Abstract
The genome of Thellungiella parvula, a halophytic relative of Arabidopsis (Arabidopsis thaliana), is being assembled using Roche-454 sequencing. Analyses of a 10-Mb scaffold revealed synteny with Arabidopsis, with recombination and inversion and an uneven distribution of repeat sequences. T. parvula genome structure and DNA sequences were compared with orthologous regions from Arabidopsis and publicly available bacterial artificial chromosome sequences from Thellungiella salsuginea (previously Thellungiella halophila). The three-way comparison of sequences, from one abiotic stress-sensitive species and two tolerant species, revealed extensive sequence conservation and microcolinearity, but grouping Thellungiella species separately from Arabidopsis. However, the T. parvula segments are distinguished from their T. salsuginea counterparts by a pronounced paucity of repeat sequences, resulting in a 30% shorter DNA segment with essentially the same gene content in T. parvula. Among the genes is SALT OVERLY SENSITIVE1 (SOS1), a sodium/proton antiporter, which represents an essential component of plant salinity stress tolerance. Although the SOS1 coding region is highly conserved among all three species, the promoter regions show conservation only between the two Thellungiella species. Comparative transcript analyses revealed higher levels of basal as well as salt-induced SOS1 expression in both Thellungiella species as compared with Arabidopsis. The Thellungiella species and other halophytes share conserved pyrimidine-rich 5′ untranslated region proximal regions of SOS1 that are missing in Arabidopsis. Completion of the genome structure of T. parvula is expected to highlight distinctive genetic elements underlying the extremophile lifestyle of this species.
The crucifer genus Thellungiella includes a number of species that have recently been recognized as models for identifying processes, pathways, and genes of importance in plant abiotic stress tolerance (Bressan et al., 2001; Xiong and Zhu, 2002; Taji et al., 2004; Amtmann et al., 2005; Griffith et al., 2007). In particular, Thellungiella salsuginea, previously known as Thellungiella halophila (Al-Shehbaz and O’Kane, 1995; Amtmann, 2009), has been studied for its extreme salt, cold, and freezing tolerance and for its efficient mobilization of resources in poor or degraded soils (Kant et al., 2008). Another species, Thellungiella parvula, has been reported to have slightly higher salt and drought tolerance, being otherwise comparable to T. salsuginea in cold and freezing response characteristics (Orsini et al., 2010). The stress tolerance behavior of both species is appropriate to their natural habitats: of the several T. salsuginea ecotypes that have been collected, all are from stress-prone locations, such as the delta of the Yellow River in China, the Yukon Territory of Canada, and the Rocky Mountains of the United States. Similarly, the T. parvula ecotype used in this study originated from a salt flat in central Anatolia, Turkey.
Because of their close phylogenetic relationships to Arabidopsis (Arabidopsis thaliana; O’Kane and Al-Shehbaz, 2003) and because many transcripts have nucleotide sequence identities with the better known model in the 90% range (Wang et al., 2004; Taji et al., 2008), focusing on Thellungiella spp. in order to derive a molecular-level understanding of adaptation to challenging environments appears particularly appropriate. Both Thellungiella species are comparable to Arabidopsis in terms of growth rates and size under similar growth conditions and in terms of fertility, seed number, and ease of transformation by flower dipping (Inan et al., 2004).
A number of tools for comparative molecular characterizations have been developed for T. salsuginea, including tagged mutants, EST collections, and full-length cDNA collections (Inan et al., 2004; Taji et al., 2004, 2008; Wang et al., 2004, 2006; Gong et al., 2005; Wong et al., 2005). To date, results of stress challenge studies have identified transcriptome responses that are broadly similar to those shown by Arabidopsis. Some universally stress-responsive genes, however, appear to be highly expressed in Thellungiella even in the absence of challenges. Others are induced only at much higher levels of stress than their isologs in Arabidopsis. In addition, altered gene expression patterns appear to be more specific to the stress that is experienced. This contrasts with transcriptional responses in Arabidopsis, which are much less discriminating and typically are global (Gong et al., 2005; Kant et al., 2006).
In Arabidopsis, salt tolerance, albeit weak, is based on the action of a complex of two proteins (SALT OVERLY SENSITIVE3 [SOS3], a sensor, and SOS2, a protein kinase) involved in sensing and responding to the influx of sodium ions. This complex activates a number of proteins (Halfter et al., 2000; Cheng et al., 2004; Quan et al., 2007; Lin et al., 2009), including SOS1, a plasma membrane-located sodium/proton antiporter. Activation of SOS1 mediates the export of the Na+ ions, which may also be redistributed throughout the plant (Qiu et al., 2002; Quintero et al., 2002; Shi et al., 2002, 2003). The SOS complex exists in many other species, including T. salsuginea, suggesting that it is a general feature of plants. Recently, the importance of SOS1 in stress tolerance in T. salsuginea has been established using RNA interference to generate lines with reduced SOS1 transcript and protein abundances. These lines showed salt sensitivity nearly as pronounced as that exhibited by Arabidopsis (Oh et al., 2009).
Like Arabidopsis, both T. parvula and T. salsuginea have relatively small genomes. The Thellungiella species have seven pairs of chromosomes (Orsini et al., 2010). For T. salsuginea, the diploid genome size is about twice that of Arabidopsis, approximately 260 Mb (Inan et al., 2004). We now estimate, based on flow cytometry, a diploid genome size of approximately 180 Mb for T. parvula. These values, in combination with the Arabidopsis reference genome structure against which sequences can be compared, make genomic sequence analysis approachable.
Here, we report on our initial analyses of the T. parvula genome structure based on sequencing using the Roche-454 platform. Our objectives were to shed light on differences in the overall gene structure and composition of the T. parvula genome in comparison with those of Arabidopsis and T. salsuginea. We compare regions in the T. parvula genome with two bacterial artificial chromosome (BAC) sequences from orthologous regions in T. salsuginea (Deng et al., 2009; Nah et al., 2009) as well as with orthologous segments in the Arabidopsis genome and, in a three-species comparison, analyze gene and genome structures. We include an analysis of promoter structures, the presence and organization of repeats, and transcript expression for one of the genes in this region, SOS1, in an attempt to distinguish characteristics of the glycophytic and halophytic versions of this important salt tolerance determinant.
