heterologous gene transfer have come from meeting global health needs, namely, the treatment of malaria. In a landmark study, production of the antimalarial drug artemisinin required incorporation of amorphadiene synthase and cytochrome P450 monoxygenase from the plant Artemisia annua [14]. These genes were incorporated into a strain of S. cerevisiae modified to produce large quantities of the amorphadiene precursor, farnesyl pyrophosphate, by a complete overhaul of the mevalonate pathway.
Beyond heterologous gene transfer, protein engineering is being explored to create nonnatural enzymes that can be combined with biosynthetic pathways to create novel biochemical products. New enzymes can be created through either mutagenesis of existing enzymes or rational design of new enzymes. For example, Liao and colleagues expanded their work on the biosynthesis of alcohols by utilizing promiscuous enzyme activity [17]. By altering substrate specificity of an enzyme through site-directed mutagenesis, they were able to synthesis enzymes to make nonnatural alcohols with possible value as biofuels. A similar approach was used to develop substrate-specific cytochrome P450 enzymes [18]. These enzymes were created through random and site-directed mutagenesis to selectively deprotect different monosaccharide substrates for polysaccharide synthesis. In addition, protein engineering has advanced to a point where design of new enzymes from scratch is possible. An enzyme to perform a Diels-Alder reaction was created through computational design and experimental fine-tuning to achieve substrate specificity and stereoselectivity [19]. The designed enzyme was then altered to demonstrate the ability to perform similar reactions with different substrates. Such work demonstrates a powerful new approach to achieving molecular transformations that cannot be efficiently performed by either nature or conventional chemistry.
As the number of nonnative and unnatural genes transferred into a host increases to include entire pathway branches, a critical consideration is seamless integration with the organism's regular metabolic functions. One issue with metabolic network transfer is that the global energy resources of the cell must support both synthetic modules and the host cell. Despite this challenge, as well as other hurdles associated with constructing, testing, and balancing large networks of molecular pathways (where design complexity can be intimidating), some examples have shown success. Two notable applications include production of high butanol titers through the transfer of the Clostridium acetobutylicum butanol pathway to a variety of bacterial hosts [20] and production of valencene in yeast by transferring an isoprenoid pathway consisting of 7 E. coli genes [21].
In addition to manipulating and extending native pathways, it is also possible to engineer novel biochemical pathways that have never been observed in biology. For example, Moon et al. designed a synthetic pathway using genes from three sources: myo-inositol-1-phosphate synthase from S. cerevisiae, myo-inositol oxygenase from mice, and uronate dehydrogenase from Pseudomonas syringae [22]. The novel combined pathway was inserted into E. coli to convert myo-inositol into glucaric acid, a value-added chemical used as a dietary supplement, therapeutic, and potential feedstock for polymers. Similarly, Hawkins and Smolke synthesized benzylisoquinoline alkaloids in S. cerevisiae through the combination of genes from three different plant species as well human P450 enzyme [23]. These examples illustrate that complex molecules of therapeutic interest may be produced by combining existing enzymes into novel assemblies to create novel biosynthetic pathways.
Protein scaffolds that hold tagged pathway enzymes in close proximity have also been used to improve biochemical transformations [24]. In this model, scaffolds have been designed for biosynthesis pathways including mevalonate and glucaric acid pathways, and spatial colocalization of pathway enzymes has lead to increased productivity [24]. The use of scaffolds increases the local concentration of pathway intermediates near the desired downstream enzymes, which improves overall pathway kinetics, avoids the accumulation of hazardous intermediates, and minimizes consumption of intermediates by competing pathways [25, 26]. Such synthetic biology tools provide novel mechanisms for manipulating metabolic pathways and complement traditional pathway optimization.
Characterizing transcriptional and translational architectures and developing technologies for manipulating these processes provide other useful approaches for engineering microbial biotransformations. For instance, new genetic elements have been created to carry out circuit-like regulation that mimic toggle switches, AND gates, and oscillators [27–29]. Inserting such regulatory elements into cellular devices could lead to high-resolution control and novel functional behavior. Posttranscriptional regulation is also possible. Translation efficiency, for example, can be adjusted by modifying ribosome binding site affinity [30].
