Abstract
Summary: The GeneMANIA Cytoscape plugin brings fast gene function prediction capabilities to the desktop. GeneMANIA identifies the most related genes to a query gene set using a guilt-by-association approach. The plugin uses over 800 networks from six organisms and each related gene is traceable to the source network used to make the prediction. Users may add their own interaction networks and expression profile data to complement or override the default data.
Availability and Implementation: The GeneMANIA Cytoscape plugin is implemented in Java and is freely available at http://www.genemania.org/plugin/.
Contact: gary.bader@utoronto.ca; quaid.morris@utoronto.ca
1 INTRODUCTION
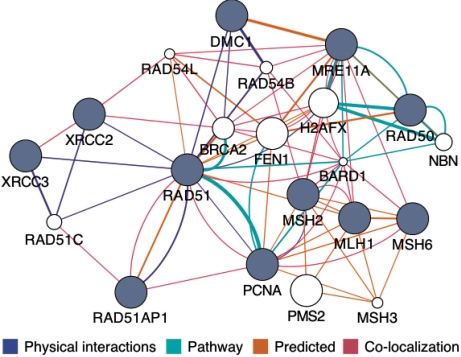
The GeneMANIA Cytoscape plugin is a standalone tool for making fast and efficient gene function predictions. The plugin implements the GeneMANIA algorithm (Mostafavi et al., 2008), which uses a guilt-by-association approach to derive predictions from a combination of potentially heterogeneous data sources. GeneMANIA has been shown to be as good or better in speed and accuracy compared with other gene function prediction algorithms in a competition based on mouse functional association network data (Pena-Castillo et al., 2008). The plugin extends the Cytoscape network visualization and analysis platform (Shannon et al., 2003) and the functionality of the GeneMANIA gene function prediction website (Warde-Farley et al., 2010) to enable computational biologists and biologists to conduct queries using any number of genes and networks as long as their machine has enough memory. The resulting predicted network of functional relationships among query and predicted genes is then available as an annotated Cytoscape network for further analysis (Fig. 1).
Fig. 1.
The GeneMANIA Cytoscape plugin analysis view showing an example prediction. A set of known DNA repair genes were provided as a query (gray nodes) and a number of additional DNA repair genes were predicted to be related (white nodes).
The plugin uses a large dataset of functional association networks, which includes over 800 networks for six organisms: Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Homo sapiens and Saccharomyces cerevisiae. The networks are grouped into six categories: co-expression, co-localization, genetic interaction, physical interaction, predicted and shared protein domain. The data come from a wide range of sources including individual studies and large databases such as BIOGRID (Breitkreutz et al., 2008), GEO (Barrett et al., 2009), I2D (Brown and Jurisica, 2005) and Pathway Commons (http://www.pathwaycommons.org). Networks are updated regularly. The plugin automatically checks for these updates and prompts the user to download and install new networks and organisms as they become available.
2 IMPLEMENTATION
The GeneMANIA Cytoscape plugin integrates association networks from multiple sources into a single composite network using a conjugate gradient optimization algorithm as described in Mostafavi et al. (2008). Networks are weighted according to query dependent criteria. Each source network is stored on disk as a sparse weighted adjacency matrix, where weight corresponds to gene interaction strength. Matrices are loaded as needed and selectively cached in memory for subsequent queries. This compact matrix representation is used directly by the conjugate gradient algorithm resulting in very fast predictions, given sufficient memory.
A Lucene index stores the mappings between gene symbols and their positions within the sparse matrices. This enables fast gene validation and synonym detection. The index also stores all gene and network metadata including associated publications and hyperlinks to external data sources.
3 FEATURES
Ease of Use: the GeneMANIA Cytoscape plugin has a user-friendly graphical user interface, which makes powerful prediction tools and data accessible to typical biologists. It can be installed using Cytoscape's menu-driven plugin manager. Upon first use, a user must download the latest version of GeneMANIA data for the organisms they are interested in. A simple graphical interface aids in this process, which can be time consuming depending on organism choice (e.g. all data for human is currently 1.4 GB compressed). However, once the database is downloaded, it does not need to be downloaded again, unless there is an update.