RESULTS
T. parvula genome sequences were determined by scaffold generating de novo assemblies constructed with Roche-454 GS-FLX Titanium single and paired-end sequencing. The average read lengths were 360 bp or greater in each paired-end library sequenced, a significant improvement compared with the predecessor version GS-FLX. Further sequencing and a draft genome assembly are in progress. We deduced a minimal genome size of 163.6 Mb, based on the total length of the preliminary assembly contigs. An independent estimate to the genome size of about 180 Mb was obtained through flow cytometric analysis of propidium iodide-stained nuclei (Supplemental Fig. S1).
Overview of T. parvula Genome Assembly and Analyses of a Scaffold
Table I summarizes the statistics of a genome draft assembly. The single end sequence assembly by Newbler and cleanup gave some 21,000 pieces that contain 163.6 Mb of contig sequences, which constitutes the minimal predicted genome size of T. parvula. Addition of paired-end libraries further assembled the contigs into 3,149 scaffolds encompassing 153.7 million bases. The scaffolds contain gaps, and incorporation of the reads from Illumina sequencing with higher coverage is in progress to fill in the gaps. Upon completion, the genome sequence will be released to the public.
Table I. Assembly statistics of the draft genome preliminary assembly.
| Criteria | Values |
| Total no. of reads used | 24.89 million |
| Total no. of bases | 5.9 Gb |
| Average read size | 360 bp |
| No. of reads aligned to contigs | 15.99 million (64%) |
| No. of reads excluded from the assembly as repeats | 6.73 million |
| No. of paired end reads | 7.16 million |
| Amount of both pairs mapped with correct orientation | 60% |
| Amount of bases Q40 or greater | 99.12% |
| Expected coverage | 22× |
| No. of contigs | 21,619 |
| Total length of contigs | 163.6 million bp |
| No. of contigs at least N50 | 1,316 |
| Contig N50 | 31,906 bp |
| Minimum contig length | 100 bp |
| Maximum contig length | 600,277 bp |
| Median contig length | 1,452 bp |
| Mean contig length | 7,648 bp |
| No. of scaffolds | 3,149 |
| No. of scaffolds at least N50 | 10 |
| Minimum scaffold length | 900 bp |
| Maximum scaffold length | 13,601,491 bp |
| Median scaffold length | 3,660 bp |
| Mean scaffold length | 48,828 bp |
| Scaffold N50 | 3,006,489 bp |
| Scaffold N90 | 28,644 bp |
| Total length of scaffolds | 153.7 million bp |
| No. of Ns in the scaffolds | 21,969 |
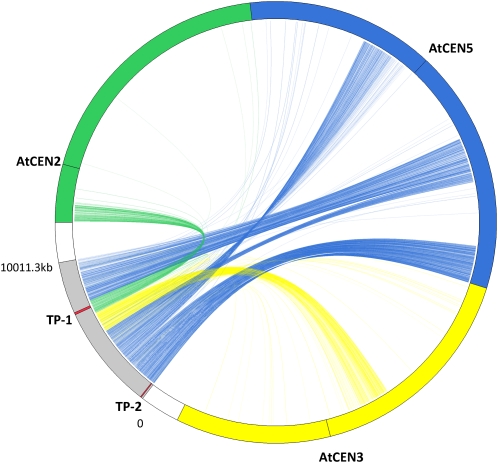
One of the larger scaffolds (scaffold 00254) of 10,011,287 bp was subjected to further analysis. Scaffold 00254 had a GC content of 35.79%, comparable to that of the Arabidopsis genome (36%). It had 346 gaps, with a mean predicted gap size of 757 bp, comprising 5.43% of the scaffold sequence. The scaffold contained overall 1.9% of repetitive sequences, including simple repeats, transposons, and retrotransposons, predicted by RepeatMasker. The distribution of repetitive sequences was not homogeneous, with the repeat contents usually very low at less than 0.5% over most of the sequences and dramatically increasing up to 15% toward the end of the scaffold sequence (Supplemental Fig. S2A). An ab initio open reading frame (ORF) prediction with FGENESH (http://linux1.softberry.com/berry.phtml) identified 2,229 ORFs, whose distribution showed a pattern opposite to the distribution of the repetitive sequences (Supplemental Fig. S2B). All 2,229 ORFs showed homology with an Arabidopsis gene locus at least for part of their sequences when subjected to BLASTn search against the Arabidopsis reference mRNA database (Supplemental Fig. S2C). When ORFs with total identity higher than 70% with Arabidopsis were considered, 73%, 15%, and 10% of the total predicted ORFs in scaffold 00254 showed the highest similarity with an Arabidopsis gene in chromosome 5, 3, and 2, respectively. To visualize the synteny between scaffold 00254 and Arabidopsis chromosomes, we provide a Circos plot linking the locations of the homologous ORFs between Arabidopsis and T. parvula. Scaffold 00254 showed extensive synteny with parts of Arabidopsis chromosomes 2, 3, and 5, with several rearrangement and inversion events, especially in the region showing colinearity with Arabidopsis chromosome 5 (Fig. 1).
Figure 1.
Synteny between a T. parvula scaffold and Arabidopsis chromosomes. A Circos plot depicting the T. parvula scaffold 00254 (gray) and Arabidopsis chromosomes 2 (green), 3 (yellow), and 5 (blue) is shown. The predicted T. parvula ORFs were connected to their putative Arabidopsis homologs, with the color of the connecting line designating the Arabidopsis chromosome, to visualize the synteny between two species. The two regions, TP-1 and TP-2, where the genomic sequences were available for three-species comparison are designated with red bands.