To fine-tune imperfect designs, genetic screening and evolutionary tools are also being developed. Sommer et al. screened a metagenomic library of arbitrary environmental DNA to identify three novel genes that convey tolerance to the biomass chemicals syringaldehyde and 2-furoic acid [31]. Similarly, Bayer et al. used a metagenomic screen of 89 suspected methyl halide transferase enzymes in E. coli to increase the yield of methyl halides from biomass to serve as precursors for chemicals and fuels [32]. Another screening technique called SCALEs (scalar analysis of library enrichments) uses libraries consisting of genomic DNA inserts of specific sizes to identify regions of a genome responsible for a particular phenotype [33]. This method has been used to identify gene regions conveying tolerance to inhibitors of metabolic pathways involving aspartic acid [34]. Such screens demonstrate approaches that can be used to discover genes responsible for other desirable or undesirable phenotypes. New robust evolutionary techniques such as multiplex automated genome engineering (MAGE) and global transcription machinery engineering (gTME) have also been developed to enable rapid fine-tuning of metabolic networks. MAGE generates genomic diversity within cells and across populations by using parallel, site-specific modification of DNA. Using MAGE, the lycopene synthesis pathway in E. coli was optimized by genetically modifying several targeted chromosome locations associated with genes of the pathway [35]. The gTME approach uses random mutagenesis of transcriptional machinery to alter the transcriptional program of an organism and has been used to develop E. coli and S. cerevisiae strains with desirable phenotypes for biofuel production, such as high ethanol tolerance [36, 37]. Evolutionary approaches provide a complementary strategy to design-based engineering that may reveal unexpected routes toward higher system performance and provide a better understanding of biological systems that may be incorporated into subsequent design strategies.
The next few years will bring increased attempts to construct and program large-scale user-defined pathways, and even whole organisms. Recently, researchers at the J. Craig Venter Institute (JCVI) assembled, modified, and implanted a complete Mycoplasma mycoides donor genome into a related Mycoplasma recipient cell [38]. This technological milestone marks the dawn of synthetic genomics. While this achievement is an important step towards designer organisms, applications of this approach remain challenging. For example, if one knew how to design a cell that efficiently converts sunlight, water, and CO2 into fuel, then JCVI's technology could enable the synthesis and transplantation of that chemically synthesized genome into a recipient cell. However, we do not yet know how to design such a genome from scratch, and simply screening astronomical numbers of synthetic genome variants is impractical or even impossible. In addition, the costs of genome construction currently prohibit most researchers from applying this approach. In the long run, however, there is no doubt that JCVI's technology will join a suite of other whole-genome engineering techniques to accelerate the development of microbial factories for producing fuels, pharmaceuticals, green biochemicals, and novel materials.
2.2. Cellular Devices and Therapies
For applications in medicine, synthetic biology may also be used to create new cellular functions, such that the engineered cell itself—not just a product that it produces—serves a therapeutic role. In this context, a cell can be viewed as a device that receives inputs, processes this information, and produces outputs all based upon its genetic programming. Therefore, novel cellular functions may be constructed through strategies that create new connections between existing and novel mechanisms for input (e.g., receptors), processing (e.g., intracellular signaling cascades and regulatory interactions), and output (e.g., the production of bioactive molecules or the induction of specialized cellular effector functions). Every such strategy must take into account both bottom-up considerations (including the choice of biological parts and their configuration) and top-down considerations (including the choice of cell type to be engineered and the interactions—both desirable and undesirable—of the engineered components with native cellular functions) (Figure 2). Each of these design choices is guided by the type of cellular function required for a given application.
For some strategies, a cell may be reprogrammed to change the way it relates existing inputs to existing outputs by rewiring intracellular signal processing. In one example of this approach, signaling through the ErbB2 receptor tyrosine kinase, which mediates mitogenic or transformative signaling in tumor cells, was redirected into a proapoptotic pathway [39]. This was accomplished by engineering novel intracellular signaling proteins, in which SH2 or PTB phosphotyrosine-recognition domains from Grb2 or ShcA, respectively, (which interact with ErbB2), were fused to the death effector domain of Fadd. Such intracellular signaling proteins can be engineered to modulate multiple functional characteristics, including autoregulation, ligand specificity, and signaling dynamics using modular functional domains [40, 41] and various scaffold motifs and configurations [42]. An alternative approach is one often used in basic research, whereby the extracellular ligand-binding domain of one receptor is fused to the intracellular signaling domain of another receptor to generate a chimeric receptor. Strategies such as these could be used to generate cellular therapies for situations in which the natural input/output relationship is dysfunctional, such as tumor-mediated conversion of tumor-reactive T cells to an immunosuppressive phenotype. An important consideration when using this approach is that the engineered signaling pathway does not replace native signaling pathways, but rather engineered and native pathways are coupled by the shared use of native receptors or downstream components.
