Abstract
Summary: SpotXplore is a plugin for Cytoscape for extraction and visualization of differentially expressed subnetworks (hotspots) from gene networks. The hotspot-based visualization approach enables interactive exploration of regulatory interactions in differentially expressed gene sets, and it allows a researcher to explore gene expression in direct relation to the affected cellular gene network. The hotspots provide a view beyond the commonly used metabolic pathways and gene ontologies.
Availability: http://www.win.tue.nl/∼mwestenb/spotxplore/
Contact: m.a.westenberg@tue.nl
Supplementary information: Example data and user manual are available at SpotXplore's web site.
1 INTRODUCTION
Interpretation of gene expression from time series experiments or multiple perturbation studies is a challenging task. Insight can be obtained by analyzing the data in the context of molecular interaction networks, such as gene regulatory networks or metabolic pathways, which requires visualization in addition to statistical analysis tools. Many tools for visualizing the various types of biological networks have been developed (Suderman and Hallett, 2007). Nevertheless, there is a need for tools that incorporate dynamics of gene expression in the networks by offering both data analysis and visualization. Only then, a researcher does not have to switch between various applications to obtain a comprehensive understanding of the biological processes affected in the experiments.
The generic biological network visualization tool Cytoscape (Shannon et al., 2003) provides an excellent environment to implement combined analysis and visualization methods. In this article, we present SpotXplore, a plugin for analysis and visual exploration of differentially expressed subnetworks (hotspots). SpotXplore provides two methods to detect such hotspots, and allows the user to visualize the hotspots in the context of the whole network to explore regulatory interactions between differentially expressed gene sets. Our plugin also provides a method for visualization of time series data, because Cytoscape offers only basic support by coloring nodes (genes) based on a single time point. However, to detect temporal patterns in gene expression, it is necessary to consider all time points, which requires simultaneous visualization of multiple expression values. For this purpose, we re-implemented and adapted the visualization approach of our stand-alone tool GENeVis (Westenberg et al., 2007), which allows embedded visualization of all time points in the nodes. To our knowledge, only the VistaClara plugin (Kincaid et al., 2008) offers a similar visualization by drawing time points under the nodes. Our approach is different, because it also encodes statistical properties of the time points in the visualization, which is important for data analysis.
SpotXplore's main contribution is the hotspot-based visualization approach, which enables interactive exploration of regulatory interactions in differentially expressed gene sets, allowing a researcher to explore gene expression in direct relation to the affected cellular gene network.
2 METHODS AND IMPLEMENTATION
SpotXplore is a plugin for Cytoscape, which has been tested in versions 2.6.3 and 2.7.0 on multiple platforms (Windows, Linux, MacOS). It requires a gene network and gene expression data already loaded in Cytoscape. Gene networks can be obtained from databases, such as DBTBS (Makita et al., 2004), or by constructing a co-regulation network from the expression data. The user manual, available as Supplementary Material, provides details about the required and optional node and edge attributes.
2.1 Gene expression visualization
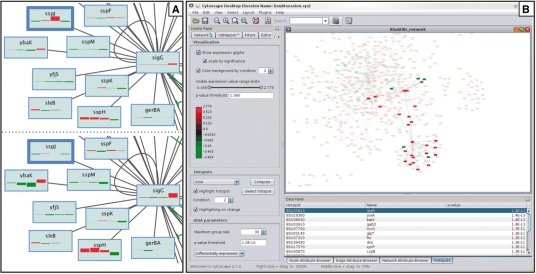
SpotXplore assumes that gene expressions are given as log-transformed ratios between a test condition and a reference condition. Each node (representing a gene) can be colored according to the expression ratio. Additionally, all gene expression ratios for a certain gene, i.e. all time points or all conditions, can be shown simultaneously as small glyphs embedded in the rectangular box representing the gene. Each glyph is a small rectangle, colored according to expression ratio and scaled in height according to confidence value (e.g. a P-value) (Westenberg et al., 2007). This graphical representation is inspired by perception research, from which it is known that color and size are perceptually easy to separate and interpret independently. Scaling by significance gives reliable time points more visual emphasis and suppresses noisy expression ratios (Fig. 1A).
Fig. 1.
SpotXplore applied to a Bacillus subtilis gene network and a four time point microarray experiment (Supplementary Materials). (A) Part of the SigG regulon. Expression ratio and P-values of four time points are displayed in the nodes. Time point boxes are colored according to expression ratio (red–green). Top: box height scaled linearly by expression ratio. Bottom: box height scaled logarithmically by P-value. Scaling by expression ratio may give the impression that the gene sspJ (highlighted by a wide border) is strongly upregulated at time point 3. However, the corresponding P-value is 1.0. Scaling by significance value suppresses these insignificant data points (see bottom). Conversely, it enhances data points that do show significant differential expression. (B) Screenshot of SplotXplore. Left panel: visual appearance controls, color legend and hotspot detection algorithm parameters. Bottom right panel: table of detected hotspots. The hotspot rbsR at time point 2 is selected and its members highlighted (top right panel). The remaining nodes (genes that are not differentially expressed) and edges are drawn translucent.
2.2 Hotspot detection
Hotspots are network components that are differentially expressed as a function of experimental parameters. We provide two algorithms to detect hotspots: Graph-based iterative Group Analysis (Breitling et al., 2005) and a two-pass algorithm that requires both expression ratios and expression levels for its computation (Luscombe et al., 2004). Each algorithm computes hotspots per condition or time point. SpotXplore runs the selected algorithm for all conditions, and stores the detected hotspots for later exploration. Additionally, hotspots can be defined by importing a set of selected nodes from Cytoscape. This allows quick manual construction of a set of hotspots, and also provides a way to use our visualization approach for hotspots detected by other Cytoscape plugins.
2.3 Hotspot visualization
SpotXplore visualizes hotspots in the context of the whole network (Fig. 1B). The conditions (or time points) can be browsed and an overview of the network activity is provided by highlighting all hotspots detected in a certain condition. An individual hotspot can be highlighted by selecting it, and can be converted into a Cytoscape selection set for further analysis.
3 CONCLUSION
SpotXplore is a plugin for the widely adopted generic Cytoscape network visualization software. It allows to interactively explore regulatory interactions in differentially expressed gene sets, occurring in collections of gene expression data (e.g. time series), obtained by microarrays or RNA-seq techniques. The approach both facilitates and significantly speeds up data analysis in comparison to a traditional gene-by-gene analysis approach of the most differentially expressed genes and manual mining of gene regulatory interaction information available at public repositories.
Conflict of Interest: none declared.
REFERENCES
- Breitling R, et al. Graph-based iterative group analysis enhances microarray interpretation. BMC Bioinformatics. 2005;5:100. doi: 10.1186/1471-2105-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid R, et al. VistaClara: an expression browser plug-in for Cytoscape. Bioinformatics. 2008;24:2112–2114. doi: 10.1093/bioinformatics/btn368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Lett. Nat. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Makita Y, et al. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–2659. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- Westenberg MA, et al. Interactive visualization of gene regulatory networks with associated gene expression time series data. In: Linsen L, et al., editors. Visualization in Medicine Life Sciences, Visualization and Mathematics. Berlin, Germany: Springer; 2007. pp. 293–312. [Google Scholar]