T. parvula Sequences Compared with Arabidopsis and T. salsuginea
Scaffold 00254 contained regions orthologous to two recently published BAC sequences of T. halophila (now T. salsuginea; Deng et al., 2009; Nah et al., 2009). Here, we focus on these regions for a three-species comparison of genome sequences (Fig. 1, red bands on scaffold 00254). The gaps in the two focus regions were seamlessly filled with genomic PCR and Sanger sequencing with perfect matches in the adjacent sequences. Nah et al. (2009) isolated a T. salsuginea BAC sequence of 193.0 kb (termed here TS-1; National Center for Biotechnology Information [NCBI] GenBank accession no. FJ386403). The orthologous Arabidopsis genomic sequence is a 146.3-kb region (AT-1) located on chromosome 2. The corresponding genomic sequence in the T. parvula genome (TP-1; HM222924) is 134.3 kb in length. The second sequence analyzed here is orthologous to a randomly selected T. salsuginea BAC clone (82.9 kb; termed TS-2 here; DQ226510) presented by Deng et al. (2009). It is compared with a 79.3-kb DNA segment from Arabidopsis chromosome 5 and an orthologous 63.7-kb segment of T. parvula (TP-2; HM222925). Taken together, the overall G+C contents of the two regions in the Thellungiella species are slightly higher than those in Arabidopsis: 35.65% (Arabidopsis), 37.6% (T. salsuginea), and 36.3% (T. parvula).
Both regions for the three species were visualized using dot-plot alignments (Supplemental Fig. S3). The comparison identifies extensive colinearity between the species, with the highest degree between the two Thellungiella species. Several local rearrangements and deletions are obvious, indicated by boxes in Supplemental Figure S3, A and B, for the TP-1 region with respect to the AT-1 and TS-1 regions. In comparison with AT-2, TP-2 showed a large sequence inversion (Supplemental Fig. S3C). In contrast, TP-2 and TS-2 showed colinearity, with several expansions in TS-2 relative to the more compact TP-2 (Supplemental Fig. S3D).
Annotation of T. parvula Genes
The ORFs of the two T. parvula sequences were initially predicted ab initio using FGENESH. Homologous Arabidopsis and T. salsuginea genes were identified using BLASTx to the protein database of both species. Final ORF sequences were determined by FGENESH+ using the homologous Arabidopsis proteins as references. Supplemental Tables S1 and S2 list the predicted ORFs for TP-1 and TP-2, respectively, in comparison with the Arabidopsis and T. salsuginea sequences. ORFs for AT-1 and AT-2 were as annotated in TAIR9, and those for TS-1 and TS-2 as described by Nah et al. (2009) and Deng et al. (2009), respectively. The average gene densities are one per 5.17, 7.15, and 4.58 kb for TP-1, TS-1, and AT-1, respectively, and one per 2.89, 3.45, and 2.78 kb for TP-2, TS-2, and AT-2, respectively.
Ka/Ks (the ratio of the rate of nonsynonymous substitutions [Ka] to the rate of synonymous substitutions [Ks]) between the orthologous genes of three species were calculated, excluding genes of ambiguous orthology (Supplemental Table S3). For the two regions, 11 and 12 sets of genes, respectively, were analyzed. All gene pairs showed Ka/Ks ratios smaller than 1, indicating evolutionary selectivity among the orthologs favoring functional conservation. In all gene pairs, estimated divergences were older when comparing Arabidopsis with the two Thellungiella species than between T. parvula and T. salsuginea, averaging 12.3 million years ago between T. parvula and Arabidopsis and 12.4 million years ago between T. salsuginea and Arabidopsis but only 8.5 million years ago between T. parvula and T. salsuginea (Supplemental Table S3).
Synteny and Similarity
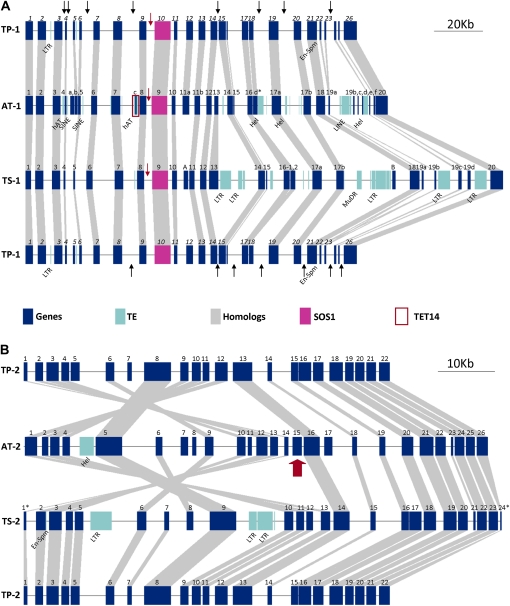
A graphical representation of the three-way comparison of distribution and spacing of genes in the two regions is given in Figure 2. TP-1 showed linear similarity in the arrangement of the genes with both AT-1 and TS-1 (Fig. 2A). One conspicuous difference was the absence of transposons in TP-1 that were present in AT-1 and TS-1; the putative deletion sites are indicated with black arrows in Figure 2A. Both TP-1 and TS-1 contained longer intergenic regions between the genes encoding SOS1 and their associated 5′ gene models compared with AT-1 (Fig. 2A, red arrows). Orthologs of At_c (TET14/AT2G01960) were missing in both TP-1 and TS-1 (Fig. 2A, red box; Supplemental Table S1). In parts of the sequence, however, TP-1 showed higher similarity with AT-1 than with TS-1. For example, orthologous gene pairs Tp05/At_b (AT2G01920) and Tp18/At_d (AT2G02061) are absent in TS-1, while Th_A and Th_B, similar to AT1G14610 and AT2G02030, respectively, were not found in either TP-1 or AT-1 (Supplemental Table S1). TP-2 showed similarity in gene order with TS-2 (Fig. 2B), but both TP-2 and TS-2 showed an inversion of genes relative to AT-2 in part of the sequences. The break point for the inversion was at the position of At15 (AT5G66410; Fig. 2B, red arrow; Supplemental Table S2). In both Thellungiella species, sequence fragments with homologies to AT5G66410 were identified at the break point of the inversion. A gene homologous to AT5G66410, however, could not be identified in the entire T. parvula genome sequence (M. Dassanayake and D.-H. Oh, unpublished data).
Figure 2.