The plugin recognizes gene identifiers, symbols and non-ambiguous synonyms from Entrez, Ensembl, RefSeq, TAIR and Uniprot. Users can supply a mixture of symbols from different sources and the plugin will attempt to map them to the corresponding gene. Users can build their query list using the auto-completion feature, which finds genes by prefix as the user types or by pasting in large gene lists from other sources, such as text files.
The plugin can produce a prediction report that lists the details of the query list, source networks and the predicted genes. The composite interaction network used for the prediction can also be exported in standard formats, e.g. XGMML, SIF and PDF.
Customization: individual networks or entire categories can be included or excluded prior to the prediction process. Users may add their own interaction networks and gene expression profiles to this set. The plugin automatically translates the networks into an optimized matrix format, reports any unrecognized gene symbols to the user and omits the corresponding interactions.
A number of weighting methods are available to adjust the degree of influence each network has on the resulting prediction. The default weighing method (‘automatic’) chooses between two different weighting methods depending on query list size. For longer gene lists, each network is weighted so that after the networks are combined, the query genes interact as much as possible with each other while interacting as little as possible with genes not in the list (‘query gene based’ weighting). For shorter gene lists, an attempt is made to reproduce Gene Ontology (GO) Biological Process co-annotation patterns (Mostafavi et al., 2010). The two non-adaptive weighting schemes also work well on small gene lists (Mostafavi et al., 2008): ‘equal by network’ weighting assigns the same weight to all networks, whereas the ‘equal by data type’ weighting ensures each network category has the same degree of influence. Network weights can also be assigned based on how well they reproduce GO co-annotation patterns for that organism in the molecular function or cellular component hierarchies.
Provenance: each prediction is annotated with all contributing source interactions. Clicking on an interaction reveals the details about its data source and links to relevant publications, if available.
Scalability: the size of the GeneMANIA network data is limited by the amount of available memory and disk space. We recommend using a system with 4 GB of total RAM when using the default list of networks and at least 6 GB of RAM for all networks.
4 EXAMPLE APPLICATION
Identification of potential biomarkers of disease is an important research area. We entered 10 samples of 200 genes each from a list of 436 well-supported potential biomarkers for pancreatic cancer (Harsha et al., 2009) into the GeneMANIA Cytoscape plugin, using all available networks. The 10 queries took 78 s each on average on an Intel Core i7 930 system with 6 GB of RAM. Of the 236 genes returned, 51.8 genes were found on average (σ = 5.51) from the 236 held-out genes. We compared these to 10 random samples of 200 genes from the human genome excluding the 436 well-supported biomarkers and found a statistically significant P-value of 3.1 × 10−12 (two-tailed independent t-statistic). This example shows how the GeneMANIA Cytoscape plugin can be used to predict additional genes that may be involved in pancreatic cancer. Selecting more predicted genes will find more related genes to the query set.
5 CONCLUSION
The GeneMANIA Cytoscape plugin is freely available at http://www.genemania.org/plugin/ and via the Cytoscape plugin manager. It also includes command-line tools for running multiple predictions in an automated fashion to facilitate performance evaluation of different algorithm parameters via cross-validation. For example, users can determine the contribution of their own networks to the performance of the algorithm using all publicly available data.
ACKNOWLEDGEMENTS
We would like to thank the GeneMANIA team for support for this project and the many Cytoscape developers for developing and maintaining a visualization and analysis platform that greatly facilitates plugin development. We would also like to thank Harsha Gowda, Kumaran Kandasamy and Akhilesh Pandey for sharing their pancreatic cancer gene lists; and Season of Usability student Elham Alizadeh and mentor Celeste Lyn Paul for guiding the evolution of the plugin's user interface.
Funding: Genome Canada through the Ontario Genomics Institute (grant number 2007-OGI-TD-05).
Conflict of Interest: none declared.
REFERENCES
- Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Harsha HC, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Morris Q. Fast integration of heterogeneous data sources for predicting gene function with limited annotation. Bioinformatics. 2010;26:1759–1765. doi: 10.1093/bioinformatics/btq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, et al. GeneMANIA: a real-time multiple association network integration algorithm for prediction gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Castillo L, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9:S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]