Abstract
Motivation: Most of the previous data mining studies based on the NCI-60 dataset, due to its intrinsic cell-based nature, can hardly provide insights into the molecular targets for screened compounds. On the other hand, the abundant information of the compound–target associations in PubChem can offer extensive experimental evidence of molecular targets for tested compounds. Therefore, by taking advantages of the data from both public repositories, one may investigate the correlations between the bioactivity profiles of small molecules from the NCI-60 dataset (cellular level) and their patterns of interactions with relevant protein targets from PubChem (molecular level) simultaneously.
Results: We investigated a set of 37 small molecules by providing links among their bioactivity profiles, protein targets and chemical structures. Hierarchical clustering of compounds was carried out based on their bioactivity profiles. We found that compounds were clustered into groups with similar mode of actions, which strongly correlated with chemical structures. Furthermore, we observed that compounds similar in bioactivity profiles also shared similar patterns of interactions with relevant protein targets, especially when chemical structures were related. The current work presents a new strategy for combining and data mining the NCI-60 dataset and PubChem. This analysis shows that bioactivity profile comparison can provide insights into the mode of actions at the molecular level, thus will facilitate the knowledge-based discovery of novel compounds with desired pharmacological properties.
Availability: The bioactivity profiling data and the target annotation information are publicly available in the PubChem BioAssay database (ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/).
Contact: ywang@ncbi.nlm.nih.gov; bryant@ncbi.nlm.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Understanding the mechanism of interaction of small molecules with their macromolecular targets is critical for drug and chemical probe development. The innovation of drug has long been recognized as time-consuming and labor-intensive, costing on average about $800 million as well as 10–12 years to bring a new drug to market (DiMasi et al., 2003). Apart from the challenges in optimizing pharmacokinetic properties and minimizing toxicities of lead compounds, the lack of publicly available/accessible biomedical assay data may represent another barrier for the success of drug discovery. Fortunately, this is changing since more public resources are emerging, offering new opportunities to chemical biology researchers for drug development. Open-access, information-rich resources include the Protein Data Bank (PDB; Berman et al., 2000), DrugBank (Wishart et al., 2006, 2008) and KEGG (Kanehisa et al., 2004), to name only a few. Without a doubt, existing public resources, as well as new ones, will evolve in future with both speed and capacity.
PubChem is a public repository for the chemical structures of small molecules and information of their biological properties (Wang et al., 2009, 2010). It was launched as a component of the NIH Molecular Libraries Roadmap Initiative (Zerhouni, 2003), with the aim to discover chemical probes via high-throughput screening (HTS) of small molecules. It also receives biological property contributions from many other organizations. As of March 17, 2010, PubChem contains more than 26 million unique compounds, among which over 870 000 have biological assay data for more than 3000 molecular targets, including proteins and genes. The public accessibility to such assay data is particularly valuable to the community, since this kind of critical information needed by drug research is typically held by pharmaceutical companies. The public availability and information-rich features altogether make PubChem an extremely valuable resource for biomedical research, as well as data mining studies (Chen and Wild, 2010; Han et al., 2008; Li et al., 2009; Rohrer and Baumann, 2009; Weis et al., 2008; Xie and Chen, 2008).
Launched by the National Cancer Institute (NCI), the Developmental Therapeutics Program (DTP) provides in vitro screening for new anticancer drugs that tested in 60 human tumor cancer cell lines (often known as the NCI-60 dataset; Shoemaker, 2006). This well-curated, publicly available dataset has been recognized as a rich resource for studying the mechanism of growth inhibition for tumor cells (Shoemaker, 2006; Weinstein et al., 1997). It has also inspired interests for developing and validating data mining tools (Paull et al., 1989; Zaharevitz et al., 2002). Bioactivity profiles derived from the NCI-60 cell lines can provide insights into the mode of actions for tested compounds (Rabow et al., 2002; Shi et al., 1998a, b, 1999; Wallqvist et al., 2003; Weinstein, et al., 1992). Structure-activity relationships (SARs) studies have also been reported for predicting or characterizing the cytotoxicity of the screened compounds in the NCI-60 dataset (Guha, 2008; Lee et al., 2008; Wang et al., 2007).