In other applications, cells may be engineered to respond to a stimulus to which the cell does not naturally respond. An early example of this approach is the use of chimeric antigen receptors (CARs) for cancer immunotherapy. In this model, ligand-binding domains from antibodies specific for tumor antigens are fused to the intracellular portion of T cell receptors in order to generate T cells capable of killing tumor cells that express the targeted antigen. Early implementations of this approach are in clinical trials, and while most benefits have been modest, some responses are quite promising [43]. In the most recent version of this strategy, third generation CARs are constructed by fusing the ligand-binding domains of tumor-specific antibodies to intracellular domains of CD28, CD137, and the TCR-ζ chain, in sequence [44]. In this construct, the TCR-ζ chain confers TCR signaling upon binding of the antibody domain to its target, while the CD28 and CD137 domains enhance TCR signaling and T cell survival in vivo. In a related approach, Xu et al. constructed tumor-responsive dendritic cells that recognize the tumor antigen, erbB2, and signal through the Toll-like Receptor 4 (TLR4) pathway to induce an inflammatory immune response against the tumor [45]. This was accomplished by fusing the ligand-binding domain of an anti-erbB2 antibody to various intracellular mediators of TLR4 signaling [45]. While each of these examples used receptors or antibodies that naturally recognize a particular ligand, it is also possible to re-engineer receptors, such as GPCRs, so that they respond to novel small molecule ligands [46]. Such an approach may be applied to generate cells whose functions are regulated by an inducible trigger, such as a small molecule drug. As an example of this strategy, T cells were engineered to express interleukin 2 (IL-2, a cytokine necessary for T cell survival) under the control of a synthetic ribozyme switch that responds to the inducer theophylline. When these cells were transferred to recipient mice, theophylline treatment induced IL-2 production and engineered T cell survival in vivo [47]. Similar ribozyme switches may be designed to respond to a variety of small molecule cues. Other RNA-based approaches can be used to program cells to perform more complicated functions, including the logical evaluation of multiple inputs, such that activation of a target gene (or genes) is conditioned upon the presence or absence of multiple extracellular cues [48].
Like mammalian cells, bacteria may also be programmed for therapeutic applications in which the bacteria itself is the therapeutic. For example, bacteria naturally home to tumor environments, so this propensity may be harnessed by engineering bacteria to selectively invade and destroy cancer cells in vivo [49]. In this model, bacteria were engineered to express the invasin protein in response to the hypoxic conditions that typify tumor microenivronments. Invasin expression enables these bacteria to invade mammalian cells expressing β1-integrins, which is a common marker on some types of cancer. This approach can be further enhanced to include the hypoxia-inducible release of cytotoxic agents to destroy the tumor cells after invasion [49]. Similarly, synthetic biology could be used to integrate more complex functions and regulatory features into other bacteria-based therapeutics. Important considerations when manipulating bacteria for therapeutic applications include both interactions with host biology and preventing the engineered bacteria from spreading beyond the patient being treated. For example, Lactococcus lactis was engineered to treat inflammatory bowel diseases by programming these microbes to secrete the immunosuppressive cytokine Interleukin-10 (IL-10) and by deleting their thymidylate synthase gene [50]. This strategy restricted bacterial growth to cultures supplemented with thymidine, which prevents these cells from replicating efficiently in the gut. This treatment has already shown promise in clinical trials [51]. Because the engineered bacteria die quickly within the gut, however, frequent treatments would be required to treat chronic disease. Synthetic biology could be used to overcome such limitations. For example, the immunosuppressive bacteria described above could be modified such that they require survival signals from the gut and quickly die outside of this environment, or such that they secrete IL-10 only when they detect disease flareups. Such modifications could allow for safer and more effective treatments while still restricting these bacteria to their targeted milieu. Synthetic biology provides the framework required to design and implement such sophisticated biotherapy strategies.