Genome organization for TP-1 (A) and TP-2 (B) in comparison with the organization of orthologous regions in Arabidopsis and T. salsuginea. A, Black arrows in TP-1 show deletions in comparison with AT-1 or TS-1. Red arrows indicate the intergenic space between SOS1 orthologs and their nearest 5′ gene model. B, The inversion break point is indicated by a red arrow. Symbols for the genes are as in Supplemental Tables S1 and S2. Symbols used for transposable elements (TEs) are LTR (retrotransposon with long terminal repeats), SINE (short interspersed transposable elements), LINE (long interspersed transposable elements), MuDR (Mutator-like DNA transposon), Hel (helitron), En-Spm (Enhancer-Suppressor/Mutator transposon), and hAT (hAT family DNA transposon).
Supplemental Figure S4 presents a detailed analysis of exon/intron structures and the overall sequence similarities. In comparing TP-1 and TP-2 with corresponding Arabidopsis and T. salsuginea sequences, the plot identified all exons of the orthologous genes; levels of identities were usually higher than 70%. Short stretches of sequence with significant (greater than 75%) identities were also present in intergenic regions, possibly designating regulatory elements or noncoding RNAs. However, we could not identify any tRNA, microRNA genes, or known small interfering RNA loci in the intergenic regions in any of the three species compared here.
In TP-1, several intergenic regions lacked homology with either Arabidopsis or T. salsuginea [Supplemental Fig. S4A, denoted (a)–(c)]. These discontinuities appear to be based either on a disruption of synteny by the insertion or deletion of genes (a) or transposable element insertion (b). The discontinuity in (c), however, is not explained. The intergenic region upstream of TpSOS1 (TP1-10) was particularly interesting as it revealed homology with T. salsuginea but not with Arabidopsis (Supplemental Fig. S4A, dashed box). In contrast, differences in the intergenic regions between TP-2 and AT-2 involved translocations in TP-2 of genes whose Arabidopsis orthologs were from chromosomes other than chromosome 5: TP2-5, -7, and -16 are orthologs of AT3G53210, AT1G77370, and AT3G47840, respectively [Supplemental Fig. 4B, denoted (a)]. TP2-16 showed similarity with the AT-2 sequence for the latter half of the exons (Supplemental Fig. 4B, dashed box), which had also been observed in the comparison of TS-2 and AT-2 sequences (Deng et al., 2009).
Paucity of Repetitive Sequences in T. parvula
The three species show significant differences in repeat composition and frequency. Repeats in the form of transposable elements, however, are underrepresented (Fig. 2A) to absent (Fig. 2B) in T. parvula compared with the other two species. In both TP-1 and TP-2, analyses using RepeatMasker and searches using other databases failed to identify a complete transposable element, while corresponding T. salsuginea and Arabidopsis sequences contained various retrotransposons and/or DNA transposable elements. In total, these elements span 17.6% (TS-1), 13.5% (TS-2), 8.7% (AT-1), and 3.0% (AT-2) of the sequences (Deng et al., 2009; Nah et al., 2009). In TP-1, small stretches of sequences, less than 100 bp, suggested similarity with Arabidopsis transposable elements (Fig. 2A); such sequences amounted to less than 0.4% of the total length. TP-2 does not present even suggestions of transposable elements (Fig. 2B).
The paucity of repeats in T. parvula is less apparent, however, when tandem repeats are considered (Table II). As observed with transposable elements, sequence 2 has fewer tandem repeats than sequence 1 in all three species. Among the short tandem repeats, the dinucleotide ATn is found at a much higher frequency than other dinucleotide, mononucleotide, trinucleotide, and tetranucleotide repeats in all three species. A survey of short tandem repeats in plants reported ATn as the most abundant short tandem repeat in plant nuclear sequences (Wang et al., 1994).
Table II. Tandem repeat distribution in the three species.
| Variable | TP-1 | AT-1 | TS-1 | TP-2 | AT-2 | TS-2 |
| No. of bases in tandem repeats | 1,661 | 1,747 | 2,521 | 417 | 1,056 | 847 |
| No. of tandem repeat loci | 26 | 28 | 49 | 7 | 11 | 10 |
| No. of short tandem repeat loci | 10 | 8 | 10 | 1 | 2 | 4 |
| No. of long tandem repeat loci | 16 | 20 | 39 | 6 | 9 | 6 |
Comparison of SOS1 Genes
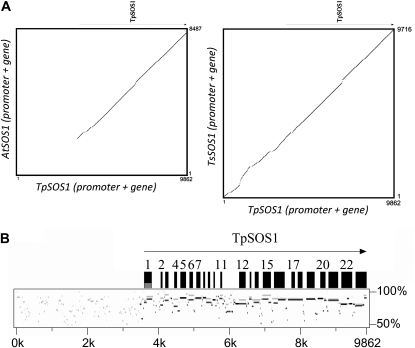
SOS1 was originally identified in Arabidopsis as an important element in the salinity stress tolerance pathway (Shi et al., 2000). Figure 3 presents a comparison of the SOS1 promoters and coding regions in the two Thellungiella species and Arabidopsis. Homology between the promoter regions of AtSOS1, on the one hand, and TpSOS1 or TsSOS1, on the other, was distinctly lacking (Fig. 3A), while PipMaker plots identified conserved motifs between the promoters of TsSOS1 and TpSOS1. Although in the SOS1 gene, TpSOS1 showed higher identity with TsSOS1 than with AtSOS1 in general, different regions exhibited different levels of similarity. For example, TsSOS1 showed higher similarity to TpSOS1 in the N-terminal regions (Fig. 3B, exons 1–15), while AtSOS1 and TsSOS1 showed similar levels of similarity to TpSOS1 in the C-terminal regions (Fig. 3B, exons 17–22).
Figure 3.
Comparison of the SOS1 omologs in T. parvula, T. salsuginea, and Arabidopsis. The genomic sequence spanning the 5′ intergenic space and SOS1 genes from T. parvula was compared with the corresponding regions in T. salsuginea and Arabidopsis using dot plot (A) and PipMaker plot (B). In B, the PipMaker plots comparing TpSOS1 with TsSOS1 (gray) and AtSOS1 (black) were superimposed to show the differences in similarity. Word size of the PipMaker plot was 7 bp.