However, due to the intrinsic cell-based nature of the NCI-60 dataset, most of the studies described above can hardly provide insights into the molecular targets for screened compounds. On the other hand, PubChem has more than 1200 publicly available HTS bioassays with over 690 defined protein targets (as of March 17, 2010). In addition, the screening laboratories under the NIH Molecular Libraries Program (MLP) share a common compound library, i.e. the Molecular Libraries Small Molecule Repository (MLSMR), which is required to be tested for each assay project if possible. As a result, the compounds in the MLSMR library are often tested in hundreds of bioassays with many of them having associated protein targets. It thus represents a rich resource for constructing the compound–target interaction network, deriving target profiles and evaluating polypharmacological properties for a large library of compounds (Chen et al., 2009). Moreover, there is a significant overlap between the MLSMR compound library and those screened in the NCI-60 cell lines. Therefore, the bioassay data in PubChem can provide experimental evidence for the interactions between the compounds in the NCI-60 dataset and their targets.
In this work, we proposed a new strategy for combining and data mining the NCI-60 dataset and PubChem HTS assays, and investigated the correlations among the bioactivity profiles, compound–target interaction network and chemical structures of small molecules. Bioactivity profiles were derived from the screening results contained in the NCI-60 dataset. Compounds were hierarchically clustered based on their bioactivity profiles. Compound–target interaction networks were constructed using the annotated bioassay data in PubChem. Strong correlations were suggested between bioactivity profiles and target networks, especially when chemical structures were related.
2 METHODS
2.1 NCI-60 dataset
The NCI-60 dataset is also available in the PubChem BioAssay database as 73 bioassays with the name of ‘NCI human tumor cell line growth inhibition assay’ under the ‘NCI/DTP’ data source. In this study, 13 bioassays were eliminated considering their relatively small number of tested compounds (less than 16 000). The screening data for the remaining 60 bioassays (will be referred to hereafter as the NCI-60) was downloaded from the PubChem FTP site (accessed on March 17, 2010). In total, 5083 unique compounds were compiled and further filtered by the following rules:
Compounds must have been tested in all of the 60 NCI cell lines with a complete spectrum of log (GI50) values, where GI50 is the compound concentration required for 50% inhibition of tumor cell growth. That is, any compound with missing log (GI50) value in one or more of the NCI-60 cell lines was discarded. 4452 compounds met with this criterion.
Compounds must demonstrate activity in at least one PubChem bioassay which has a defined protein target. This resulted in an initial set of 257 compounds with both complete bioactivity profiles and known protein targets.
Compounds must have log (GI50) values below −6 for at least 15 out of the 60 NCI cell lines.
A final set of 37 compounds matched all of the above three criteria, and were analyzed in this study.
2.2 Clustering analysis based on bioactivity profiles
End-point activity data from a single cell line may give only limited information on a compound's biological response. However, the tested activities in a broad panel of 60 cell lines (i.e. bioactivity profile) can be used to characterize the mechanism of drug action, resistance and modulation (van Osdol et al., 1994; Weinstein et al., 1992). In this study, bioactivity profiles were subjected to hierarchical clustering by using the Hierarchical Clustering Explorer (HCE, version 3.5; Seo and Shneiderman, 2002), with the complete-linkage algorithm and Euclidean distance:
| (1) |
where Ai and Bi are the log (GI50) values in the i-th NCI-60 cell line for the compound A and B, respectively.
2.3 Compound–target interaction network
A compound–target interaction network can offer a direct view of the interactions between compounds and their protein targets. The first step to construct such a network is to identify the protein targets for the compounds of interest. The detailed target annotations in the PubChem BioAssay database (assay identifier: AID) made this step very straightforward. For each of the 37 compounds in this study, the PubChem bioassays in which the compound was tested active (see each assay description for the definition of bioactivity outcome) were identified. If the bioassay was specified with a protein target, then the target was assigned to the compound and included for network construction. Note that a compound may be found active in several bioassays, so it is possible for a compound to have multiple targets associated with it. In our network, the compound and target were denoted as two different nodes, respectively. An edge was drawn to link a compound node (labeled by the PubChem compound identifier: CID) and a target node (labeled by the NCBI protein identifier: GI) if the compound is active against the target. We applied the E-Utilities tool (http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html) to get the target GI for a respective bioassay. For example, the following URL: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pcassay&db=protein&cmd=neighbor&linkname=pcassay_protein_target&id=915 will return an XML file containing the GI of the protein target for the bioassay with AID 915. An in-house script was used to extract GI from the resultant XML file. To avoid the ambiguity in target specification, several PubChem bioassays that associate with multiple GIs were excluded from analysis. The PubChem bioassays as well as the target information used for network construction are listed in Supplementary Table S1 based on the assay data in the PubChem BioAssay database as of March 17, 2010. The compound–target interaction network was visualized by using the Cytoscape (version 2.3.6; Shannon et al., 2003).