Engineering communication between cells also may be useful for therapeutic applications. Already, simple synthetic communication systems have been constructed in bacteria, yeast, and even mammalian cells. In a representative example, a yeast “sender” population was engineered to secrete the plant hormone, isopentenyladenine, and a “receiver” yeast population was made responsive to this signal by expressing the receptor protein, AtCRE1 [52]. Bidirectional communication can also be used to coordinate multicellular functions. For example, two populations of E. coli were engineered to communicate bidirectionally using P. aeruginosa quorum sensing components, such that each cell type secretes a unique “sender” signal and expresses its own reporter gene only when it detects the signal secreted by the other cell type [53]. Similar information exchange between sender and receiver cells can also be used to program spatial organization and pattern formation [54]. Other applications of intercellular communication are “polling” strategies, which can be used to coordinate and synchronize dynamic cellular functions and potentially improve performance by reducing sensitivity to variations in environmental conditions [55]. These approaches can also be extended to interkingdom communication, including bacterial, yeast, and mammalian cells [56]. When applied in mammalian cells, such approaches could eventually be used to program communication and spatial organization for the construction of synthetic organs or organ-like devices. These strategies may also provide useful tools for programming the differentiation of stem cells in a manner that does not require recapitulating the complex extracellular cues that guide natural differentiation programs. Interkingdom communication platforms could enable engineered symbiotic microbes, which might patrol the colon or skin for signs of disease, to communicate this information to their human hosts. To date, existing intercellular communication systems rely upon simple small molecule-based signaling intermediates, and sophisticated multicellular communication strategies will require the development of novel orthogonal, high-information-content signaling platforms.
Finally, synthetic biology could lead to new therapeutics through in vitro applications such as screening and diagnostics. Current drug screens are typically based upon measurements such as treatment-induced changes in the replication and viability of diseased cells relative to changes in healthy control cells. This limits the type of information about biological activity and safety that can be assessed at the screening stage. Screens constructed through synthetic biology can be used to evaluate more complicated biological effects. For example, engineered cells can be used to identify drugs that target specific pathogen functions. Such an approach was used to identify substances that block the M. tuberculosis protein, EthR, which otherwise confers resistance to the prodrug ethionamide [57]. This approach may be especially useful for pathogens that are dangerous or difficult to culture in vitro. Similar approaches could be used to construct screens that evaluate the impact of test molecules on human cellular functions that do not naturally produce an easily-assessed phenotype like apoptosis. Synthetic biology may also be useful for the construction of biological diagnostics. For example, bacterial bioreporters have been constructed to detect various compounds and toxins [58]. In most cases, this detection relies upon natural sensing mechanisms and involves expressing a reporter gene in response to the activation of an existing bacterial promoter. Synthetic biology could be employed to engineer novel sensing mechanisms and build bacterial bioreporters that perform diagnostic assessments such as detecting biochemical markers within human blood samples. These technologies could be designed to function and produce readouts without requiring external equipment, so that they could be readily deployed for mobile operations or in resource-poor areas. As we continue to develop improved technologies for engineering both human cells and the microorganisms with which they interact, synthetic biology promises to transform medicine just as it has already begun to transform biotechnology.
2.3. Expanding the Chemistry of Life: Novel Biopolymer Synthesis
To lessen undesired and unpredictable interactions that often hamper design goals, modules of nonnatural components that operate independently of the cell's natural machinery are envisioned. Such orthogonal modules offer tremendous flexibility for optimization without disrupting the cell's operating system and, as a bonus, could exploit the renewability and evolvability of biology to synthesize nonbiological materials. In nature, complex and diverse chemical tasks are carried out across a variety of length scales using biopolymers of very simple composition. The expansion of the central dogma of biology to include diverse, nonnatural analogs may lead to a set of general solutions featuring orthogonal information coding, orthogonal cellular machinery, orthogonal scaffolds, orthogonal compartments, orthogonal interactions, or orthogonal communication pathways, which could form the basis for entirely artificial living systems. As an example, even minor modifications to natural biopolymers can endow them with altered interactions and enhanced functionalities. Peptide nucleic acids (PNAs), which have backbones similar to peptides but nucleobase functional groups (as opposed to the twenty natural amino acids) [59], hybridize to short stretches of DNA, and exhibit increased resistance to both proteases and endonucleases.
To expand the chemistry of life, orthogonal modules may be constructed through evolved catalytic ensembles that enable synthesis, evolution, and organization of nonnatural or hybrid polymers. In this case, bottom-up design considerations must include template recognition, substrate recognition, catalytic activity, error correction, product folding, and transport properties (Figure 2). Top-down considerations may require that synthesis should occur under physiological conditions (temperature, buffer conditions, etc.) and that monomer building blocks must be available to the host or able to be synthesized by the host. Implicit mass transfer limitations arising from cellular structure must also be addressed.
Increasing the capacity of nucleic acids to store, recall, and propagate specific information can augment genomic information [60, 61]. Early experiments with modified nucleic acids (termed xenonucleic acids, or XNAs [62]) were motivated by the desires to understand how DNA and RNA developed in the prebiotic world [63] and to create antisense molecules that could be easily taken up by cells [64]. More recently, efforts have shifted towards modulating function by exploring diversity in the composition of bases [65], sugars [61], and phosphates [61]. Backbone modifications of natural nucleic acids generally decrease recognition by endonucleases and provide an alternative strategy for modulating XNA binding affinity to complementary strands. The most studied backbone modifications of natural nucleic acids include PNAs [59], locked nucleic acids (LNAs) [66], threose nucleic acids (TNAs) [67], phosphorothioates [68], and Morpholinos [69]. Beyond antisense applications, these XNAs have the potential to vastly expand biopolymer information storage.