Comparative Analyses of Transcription
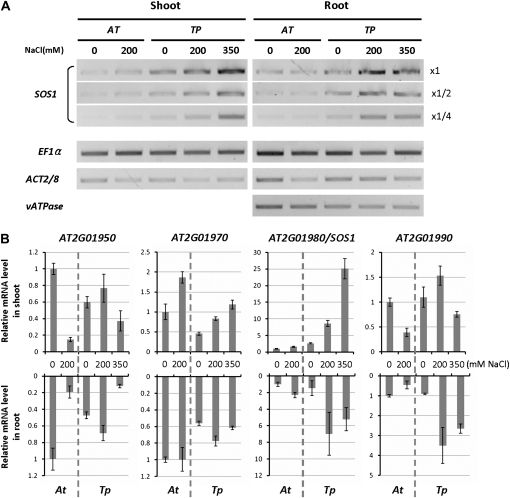
Comparison of SOS1 expression by reverse transcription (RT)-PCR previously revealed higher SOS1 mRNA levels in T. parvula than in Arabidopsis under both normal and high-salt conditions, while housekeeping genes were expressed at similar levels in both species (Oh et al., 2009). Here, we compared the expression patterns of genes surrounding SOS1 in the genome with that of SOS1, using quantitative real-time RT-PCR (Fig. 4; Supplemental Table S4). SOS1 showed 2.6- and 1.5-fold higher mRNA levels in T. parvula shoot and root tissues, respectively, than in Arabidopsis in the absence of stress. Salt stress induced SOS1 expression up to 9.5- and 4.7-fold higher in T. parvula shoots and roots, respectively, while induction was only 1.59- and 2.3-fold in Arabidopsis shoots and roots (Supplemental Table S4). Similar increases in T. parvula were found also after the addition of 350 mm NaCl, a concentration lethal for Arabidopsis ecotype Columbia. The higher SOS1 level in T. parvula compared with Arabidopsis is similar to the previous observation for SOS1 in T. salsuginea (Oh et al., 2009). For other genes tested in the vicinity of SOS1, differences in gene expression between the species were less dramatic (Fig. 4B; Supplemental Table S4).
Figure 4.
Analysis of SOS1 mRNA abundance in Arabidopsis and T. parvula SOS1. SOS1 mRNA levels were compared between Arabidopsis and T. parvula with RT-PCR (A) and quantitative real-time PCR (B). In A, dilution series of cDNA were used for SOS1 to ensure that the reaction was performed in a linear range. In B, relative mRNA levels were normalized with EF1α as a reference. Fold differences compared with the nonstressed Arabidopsis sample are shown. Error bars indicate the sd from six repeats (two biological and three analytical repeats). Expression patterns of orthologous genes around SOS1 are shown for comparison.
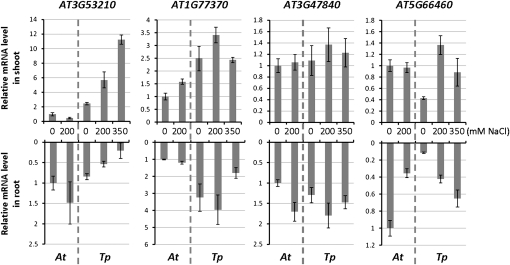
Also included in the real-time PCR analyses (Fig. 5) were three genes not located on Arabidopsis chromosome 5 but found in TP-2 as genes that disrupted the colinear order of genes: AT3G53210, AT1G77370, and AT3G47840. Analysis was extended to the AT5G66460 ortholog adjacent to AT3G47840 and induced by abscisic acid and salt stress (Zimmermann et al., 2004). The AT3G53210, AT1G77370, and AT3G47840 orthologs were found as single-copy genes in the T. parvula genome sequenced by a combination of Roche-454 and Illumina/Solexa sequencing, which is at present in the final assembly phase (M. Dassanayake and D.-H. Oh, unpublished data). As expected, quantitative real-time RT-PCR amplified single species of amplicons using T. parvula cDNA for all three genes. The most dramatic difference in expression between Arabidopsis and T. parvula was observed with AT3G53210, annotated as encoding a nodulin family protein, in which the mRNA was significantly higher in the shoots of T. parvula than in Arabidopsis, and was further induced by salt stress. AT1G77370 showed generally higher mRNA abundance in T. parvula under all conditions, while the AT5G66460 basal expression was at a lower level, in both shoot and root, than in Arabidopsis. Expression of the AT3G47840 ortholog did not show significant differences between T. parvula and Arabidopsis (Fig. 5; Supplemental Table S4).
Figure 5.
Comparison of gene expression of translocated genes in T. parvula. Shown are quantitative real-time PCR results comparing the expression of selected orthologous genes from TP-2. Normalization and error bars are as in Figure 4B.
Analyses of Sequences Upstream of SOS1
Although functions and domains of SOS1 genes have been extensively studied in a few species, little is known about transcriptional regulation and promoter evolution in this gene family. In the three species, searches for cis-elements in the region up to 2,000 bp upstream from the SOS1 translation start site (predicted for T. parvula) identified several known stress-responsive elements. These included abscisic acid-response elements, anaerobic induction elements, a MYB-binding site involved in drought inducibility, heat stress-responsive elements, low temperature-responsive elements, and defense- and generally stress- and hormone-responsive elements (Supplemental Table S5).
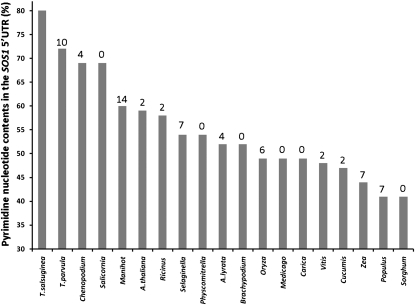
A comparison of SOS1 transcript structures of halophytes and glycophytic species revealed a surprising difference between the two groups. When analyzing the 5′ untranslated regions (UTRs) of SOS1 genes, we found differences in CT content of a stretch extending 100 bp immediately upstream of the translation start sites of all available genes in the NCBI nucleotide databases and the published plant genomes available on Phytozome (http://www.phytozome.net). The highest pyrimidine ratio was found in the two Thellungiella species, and the largest difference between two species in the list separated Salicornia, a halophyte, from Manihot, a glycophyte (Fig. 6). Within the SOS1 5′ UTR adjacent to the translation start site, there are 12 and 18 CTT repeats in T. parvula and T. salsuginea, respectively. The Arabidopsis sequence shows three CTT repeats in the homologous region. In addition, these CT-rich sequences are associated with a region that could potentially form stem-loop structures in proximity to the initiation ATG codon (Supplemental Fig. S5).