3 RESULTS AND DISCUSSION
3.1 Hierarchical clustering analysis based on bioactivity profiles
Hierarchical clustering was first carried out for the initial set of 257 compounds, which was obtained prior to the application of the third filter. The dendrogram graph of the clustering result is given in Supplementary Figure S1. The log (GI50) value of −6 was adopted as the bipartite cutoff to determine whether a compound is active (≤ −6) or inactive (> −6) in a respective NCI-60 cell line. This criterion for discriminating active compounds from inactive ones is consistent with that specified in the PubChem BioAssay database by the original NCI/DTP depositors, and as well as in other studies (Lee et al., 2008).
As shown in Supplementary Figure S1, compounds that demonstrate similar bioactivity spectra were clustered. However, a majority of compounds were clustered in proximity simply because they were inactive (shown in blue) in most of the NCI-60 cell lines. Though inactive information is also important, it is less relevant to our study, which is to investigate the correlations among the bioactivity profiles, molecular targets and chemical structures of bioactive compounds. Furthermore, previous studies have shown that the log (GI50) values in the NCI-60 dataset are skewed toward certain thresholds (Lee et al., 2008). In our case, nearly 25% of the log(GI50) values for the initial set of 257 compounds were −4. The reason is that the highest tested concentration in the NCI-60 cell lines is generally −4 (in log units), and if a compound is not sufficiently active to show 50% cell growth inhibition at this highest concentration, a upper bound of log(GI50) value of −4 is typically reported (Shi et al., 1998b). Therefore, the bioactivity profiles that contain primarily skewed inactivity data may provide biased information. To avoid such bias to certain extent, a third filter is needed to require every compound in the initial set to be active, i.e. log(GI50)≤−6, in at least 15 out of the 60 NCI cell lines. This criterion can ensure, at least to a partial extent, that the derived similarity in bioactivity profiles result from biological activity rather than inactivity. As a result, the initial compound set (257) was narrowed down to contain only 37 compounds. With a much reduced dataset, we were able to investigate the SAR and compound–target interaction network for those compounds in greater details.
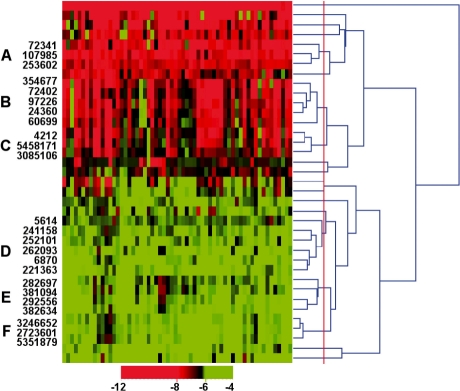
The hierarchical clustering for the 37 bioactive compounds was carried out by using exactly the same algorithm as applied to the initial compound set. The results are shown in Figure 1. The dendrogram graph indicates that compounds with biologically similar bioactivity were grouped together. By setting a relatively tight cutoff of the minimum similarity of 0.88 in the HCE, clusters containing more than one compound were obtained and labeled as A through F from top to bottom. The minimum similarity cutoff of 0.88 was chosen empirically based on visual exploration. Interesting results obtained on these clusters are given below.
Fig. 1.
Hierarchical clustering of the 37 compounds in the final set based on their bioactivity profiles in the NCI-60 cell lines. The bioactivity profile of each compound is shown in spectrum (horizontal view). A minimum similarity threshold of 0.88 (red solid line) is employed in HCE. Six clusters that contain more than one compound are marked as A through F from top to bottom. Relevant compounds (24 in total) are labeled with PubChem compound identifiers (CID).