In order for novel biopolymers to become a viable tool for synthetic biologists, sequence-defined synthesis of the polymers is required. Beyond low-efficiency solid phase synthesis techniques, enzymatic template-directed synthesis of nonnatural polymers can be performed. In one approach, naturally occurring DNA and RNA polymerases have been identified that accept nonnatural substrates, thus allowing the templated synthesis of TNA from DNA [70]. In addition, these polymerases can be evolved in vitro to improve processivity and decrease error rates with nonnatural monomer units [71]. Improved understanding of structure-function relationships in polymerases has further resulted in the rational modification of a natural transcriptase to a polymerase that accepts nonstandard nucleotides [72]. This illustrates how directed evolution can be incorporated into the design cycle. Advances in enzyme engineering and our understanding of nature's design rules will speed progress toward engineered polymerases and enable us to probe the feasibility of enzymatic XNA synthesis and application.
In another approach, the cell's protein polymerase, the ribosome, could be engineered to selectively accept and polymerize synthetic monomers with high template fidelity. Driven by the promise of new therapeutics, evolvable abiological polymers, and biophysical probes, there has long been interest in harnessing natural translation machinery to synthesize polymers of nonnatural building blocks [73]. In concept, ribosome engineering may provide a more general, modular solution to novel biopolymer synthesis due to the use of transfer RNAs (tRNAs) that serve as adapters between the genetic code and the biopolymer product. Incorporating nonnatural monomer units on the ribosome depends on (i) reassignment of codons to nonnatural monomer units and (ii) acceptance of these units in the catalytic center. Remarkably, dozens of nonnatural amino acids have already been site-specifically incorporated into polypeptides by the ribosome. This was accomplished by engineering synthetic routes toward aminoacylation of tRNAs [74, 75] and orthogonal synthetase-tRNA pairs [76] and showing that natural ribosomes could accept nonnatural substrates. Even nonpeptide backbones, such as N-substituted amino acids [77] and hydroxy acids [78] have been incorporated by the ribosome, although with varying degrees of success. When using orthogonal synthetase-tRNA pairs, one potential issue is nonspecific incorporation that results from competing natural aminoacyl-tRNAs. To obtain greater control of homogeneous sequence composition, genetic code reprogramming [79, 80] or synthetic genetic systems (based on XNAs or modified bases) can be applied.
Although a variety of nonnatural monomer units have been incorporated by native ribosomes, it is clear that these substrates are inherently suboptimal. The tuning of polymerase machines to their natural substrates is exquisite [81]. Therefore, a major design challenge lies not only in the bottom-up aspect of incorporating nonnatural building blocks, but also in reversing the evolutionary optimization of finely tuned machinery and aligning it onto an entirely new path. To address this limitation, the ribosome and its machinery can be modified to enable increased promiscuity and efficiency at polymerizing nonnatural monomer units. In one case, EF-Tu was evolved to more efficiently incorporate amino acids containing bulky aromatic groups [82]. In examples that modify the ribosome directly, ribosomal RNA has been mutated or chemically modified to enable more efficient nonnatural substrate incorporation [83, 84]. Although the ribosome has been shown to be tolerant of modifications, there are relatively few examples because constraints imposed by living cells (e.g., dominant lethality) have hindered efforts to engineer ribosomes.
Specialized ribosomes overcome the dominant lethality constraint and provide an exemplary illustration of orthogonal performance. Originally pioneered by de Boer and colleagues [85], anti-Shine-Dalgarno sequences can be engineered into 16S ribosomal RNA to enable a group of ribosomes (orthogonal from the host) to translate specific mRNA populations. This has shown advantages for performing logic operations [85, 86], synthesizing proteins with nonnatural amino acids, such as p-benzoyl-(L)-phenylalanine [87], and efficiently incorporating quadruplet codons [88]. Orthogonal ribosomes will provide useful tools in programming synthetic function and expanding the genetic code to make nonnatural products for biotechnology.