Figure 6.
Frequency of pyrimidine nucleotides in SOS1 5′ UTR sequences of diverse organisms. CT content values (%) are sorted from large to small, and the percentage difference between two consecutive columns is shown above each column. Additional sequences are Chenopodium quinoa (gi:154269387), Salicornia brachiata (gi:214028395), Populus trichocarpa gene POPTR_0010s11130, Manihot esculenta cassava39868.m1, Ricinus communis 29780.t000067, Medicago truncatula Medtr2g043140, Cucumis sativus Cucsa.026500, Arabidopsis lyrata SOS1, Oryza sativa LOC_Os12g44360, Carica papaya evm.TU.supercontig_2.63, Vitis vinifera gene GSVIVT00030673001, Sorghum bicolor Sb08g023290, Zea mays GRMZM2G098494, Brachypodium distachyon Bradi4g00290, Selaginella moellendorfii, Physcomitrella patens, and Chlamydomonas reinhardtii Au9.Cre24.g769600. One hundred nucleotides upstream of the translation start site was taken unless a shorter 5′ UTR was defined for a given species.
DISCUSSION
Abiotic stress tolerance and sensitivity are unequally distributed among plant species. Although most species are stress sensitive, there are, interestingly, also stress-tolerant species in most families. Moreover, there is significant plasticity in environmental adaptation; even many species that are typically viewed as stress sensitive have salt-tolerant ecotypes. Thus, it seems highly likely that the basic machinery for adaptation to a stressful environment is universally endowed in plant genomes. It is unclear, however, whether the potential for adaptation has failed to develop in some environments or whether it has secondarily been lost in less demanding environments or through human invention. Given the importance of fresh water for agriculture and the increasing salinization of many irrigated lands, especially in dry, subtropical areas, it is important that we find the signposts indicating salinity stress tolerance and that we know about the essential processes and genes (Cheeseman, 1988; Hasegawa et al., 2000; Zhu, 2001; Flowers, 2004; Flowers and Colmer, 2008)
In this paper, we address these goals using high-throughput sequencing. Our strategy was to employ closely related plant species that exhibit very different salt stress tolerance characteristics. Although genome sequencing by Joint Genome Initiative is reported to have been completed, the available genomic resources for T. salsuginea are still limited. We have concentrated on two BAC clones for T. salsuginea (Deng et al., 2009; Nah et al., 2009) and the orthologous regions in Arabidopsis and T. parvula. A three-way comparison revealed similarities in the synteny and gene structures among the two genome segments. Synteny, as expected, is more pronounced among the Thellungiella species. Orthologous genes showed functional conservation, as supported by the Ka/Ks values. However, there were differences in genome features and sequences between the Thellungiella species and Arabidopsis and also within the Thellungiella genus. Some of these may be expected to be related to physiological differences that separate the three species.
Differences between the Thellungiella Species and Arabidopsis
Based on the Ks values of orthologous genes, the divergence of T. parvula and T. salsuginea from Arabidopsis can be placed at approximately 12 million years ago, and the Thellungiella species separated approximately 8 million years ago. The observed genome structures, inversions, and breaks in the colinearity are consistent with the time of divergence.
An obvious difference with respect to Arabidopsis and a specific feature of the Thellungiella species identified genome sequences around the SOS1 gene, one of the well-established salt tolerance determinants (Shi et al., 2000). Irrespective of extensive synteny of the ORFs and the conservation of gene structures for SOS1 between Arabidopsis and T. parvula, colinearity falls apart starting upstream of the first exon in SOS1 (Fig. 3). A hypothesis based on the analysis of SOS1 expression in T. salsuginea (Oh et al., 2009) suggested different expression strength or, possibly, transcript stability. This is further supported in T. parvula by the analysis of SOS1 mRNA levels in comparison with Arabidopsis (Fig. 4). Coupled with the significant sequence similarity in the SOS1 5′ UTR of halophytic species (Fig. 6), a closer examination of this transcript structural feature is appropriate because it may point to a fundamental glycophyte-halophyte distinguishing character that is less or not apparent when promoter cis-elements are compared. The hypothesis of a halophytic transcript structure is particularly attractive because of the very similar pyrimidine tracts, CTn, present in the promoters of Chenopodium quinoa and Salicornia brachiata, which are both recognized as halophytes (Sanchez et al., 2003; Lieth et al., 2008). The possibility of TsSOS1 and TpSOS1 5′ UTR sequences involved in the formation of hairpin structures with free energies of −4.0 and −5.40 kcal mol−1, respectively, close to the protein initiation codon enforces the notion that this structure appears to be important (Supplemental Fig. S5). Very similar stem-loop structures also characterize the 5′ UTR of the two other halophytes for which this sequence information is available.
Analyzing the conservation of (CnTn)n repeats on the 5′ UTR sequences in all stress-related and highly expressed genes is beyond the scope of this study. However, we investigated the emerging apparent pattern in genes with high expression levels or highly induced under abiotic stress in Arabidopsis and the two Thellungiella species. The availability of publicly available 5′ UTR sequences from T. salsuginea full-length transcripts limited candidate genes to a small collection. Nevertheless, a general pattern appears to be preserved in highly expressed genes, and the prevalence of CT-rich regions is more prominent in the two Thellungiella species compared with Arabidopsis. Arabidopsis heat shock protein 70 (AT3G09440) is known to be highly expressed under abiotic stress, including salt stress, both in Arabidopsis and T. salsuginea (Taji et al., 2004; Swindell et al., 2007). There is a pyrimidine-rich region close to the ATG initiation codon in the putative homologs of Thellungiella 5′ UTR sequences (Supplemental Fig. S6). Similar observations were found for cytosolic cyclophilin ROC3 (peptidyl-prolyl cis-trans isomerase; AT2G16600) and a transcription factor B3 (AT4G21550). Cyclophilins such as ROC3 are highly abundant cytosolic proteins known to be induced under abiotic stress (Chou and Gasser, 1997; Romano et al., 2004), and in addition to broad-range stress-responsive expression, the transcription factor AT4G21550 is also induced in specific developmental stages (Suzuki et al., 2007). All nine 5′ UTR sequences can form stem-loop structures close to the initiation codon.