3.2 Highly similar structures with highly similar bioactivity profiles (Cluster B)
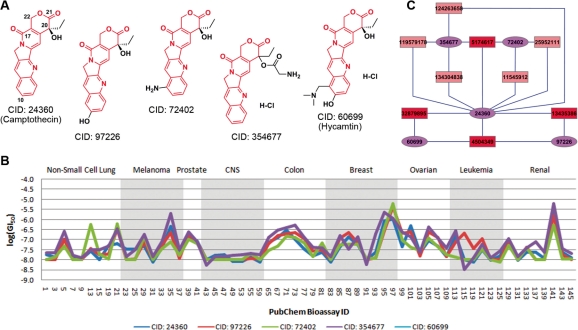
Five compounds (CID: 24360, 97226, 72402, 354677 and 60699) were identified from cluster B. Their 2D chemical structures are depicted in Figure 2A. Compared to the compounds in other clusters, they gave the highest structural similarity as calculated by using the Tanimoto metric and PubChem fingerprint (ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt). The average inter-compound structural similarity for these compounds was 0.887. Therefore, it may not be surprising to see that they also exhibited similar biological responses in the NCI-60 cell lines (Fig. 2B). Indeed, similar bioactivity profiles were observed for these compounds, indicating strong and consistent inhibitory activity for a considerable number of cell lines. The results from Figure 2A and B suggest that this group of compounds demonstrated strong SAR.
Fig. 2.
The five camptothecin analogs identified from cluster B. (A) 2D chemical structures, (B) bioactivity profiles in the NCI-60 cell lines on nine different organs and (C) compound–target interaction network (see Fig. 4 for general description).
Further analysis on chemical structures shows that these compounds are the analogs of camptothecin, a selective inhibitor of the topoisomerases I (TOP1). Among the five compounds, two (CID: 24360 and 60699) are well-known inhibitors of TOP1 (Pizzolato and Saltz, 2003; Wethington et al., 2008). The former is camptothecin itself, while the latter has recently been approved by the FDA (trade name Hycamtin) in 2007 for oral use to treat ovarian cancer (http://en.wikipedia.org/wiki/Hycamtin, accessed on April 12, 2010), which is consistent with its activity in the ovarian cell lines (Fig. 2B). Considering the significant similarity in both chemical structures and bioactivity profiles, we proposed that the other three compounds (CID: 97226, 72402 and 354677) might be novel candidates of TOP1 inhibitors. Nevertheless, one must always keep in mind that this may only be confirmed if the binding mechanism is understood. In this case, the binding modes of the two known inhibitors (CID: 24360 and 60699) have already been previously clarified (Staker et al., 2002, 2005). The X-ray crystal structures of the enzyme–inhibitor complexes indicate that the oxygen atoms connected to the positions 10, 17, 20, 21 and 22 (Fig. 2A) are critical for the binding process by forming several hydrogen bonding interactions directly or indirectly (through water salt bridges) with relevant residues on TOP1. These key interacting sites are basically preserved in other three compounds (CID: 97 226, 72 402 and 354 677). Therefore, it further suggests that these compounds might be true TOP1 inhibitors, as supported by previous studies (Ping et al., 2006; Rapisarda et al., 2002; Wethington et al., 2008).
As mentioned in the Section 1, PubChem can provide rich information of the compound–target associations for a number of tested compounds in the NCI-60 dataset. By combining such data from both repositories, it is possible for us to characterize the bioactivity of tested compounds at cellular level and molecular level simultaneously. The compound–target interaction network drawn from the available PubChem HTS bioassays for the five compounds in cluster B is shown in Figure 2C.
As one can see, these five compounds were closely packed by sharing some common or relevant protein targets. Among the three compounds (CID: 24360, 354677 and 72402), the first compound shared four common protein targets (GI: 119579178, 134304838, 124263658 and 5174617) with the second compound. In addition, it also shared three common protein targets (GI: 11545912, 25952111 and 5174617) with the third compound. Moreover, these three compounds shared a common protein target (GI: 5174617). This observation again demonstrates the effectiveness of the similarity principle. While it remains to be further evaluated when sufficient data is available, we propose that compounds sharing significant similarity in both chemical structures and bioactivity profiles may have a higher chance for sharing similar patterns of interactions in the compound–target interaction network, comparing to those with only structural similarity. The remaing two compounds (CID: 97226 and 60699) appear to be apart from the above three compounds in the common (or shared) compound–target interaction network. This is mainly because they had not been tested on the same targets, against which the previous three compounds were tested, and thus gave insufficient information on their interactions with those relevant protein targets. However, as indicated in Figure 2C, they had demonstrated similar activity to the compound CID: 24360 against several common protein targets in a pairwise manner, which may provide links to the other two compounds (CID: 354677 and 72402).