Cell-free systems are being used to remove the most stringent top-down design constraints confronted when attempting to expand the chemistry of life [89–92]. Cellular overhead is reduced (e.g., genes of unknown functions, metabolic redundancies, and unintended interactions), transport barriers are removed, cellular viability is not required, and resources are more efficiently directed towards user-defined objectives. An additional advantage is direct access to the system of interest, similar to opening the hood of a car and accessing the engine. Cell-free protein synthesis systems [93–95] are one of the most prominent examples of cell-free biology and can be employed to create large amounts of nonnatural peptide [96]. Complex systems of biochemical transformations can also be built [97]. Looking forward, both in vitro and in vivo efforts towards novel catalytic ensembles promise to meld the complexity and diversity of chemistry with the accuracy and reproducibility of biology. The main rewards will be increased understanding of biology and new classes of evolvable molecules, which may find use as therapeutics or novel materials.
3. Moving Forward
Given the complexity that characterizes living systems developed through eons of evolution, the challenge to transform biology into an engineerable system is formidable. Considering biological design from both top-down and bottom-up perspectives provides a useful framework for proceeding towards this goal. At each scale of biological organization, this approach guides both the design and analysis of novel systems, as well as our investigations into the fundamental mechanisms that impact the performance of engineered biological systems. Synthetic biology is already changing the way we think about producing useful molecules, engineering our own biology, and expanding the chemistry of life. Making progress on these and other fronts will require both interdisciplinary approaches and new pedagogical strategies for training a new cadre of whole-brain thinkers. If successful, these efforts may revolutionize our ability to meet pressing societal needs through a new paradigm of biology by design.
Acknowledgments
The authors gratefully acknowledge funding from the National Institutes of Health (Grant Number R00GM081450), the National Academies Keck Futures Initiative (Grant Numbers NAFKI-SB5 and NAKFI-SB6 to M. Jewett and J. Leonard, resp.), and the National Science Foundation (Grant Numbers MCB-0943393 and PHY-0943390 to M. Jewett and J. Leonard, resp.). B. R. Fritz, L. E. Timmerman, and N. M. Daringer contributed equally.
References
- 1.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Güell M, van Noort V, Yus E, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326(5957):1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 3.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature Reviews Genetics. 2010;11(5):367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benner SA, Sismour AM. Synthetic biology. Nature Reviews Genetics. 2005;6(7):533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker D, Church G, Collins J, et al. Engineering life: building a FAB for biology. Scientific American. 2006;294(6):44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 6.Endy D. Foundations for engineering biology. Nature. 2005;438(7067):449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 7.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nature Reviews Molecular Cell Biology. 2009;10(6):410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nature Biotechnology. 2006;24(5):545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 9.Carothers JM, Goler JA, Keasling JD. Chemical synthesis using synthetic biology. Current Opinion in Biotechnology. 2009;20(4):498–503. doi: 10.1016/j.copbio.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Kwok R. Five hard truths for synthetic biology. Nature. 2010;463(7279):288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 11.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252(5013):1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen J. Metabolic engineering. Applied Microbiology and Biotechnology. 2001;55(3):263–283. doi: 10.1007/s002530000511. [DOI] [PubMed] [Google Scholar]
- 13.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 14.Ro D-K, Paradise EM, Quellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 15.Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 16.Atsumi S, Wu T-Y, Eckl E-M, Hawkins SD, Buelter T, Liao JC. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Applied Microbiology and Biotechnology. 2010;85(3):651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(52):20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis JC, Bastian S, Bennett CS, et al. Chemoenzymatic elaboration of monosaccharides using engineered cytochrome P450BM3 demethylases. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(39):16550–16555. doi: 10.1073/pnas.0908954106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel JB, Zanghellini A, Lovick HM, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular diels-alder reaction. Science. 2010;329(5989):309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen DR, Leonard E, Yoon S-H, Tseng H-C, Yuan C, Prather KLJ. Engineering alternative butanol production platforms in heterologous bacteria. Metabolic Engineering. 2009;11(4-5):262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Maury J, Asadollahi MA, Møller K, et al. Reconstruction of a bacterial isoprenoid biosynthetic pathway in Saccharomyces cerevisiae. FEBS Letters. 2008;582(29):4032–4038. doi: 10.1016/j.febslet.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 22.Moon TS, Yoon S-H, Lanza AM, Roy-Mayhew JD, Prather KLJ. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Applied and Environmental Microbiology. 2009;75(3):589–595. doi: 10.1128/AEM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nature Chemical Biology. 2008;4(9):564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dueber JE, Wu GC, Malmirchegini GR, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnology. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 25.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. Journal of Biological Chemistry. 1999;274(18):12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 26.Spivey HO, Ovádi J. Substrate channeling. Methods. 1999;19(2):306–321. doi: 10.1006/meth.1999.0858. [DOI] [PubMed] [Google Scholar]