Formation of this hairpin structure could affect transcription efficiency or the stability of the transcript, although this topic, while analyzed in the past in bacterial and viral systems (Simoes and Sarnow, 1991; Emory et al., 1992; Hellendoorn et al., 1997), has not received much attention in plants (Klaff et al., 1996). Pyrimidine-rich elements in the 5′ UTR of plants are known to have positive effects on transcription (Bolle et al., 1994), and they appear to be functionally conserved in diverse gene families across the plant kingdom. A similar pyrimidine-rich element in the 5′ UTR has been shown to confer high transcription levels without the need for other upstream cis-elements except for a TATA box in tomato (Solanum lycopersicum) hydroxy-3-methylglutaryl CoA reductase genes (Daraselia et al., 1996). Also, actin genes from Arabidopsis to Physcomitrella include 5′ UTR pyrimidine-rich stretches, which have been associated with very high levels of transcription (An et al., 1996; An and Meagher, 2010).
Finally, the possibility exists that the CTn sequence domains in halophyte species upstream of the ATG codon might be a target of DNA chromatin modifications that could exert additional expression control. Hypermethylation in Arabidopsis SUPERMAN and AGAMOUS genes is thought to be influenced through DNA secondary structures formed by pyrimidine-rich sequences (Jacobsen et al., 2000).
Differences between T. parvula and T. salsuginea
At approximately 180 Mb, the genome of T. parvula is significantly smaller than that of T. salsuginea, which is estimated as at least 260 Mb (Inan et al., 2004; Nah et al., 2009). The estimated genome based on a preliminary assembly permits us to assume approximately 163 Mb for T. parvula (Table I). Among the largest contributors to angiosperm genome size variations are repetitive DNAs and transposable elements (Ma et al., 2004; Meagher and Vassiliadis, 2005; Gaut and Ross-Ibarra, 2008). The comparison of orthologous genomic regions in this study between the two Thellungiella species showed T. salsuginea to be of consistently larger size, with the difference relating to the insertion of various transposable elements (Fig. 2). In contrast, the T. parvula genomic regions analyzed here showed a more compact gene organization than that seen in Arabidopsis, largely due to the absence of transposable elements and small repeat regions. If the prediction of a more compact structure is maintained across the entire genome, it is possible that T. parvula contains a higher total number of genes as compared with Arabidopsis. The lack of repetitive sequences extended to the bigger genomic region (scaffold 00254), with an overall repeat content as low as 1.9%. However, the T. parvula sequence also revealed regions rich in repetitive sequences; for example, the last 50-kb region of scaffold 00254 contained more than 15% repetitive sequences (Supplemental Fig. S2A), possibly indicating that this region was adjacent to a centromeric region in the corresponding T. parvula chromosome.
The genome size and complexity differences of the Thellungiella species can potentially provide explanations for their different growth habits and physiological trait differences. Differences include rosette formation in T. salsuginea but not in T. parvula and differences in stomatal conductance, stomata density, and water loss (Orsini et al., 2010). Differences also include growth parameters, with T. parvula as a fast-growing species that is adapted to high light that shows etiolation under Arabidopsis-type light conditions. In contrast, T. salsuginea shows a much slower growth rate, possibly a reflection of its adaptation to typically resource-poor habitats (Inan et al., 2004; Gong et al., 2005; Oh et al., 2009).
Toward the T. parvula Genome Sequence
T. salsuginea (salt cress) has been studied as a model halophyte, juxtaposing its physiology and genetic structure to that of Arabidopsis. Suggested mechanisms for tolerance to stress and nutrient-limiting conditions include control over stomatal conductance (Inan et al., 2004), greater discriminatory power of K+- and Na+-transport systems (Volkov et al., 2004; Volkov and Amtmann, 2006), and nitrogen utilization efficiency (Kant et al., 2008). However, a genetic basis of the differences in tolerance could only be deduced by attempts at establishing Arabidopsis-like resources and databases (Inan et al., 2004; Taji et al., 2004, 2008; Wang et al., 2004, 2006; Gong et al., 2005; Wong et al., 2005; Zhang et al., 2008; Oh et al., 2009). Merging the forthcoming T. parvula genome sequence in a three-way comparison that includes Arabidopsis and T. salsuginea (pending; http://www.jgi.doe.gov/sequencing/why/50029.html) will provide a powerful way to probe for the essence of halophytism, in particular if other crucifer species and ecotypes of well-studied species are included (Weigel and Mott, 2009; Orsini et al., 2010).
Halophytism is an intriguing ecophysiological adaptation to saline environments. However, defining its essence in molecular or genetic terms has been elusive, due in a large part to the observation that many different strategies have evolved by which species became tolerant or resistant to the stress. What appears to emerge is the existence of a small number of novel genes (Vinocur and Altman, 2005), while novel uses of kingdom-wide existing genes and proteins seem to provide the bulk of the mechanisms that form the basis of the stress-tolerance phenotype. Increasingly, halophyte transcriptomes are being analyzed and contrasted to glycophytic relatives (Kore-eda et al., 2004; Gong et al., 2005; Zouari et al., 2007; Dassanayake et al., 2009; Guo et al., 2009; Chelaifa et al., 2010). With the advent of numerous ongoing whole genome sequencing projects and comparisons throughout the plant kingdom, a multifaceted approach becomes possible. The result will be the integration of results on genome organization, gene and transcript structures, noncoding regulatory RNAs, regulatory mechanisms, biochemical complexity, and hormone- or metabolite-based signaling networks. From integration will emerge understanding of the processes that enabled individual species in all plant orders to use common genes in the evolution of a halophytic way of life.