It should be mentioned that the abundant information of compound–target association in PubChem bioassays may also contain experimental noises such as promiscuous results. While we cannot rule out the possibilities of the promiscuous effects or other artifacts in our analysis, we found that some of the compound–target associations were supported by previous studies. For example, for the two compounds (CID: 24360 and 60699) in cluster B, they both exhibited activity against two common targets with one of them, hypoxia-inducible factor 1α (HIF-1α, GI: 32879895), having been reported as the biological target for these two compounds (Klausmeyer et al., 2007; Rapisarda et al., 2002). These investigations support that our findings result from experimental signals rather than noises. It is noticeable that the compound CID: 354677 is also a known HIF-1α inhibitor (Rapisarda et al., 2002), though it had not been included in the compound–target network due to insufficient data in PubChem (Fig. 2C). This again demonstrates that similarity in bioactivity profiles and patterns of interactions with relevant targets can be used to identify novel compounds for a certain target. Nevertheless, further experiments will be needed to validate some of the potentially novel compounds identified by the MLP project.
3.3 Moderately similar structures with highly similar bioactivity profiles (Cluster F)
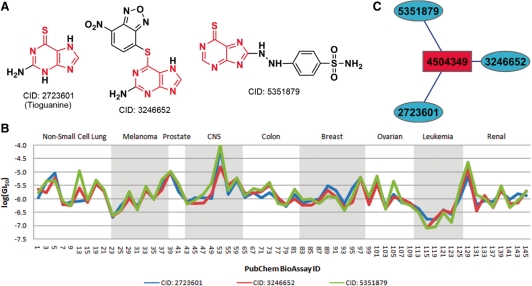
Three compounds (CID: 2723601, 3246652 and 5351879) bearing partially structural similarity were identified from cluster F. Their 2D chemical structures and bioactivity profiles in the NCI-60 cell lines are given in Figure 3A and B, respectively. These three compounds stood out from the rest because they exhibited the maximal intra-cluster similarity in their bioactivity profiles (Fig. 3B). In fact, cluster F was the first merged sub-tree during the hierarchical clustering process (Fig. 1). Compared to cluster B, the observation in cluster F may be even more interesting as the inter-compound structural similarities were significantly lower than those of cluster B. For example, the two compounds (CID: 2723601 and 3246652), which produced the highest structural similarity (0.462) among this cluster, indicates only a moderate level of similarity in their chemical structures. Another interesting observation seen from Figure 3B is that these three compounds show effective but still selective activity in the six leukemia cell lines, suggesting their potential treatment to leukemia. Indeed, one compound (CID: 2723601) is an approved small-molecule drug used in the therapy of several forms of leukemia (http://www.drugbank.ca/drugs/DB00352, last accessed date October 7, 2010).
Fig. 3.
The three compounds identified from cluster F. (A) 2D chemical structures, (B) bioactivity profiles in the NCI-60 cell lines on nine different organs and (C) compound–target interaction network (see Fig. 4 for general description).
The compound–target interaction network for the three compounds in cluster F is given in Figure 3C. A single, common protein target (β-globin, GI: 4504349) was shared by all three compounds, demonstrating again a strong correlation between the similarity in bioactivity profiles and that in the patterns of interactions with relevant protein targets. According to the PubChem BioAssay database, the compound CID: 2723601 was tested active in the bioassay (AID: 910), while the other two compounds (CID: 3246652 and 5351879) were tested active in another bioassay (AID: 925). Despite two separate bioassays, they were actually part of a series of assays in an attempt to seek for the modulators of hemoglobin β-splicing (Supplementary Table S1). A further analysis shows that though the overall structural similarity was relatively low, these three compounds possessed a common fragment of thioguanine, which may play a key role for the compounds to exhibit activity in modulating the hemoglobin splicing and some other biological processes. This example suggests that similarity in bioactivity profiles derived from a broad panel of assays, together with the common features in chemical structures, can also indicate similarity in the mode of action for respective compounds, and can be used as a basis to determine information such as molecular targets or biological pathways for uncharacterized compounds.
3.4 Results for clusters A, C, D and E
Unlike the compounds in clusters B and F, where strong correlations among chemical structures, bioactivity profiles and patterns of interactions with relevant protein targets can be observed, the compounds in the other four clusters did not fall in the same category for various reasons.