- 27.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular and gate. Molecular Systems Biology. 2007;3, article 133 doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 29.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457(7227):309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 30.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotechnology. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommer MOA, Church GM, Dantas G. A functional metagenomic approach for expanding the synthetic biology toolbox for biomass conversion. Molecular Systems Biology. 2010;6, article 360 doi: 10.1038/msb.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayer TS, Widmaier DM, Temme K, Mirsky EA, Santi DV, Voigt CA. Synthesis of methyl halides from biomass using engineered microbes. Journal of the American Chemical Society. 2009;131(18):6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 33.Lynch MD, Warnecke T, Gill RT. SCALEs: multiscale analysis of library enrichment. Nature Methods. 2007;4(1):87–93. doi: 10.1038/nmeth946. [DOI] [PubMed] [Google Scholar]
- 34.Bonomo J, Lynch MD, Warnecke T, Price JV, Gill RT. Genome-scale analysis of anti-metabolite directed strain engineering. Metabolic Engineering. 2008;10(2):109–120. doi: 10.1016/j.ymben.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Wang HH, Isaacs FJ, Carr PA, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314(5805):1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 37.Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metabolic Engineering. 2007;9(3):258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Gibson DG, Glass JI, Lartigue C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 39.Howard PL, Chia MC, Del Rizzo S, Liu F-F, Pawson T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(20):11267–11272. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301(5641):1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 41.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Current Opinion in Structural Biology. 2004;14(6):690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319(5869):1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 43.Marcu-Malina V, van Dorp S, Kuball J. Re-targeting T-cells against cancer by gene-transfer of tumor-reactive receptors. Expert Opinion on Biological Therapy. 2009;9(5):579–591. doi: 10.1517/14712590902887018. [DOI] [PubMed] [Google Scholar]
- 44.Milone MC, Fish JD, Carpenito C, et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Molecular Therapy. 2009;17(8):1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Darcy PK, Kershaw MH. Tumor-specific dendritic cells generated by genetic redirection of Toll-like receptor signaling against the tumor-associated antigen, erbB2. Cancer Gene Therapy. 2007;14(9):773–780. doi: 10.1038/sj.cgt.7701073. [DOI] [PubMed] [Google Scholar]
- 46.Conklin BR, Hsiao EC, Claeysen S, et al. Engineering GPCR signaling pathways with RASSLs. Nature Methods. 2008;5(8):673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnology. 2007;25(7):795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 49.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006;355(4):619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 50.Steidler L, Neirynck S, Huyghebaert N, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nature Biotechnology. 2003;21(7):785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- 51.Braat H, Rottiers P, Hommes DW, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clinical Gastroenterology and Hepatology. 2006;4(6):754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 52.Chen M-T, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nature Biotechnology. 2005;23(12):1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 53.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(44):17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434(7037):1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 55.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463(7279):326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(25):10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber W, Schoenmakers R, Keller B, et al. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):9994–9998. doi: 10.1073/pnas.0800663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Meer JR, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nature Reviews Microbiology. 2010;8(7):511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 59.Nielsen PE. PNA technology. Molecular Biotechnology. 2004;26(3):233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 60.Brudno Y, Liu DR. Recent progress toward the templated synthesis and directed evolution of sequence-defined synthetic polymers. Chemistry & Biology. 2009;16(3):265–276. doi: 10.1016/j.chembiol.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loakes D, Holliger P. Polymerase engineering: towards the encoded synthesis of unnatural biopolymers. Chemical Communications. 2009;(31):4619–4631. doi: 10.1039/b903307f. [DOI] [PubMed] [Google Scholar]
- 62.Herdewijn P, Marlière P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chemistry & Biodiversity. 2009;6(6):791–808. doi: 10.1002/cbdv.200900083. [DOI] [PubMed] [Google Scholar]