The three compounds in cluster A did not show significant similarity in either chemical structures or bioactivity profiles based on the data available (Supplementary Fig. S2), yet the results are still interesting despite the lack of overall coherence in the compound–target interaction network. For example, two compounds (CID: 107985 and 253602), regardless of the difference in chemical scaffolds, were identified as showing inhibitory activities for the heat shock factor 1 (HSF1, GI: 62740231), which is in agreement with previous findings that both compounds are involved in the heat shock response pathway (Park and Liu, 2001; Westerheide et al., 2006). Likewise, the four compounds in cluster E generally show low similarity in their bioactivity profiles (Supplementary Figure S3). Moreover, structural similarity is also missing among compounds, making the current bioactivity profiles less useful in discovering novel compounds for the given targets. Nevertheless, it remains interested to investigate the relationships among the compounds and their target networks in future when more bioassay data become available in PubChem.
The chemical structures and bioactivity profiles for the three compounds identified from cluster C are listed in Supplementary Figure S4. These three compounds exhibited certain structural similarity by sharing a common fragment of di-ketone (Supplementary Figure S4A), which may be responsible for the notable similarity in their bioactivity profiles (Supplementary Figure S4B). This observation resembled the results of cluster F, where compounds moderately similar in chemical structures with common fragment demonstrated significant similarity in bioactivity profiles. As for the compound–target interaction network (Supplementary Figure S4C), however, only one compound (CID: 4212) in cluster C had been extensively assayed on multiple protein targets, against which the other two compounds were not tested. Therefore, target networks cannot be compared directly due to lack of experimental support. Nevertheless, for the compound CID: 548171, considering its notably high similarities to the compound CID: 4212 in both chemical structures and bioactivity profiles (Supplementary Figure S4A and B), it may also interact with certain protein targets shared by the compound CID: 4212. This hypothesis for predicting partially characterized compound according to well-characterized ones remains highly interested to be verified by future experiments.
As for the six compounds identified from cluster D, though there was no obvious similarity either in chemical structures or bioactivity profiles, they seemed to considerably show several common patterns of interactions with relevant protein targets (Supplementary Figure S5). For example, four (CID: 262093, 5614, 221363 and 252101) out of the six compounds in cluster D were found active against several protein targets belonging to various cytochrome p450 families and/or subfamilies, suggesting that they may be effective in the p450-regulated pathways. This observation suggests that the compound–target interaction network derived from PubChem bioassays may be useful to identify a set of related compounds involving in the same/similar biological pathway.
3.5 Overview of compound–target interaction network
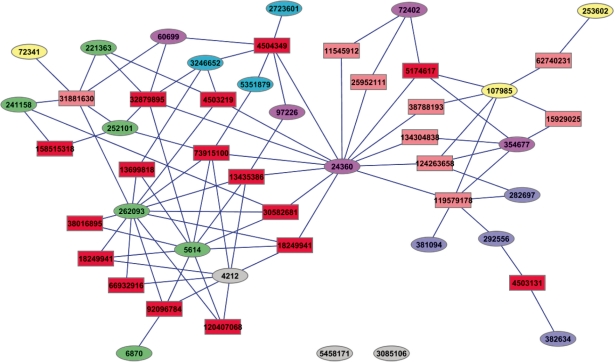
The compound–target interaction networks analyzed in the above sections were drawn for the compounds within the same hierarchical cluster of bioactivity profiles. It remained highly interested to investigate how compounds can be organized solely by their patterns of interactions with relevant protein targets, and how that can be compared to the hierarchical clusters derived from the bioactivity profiles analysis. To this end, a complete diagram of compound–target interaction network, as shown in Figure 4, was built for all the compounds identified from the six clusters (i.e. A to F) using the abundant information of compound–target association in PubChem bioassays.
Fig. 4.
The complete diagram of the compound–target interaction network for the 24 compounds identified from the six clusters (i.e. A to F) obtained by hierarchical clustering. Compounds are denoted as ellipses, which are labeled with PubChem compound identifier (CID) and colored according to the clusters they belong to. Targets are denoted as rectangles, which are labeled with NCBI protein identifier (GI) and colored with dark or light red if the corresponding assay is a confirmatory or primary bioassay in PubChem, respectively. The edge linking an ellipse and a rectangle indicates that there is an interaction if the current compound is found active against the target of interest. No edge is allowed between either two ellipses or two rectangles. For simplicity, target nodes that have only single connecting compound node are not shown.
In general, the network was rather complex and presented a great challenge for data analysis as a considerable number of compounds had demonstrated interactions against multiple protein targets. Though these interactions remained to be further evaluated by identifying and excluding noises in the current assay data, the multitude of compound–target associations may reveal the promiscuous properties for certain compounds at the first glance and may facilitate the investigation of the polypharmacological properties of small molecules.
Despite the observed complexity, the compounds shown in Figure 4 can still be roughly grouped by their patterns of interactions with relevant protein targets. For instance, the six compounds obtained from the bioactivity profile cluster D (colored in green) tended to pack into a group, which was well supported by the fact that there were so many common interacting targets shared by two or more compounds (Supplementary Figure S5). Similarly, the three compounds from the bioactivity profile cluster F (colored in cyan) can also be identified as a group due to a commonly shared protein target (GI: 4504349). Therefore, it is interesting to observe that the groups of compounds identified from the target network were, to certain extent, consistent with those obtained by the clustering analysis based on bioactivity profiles. This observation indicates that there could be strong correlations between a compound's bioactivity profile (cellular level) and its pattern of interactions with relevant protein targets (molecular level). The compounds in the above two clusters exhibited much larger variances (i.e. higher specificity) in their bioactivity profiles, which may contribute to their relatively converged patterns of interactions with relevant protein targets. In contrast, some compounds presented in bioactivity profile cluster A (colored in yellow) showed generalized toxicity with low selectivity and specificity (Supplementary Figure S2B), making them difficult to be identified as a group from Figure 4. This analysis was done using a binary bioactivity outcome when considering the compound–target association. Further analysis may be performed in future work by incorporating the quantitative potency data (e.g. IC50) of each compound to provide more insights.
4 CONCLUSIONS
By taking advantages of the publicly available data from both PubChem HTS bioassays and NCI-60 human tumor cancer cell line screens, we have investigated the correlations among the bioactivity profiles, molecular targets and chemical structures of small molecules. Hierarchical clustering of tested compounds was carried out based on their bioactivity profiles derived from the NCI-60 cell line screens, and several interesting clusters were identified. First, the correlation between bioactivity profiles and chemical structures was analyzed and strong SAR was suggested. For example, the compounds in cluster B, which were highly similar in chemical structures, also demonstrated notable similarity in their bioactivity profiles. Even more interesting observations were given by cluster F, where compounds were only moderately similar in chemical structures and produced extremely significant similarity in bioactivity profiles. Second, analysis on the compound–target interaction network was performed and showed clear correlations between the bioactivity profiles of compounds and their patterns of interactions with relevant protein targets, especially when chemical structures were related. Furthermore, a complete compound–target interaction network, which was drawn for all the compounds identified from the six clusters, produced roughly the same groups of compounds as that obtained by hierarchical clustering analysis based on bioactivity profiles. This study shows that strong correlations can be observed between similarity in bioactivity profiles (cellular level) and that from the patterns of interactions with relevant protein targets (molecular level), and suggests that novel compound candidates with desired pharmacological properties can be identified by comparing their bioactivity profiles and/or compound–target interaction network to well-characterized compounds.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Institutes of Health Fellows Editorial Board (FEB) for article revision.
Funding: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of Interest: none declared.