- 63.Benner SA. Understanding nucleic acids using synthetic chemistry. Accounts of Chemical Research. 2004;37(10):784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 64.Freier SM, Altmann K-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Research. 1997;25(22):4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geyer CR, Battersby TR, Benner SA. Nucleobase pairing in expanded Watson-Crick-like genetic information systems. Structure. 2003;11(12):1485–1498. doi: 10.1016/j.str.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 66.Kaur H, Babu BR, Maiti S. Perspectives on chemistry and therapeutic applications of locked nucleic acid (LNA) Chemical Reviews. 2007;107(11):4672–4697. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- 67.Ichida JK, Zou K, Horhota A, Yu B, McLaughlin LW, Szostak JW. An in vitro selection system for TNA. Journal of the American Chemical Society. 2005;127(9):2802–2803. doi: 10.1021/ja045364w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu T, Orgel LE. Disulfide-linked oligonucleotide phosphorothioates: novel analogues of nucleic acids. Journal of Molecular Evolution. 1991;32(4):274–277. doi: 10.1007/BF02102183. [DOI] [PubMed] [Google Scholar]
- 69.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense and Nucleic Acid Drug Development. 1997;7(3):187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 70.Chaput JC, Szostak JW. TNA synthesis by DNA polymerases. Journal of the American Chemical Society. 2003;125(31):9274–9275. doi: 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]
- 71.Fa M, Radeghieri A, Henry AA, Romesberg FE. Expanding the substrate repertoire of a DNA polymerase by directed evolution. Journal of the American Chemical Society. 2004;126(6):1748–1754. doi: 10.1021/ja038525p. [DOI] [PubMed] [Google Scholar]
- 72.Sismour AM, Lutz S, Park J-H, et al. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Research. 2004;32(2):728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Schultz PG. Expanding the genetic code. Angewandte Chemie International Edition. 2005;44(1):34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 74.Hecht SM, Alford BL, Kuroda Y, Kitano S. “Chemical aminoacylation” of tRNA’s. Journal of Biological Chemistry. 1978;253(13):4517–4520. [PubMed] [Google Scholar]
- 75.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nature Methods. 2006;3(5):357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 76.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292(5516):498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 77.Kawakami T, Murakami H, Suga H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chemistry & Biology. 2008;15(1):32–42. doi: 10.1016/j.chembiol.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 78.Fahnestock S, Rich A. Ribosome-catalyzed polyester formation. Science. 1971;173(994):340–343. doi: 10.1126/science.173.3994.340. [DOI] [PubMed] [Google Scholar]
- 79.Ohta A, Murakami H, Higashimura E, Suga H. Synthesis of polyester by means of genetic code reprogramming. Chemistry & Biology. 2007;14(12):1315–1322. doi: 10.1016/j.chembiol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 80.Ohta A, Yamagishi Y, Suga H. Synthesis of biopolymers using genetic code reprogramming. Current Opinion in Chemical Biology. 2008;12(2):159–167. doi: 10.1016/j.cbpa.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 81.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Molecular Cell. 2008;31(1):114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. Journal of the American Chemical Society. 2007;129(46):14458–14462. doi: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- 83.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Enhanced D-amino acid incorporation into protein by modified ribosomes. Journal of the American Chemical Society. 2003;125(22):6616–6617. doi: 10.1021/ja035141q. [DOI] [PubMed] [Google Scholar]
- 84.Chirkova A, Erlacher M, Micura R, Polacek N. Chemically engineered ribosomes: a new frontier in synthetic biology. Current Organic Chemistry. 2010;14(2):148–161. [Google Scholar]
- 85.Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(14):4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. Journal of the American Chemical Society. 2005;127(50):17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 87.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nature Biotechnology. 2007;25(7):770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 88.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464(7287):441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 89.Forster AC, Church GM. Towards synthesis of a minimal cell. Molecular Systems Biology. 2006;2, article 45 [Google Scholar]
- 90.Forster AC, Church GM. Synthetic biology projects in vitro. Genome Research. 2007;17(1):1–6. doi: 10.1101/gr.5776007. [DOI] [PubMed] [Google Scholar]
- 91.Hold C, Panke S. Towards the engineering of in vitro systems. Journal of the Royal Society Interface. 2009;6(Suppl 4):S507–S521. doi: 10.1098/rsif.2009.0110.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jewett MC, Forster AC. Update on designing and building minimal cells. Current Opinion in Biotechnology. 2010;21(5):697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Molecular Systems Biology. 2008;4, article 220 doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnology and Bioengineering. 2004;86(1):19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 95.Barrett OPT, Chin JW. Evolved orthogonal ribosome purification for in vitro characterization. Nucleic Acids Research. 2010;38(8):2682–2691. doi: 10.1093/nar/gkq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goerke AR, Swartz JR. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnology and Bioengineering. 2009;102(2):400–416. doi: 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- 97.Bujara M, Schümperli M, Billerbeck S, Heinemann M, Panke S. Exploiting cell-free systems: implementation and debugging of a system of biotransformations. Biotechnology and Bioengineering. 2010;106(3):376–389. doi: 10.1002/bit.22666. [DOI] [PubMed] [Google Scholar]