REFERENCES
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wild DJ. PubChem BioAssays as a data source for predictive models. J. Mol. Graphics Model. 2010;28:420–426. doi: 10.1016/j.jmgm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Chen B, et al. PubChem as a source of polypharmacology. J. Chem. Inf. Model. 2009;49:2044–2055. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, et al. The price of innovation: new estimates of drug development costs. J. Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Guha R. Flexible web service infrastructure for the development and deployment of predictive models. J. Chem. Inf. Model. 2008;48:456–464. doi: 10.1021/ci700188u. [DOI] [PubMed] [Google Scholar]
- Han L, et al. Developing and validating predictive decision tree models from mining chemical structural fingerprints and high-throughput screening data in PubChem. BMC Bioinf. 2008;9:401. doi: 10.1186/1471-2105-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausmeyer P, et al. Identification of a new natural camptothecin analogue in targeted screening for HIF-1α inhibitors. Planta Med. 2007;73:49–52. doi: 10.1055/s-2006-951767. [DOI] [PubMed] [Google Scholar]
- Lee AC, et al. Data mining the NCI60 to predict generalized cytotoxicity. J. Chem. Inf. Model. 2008;48:1379–1388. doi: 10.1021/ci800097k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. A novel method for mining highly imbalanced high-throughput screening data in PubChem. Bioinformatics. 2009;25:3310–3316. doi: 10.1093/bioinformatics/btp589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Liu AY. JNK phosphorylates the HSF1 transcriptional activation domain: role of JNK in the regulation of the heat shock response. J. Cell. Biochem. 2001;82:326–338. doi: 10.1002/jcb.1163. [DOI] [PubMed] [Google Scholar]
- Paull KD, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Ping Y-H, et al. Anticancer effects of low-dose 10-hydroxycamptothecin in human colon cancer. Oncol. Rep. 2006;15:1273–1279. [PubMed] [Google Scholar]
- Pizzolato JF, Saltz LB. The camptothecins. Lancet. 2003;361:2235–2242. doi: 10.1016/S0140-6736(03)13780-4. [DOI] [PubMed] [Google Scholar]
- Rabow AA, et al. Mining the National Cancer Institute's tumor-screening database: identification of compounds with similar cellular activities. J. Med. Chem. 2002;45:818–840. doi: 10.1021/jm010385b. [DOI] [PubMed] [Google Scholar]
- Rapisarda A, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- Rohrer SG, Baumann K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J. Chem. Inf. Model. 2009;49:169–184. doi: 10.1021/ci8002649. [DOI] [PubMed] [Google Scholar]
- Seo J, Shneiderman B. Interactively exploring hierarchical clustering results. IEEE Computer. 2002;35:80–86. [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LM, et al. Mining the NCI anticancer drug discovery databases: genetic function approximation for the QSAR Study of anticancer ellipticine analogues. J. Chem. Inf. Comput. Sci. 1998a;38:189–199. doi: 10.1021/ci970085w. [DOI] [PubMed] [Google Scholar]
- Shi LM, et al. Mining the National Cancer Institute anticancer drug discovery database: cluster analysis of ellipticine analogs with p53-inverse and central nervous system-selective patterns of activity. Mol. Pharmacol. 1998b;53:241–251. doi: 10.1124/mol.53.2.241. [DOI] [PubMed] [Google Scholar]
- Shi LM, et al. Mining and visualizing large anticancer drug discovery databases. J. Chem. Inf. Comput. Sci. 1999;40:367–379. doi: 10.1021/ci990087b. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Staker BL, et al. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- Staker BL, et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Nat. Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Osdol WW, et al. Use of the Kohonen self-organizing map to study the mechanisms of action of chemotherapeutic agents. J. Natl Cancer Inst. 1994;86:1853–1859. doi: 10.1093/jnci/86.24.1853. [DOI] [PubMed] [Google Scholar]
- Wallqvist A, et al. Linking the growth inhibition response from the National Cancer Institute's anticancer screen to gene expression levels and other molecular target data. Bioinformatics. 2003;19:2212–2224. doi: 10.1093/bioinformatics/btg302. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Chemical data mining of the NCI human tumor cell line database. J. Chem. Inf. Model. 2007;47:2063–2076. doi: 10.1021/ci700141x. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, et al. Neural computing in cancer drug development: predicting mechanism of action. Science. 1992;258:447–451. doi: 10.1126/science.1411538. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Weis DC, et al. Data mining PubChem using a support vector machine with the signature molecular descriptor: classification of factor XIa inhibitors. J. Mol. Graphics Model. 2008;27:466–475. doi: 10.1016/j.jmgm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, et al. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J. Biol. Chem. 2006;281:9616–9622. doi: 10.1074/jbc.M512044200. [DOI] [PubMed] [Google Scholar]
- Wethington SL, et al. Key role of topoisomerase I inhibitors in the treatment of recurrent and refractory epithelial ovarian carcinoma. Expert Rev. Anticancer Ther. 2008;8:819–831. doi: 10.1586/14737140.8.5.819. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X-Q, Chen J-Z. Data mining a small molecule drug screening representative subset from NIH PubChem. J. Chem. Inf. Model. 2008;48:465–475. doi: 10.1021/ci700193u. [DOI] [PubMed] [Google Scholar]
- Zaharevitz DW, et al. COMPARE: a web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graphics Model. 2002;20:297–303. doi: 10.1016/s1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- Zerhouni E. The NIH roadmap. Science. 2003;302:63–72. doi: 10.1126/science.1091867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.