MATERIALS AND METHODS
Plant Material
The ecotype of Thellungiella parvula used here was collected around a saline lake in the Tuz Golu region occupying a depression on the central plateau of Turkey at an elevation of 905 m above sea level. For most of the year, the shallow (1–2 m) saline lake covers an area of approximately 1,500 km2. Water density is 1.225 g cm−3 (32.4% salt). The lake has no outlet; only a few surface streams feed into it, which may run dry in summer. During the summer months (with temperatures of greater than 40°C), the lake can evaporate completely. Precipitation in the area averages 250 mm per year. An inbred line was used for the genome sequencing and analyses.
Plant Growth and Genome Size
Genomic DNA was extracted from 10-d-old seedlings of T. parvula using the Genomic DNA Preparation Kit (Illumina). For gene expression analyses, total RNAs were isolated from 10-d-old seedlings of T. parvula and Arabidopsis (Arabidopsis thaliana) at the stage when the first true leaves emerge. Salt stress treatment and sample harvest were done as described by Oh et al. (2009). Genome size was determined by flow cytometry using a Becton-Dickinson LSR II (BD Biosciences). Homogenates of Arabidopsis and T. parvula were prepared by chopping leaves, and nuclei were stained with 40 μg mL−1 propidium iodide (Galbraith et al., 1983). Samples were analyzed at an event rate of 200 nuclei s−1. The configuration for propidium iodide detection comprised 488-nm laser excitation and a 630/22-nm band-pass filter. Chromosome numbers were determined in root tips after staining of mitotic cells (M. Dassanayake and D.-H. Oh, unpublished data).
DNA Libraries, DNA Sequencing, and Sequence Assembly
T. parvula DNA was isolated and prepared for Roche-454 GS FLX Titanium series long-read and paired-end sequencing and initial flowgram data processing following standard protocols (Roche-454 Life Sciences). Roche-454 sequences from eight single-ended genomic shotgun libraries and the respective titration plates were first assembled using Newbler (version 2.0.01.14) with a 40-bp minimum overlap and 90% identity. Sequences from one 2.5-kb, three 3-kb, two 8-kb, and one 20-kb paired-end libraries were added to the existing contigs to build scaffolds. A total of 24.89 million reads with a total of 5.9 Gb and an average read length of 360 bp were used for the de novo genome assembly with an inferred read error of 1.25%. A total of 99.2% of the bases used had a quality score of Q40 or greater. The two T. parvula genomic regions corresponding to the two BAC clones of Thellungiella salsuginea were selected using BLASTn. The sequence gaps in these regions were filled by PCR with specific primers targeted to flanking regions of the gaps in the assembly. Sanger sequencing of genomic PCR products was used to fill sequencing gaps and to confirm the sequences adjacent to the gaps.
Sequence Analysis and Annotation
The ORFs in genomic sequences were predicted ab initio with FGENESH (http://www.softberry.com) adopting Arabidopsis parameters. The predicted ORFs were annotated using the NCBI nonredundant databases and the TAIR9 database (http://www.arabidopsis.org/) with BLASTN and BLASTP programs. When analyzing TP-1 and TP-2, the ORF predictions were refined using FGENESH+ with Arabidopsis orthologous proteins as references. Repeat elements were identified by RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) and the program Tandem Repeats Finder version 4.04 (Benson, 1999). The homologous sequence comparisons were made with PipMaker (Schwartz et al., 2000) and dot-plot analysis (Sonnhammer and Durbin, 1995).
Quantitative RT-PCR
Primers were designed to have identical sequences and amplifying amplicons of identical size from T. parvula and Arabidopsis cDNAs (Supplemental Table S6). Quantitative real-time RT-PCR was performed as described previously (Poroyko et al., 2007; Oh et al., 2010). For the reference genes, orthologs of EF1α (AT5G60390) and cytochrome c (AT5G40810) from the T. parvula transcriptome (M. Dassanayake and D.-H. Oh, unpublished data) were used and gave similar results. Represented in Figures 4 and 5 are results normalized to EF1α from six repeats (two biological and three analytical repeats).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HM222924 and HM222925.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of relative DNA contents by flow cytometry.
Supplemental Figure S2. Analysis of scaffold 00254.
Supplemental Figure S3. Comparison between T. parvula genomic sequences TP-1 and TP-2 with three orthologous genomic regions in Arabidopsis and T. salsuginea.
Supplemental Figure S4. Syntenic relationships and homologies of sequences represented by percent identity plots.
Supplemental Figure S5. Hairpin structures in the 5′ UTR of the halophytic Thellungiella species.
Supplemental Figure S6. Comparison of CT tracts in the 5′ UTRs of various genes among the three species.
Supplemental Table S1. ORFs identified in TP-1, AT-1, and TS-1.
Supplemental Table S2. ORFs identified in TP-2, AT-2, and TS-2.
Supplemental Table S3. Comparison of Ka/Ks ratios in orthologous genes and the estimated divergence time of three species.
Supplemental Table S4. Comparison of relative gene expression levels by quantitative PCR.
Supplemental Table S5. Known cis-elements in SOS1 promoter regions up to 2,000 bp upstream of the translation start site.
Supplemental Table S6. List of PCR primers.
Supplementary Material
Acknowledgments
We thank Keith Frazier and Jyothi Thimmapuram (Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign) for advice and generous help.
References
- Al-Shehbaz IA, O’Kane SL., Jr (1995) Placement of Arabidopsis parvula in Thellungiella (Brassicaceae). Novon 5: 309–310 [Google Scholar]
- Amtmann A. (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Bohnert HJ, Bressan RA. (2005) Abiotic stress and plant genome evolution: search for new models. Plant Physiol 138: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y, Meagher R. (2010) Strong expression and conserved regulation of ACT2 in Arabidopsis thaliana and Physcomitrella patens. Plant Mol Biol Rep 28: 481–490 [Google Scholar]
- An YQ, Huang S, McDowell JM, McKinney EC, Meagher RB. (1996) Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell 8: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Sopory S, Lübberstedt T, Herrmann RG, Oelmüller R. (1994) Segments encoding 5′-untranslated leaders of genes for thylakoid proteins contain cis-elements essential for transcription. Plant J 6: 513–523 [DOI] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK. (2001) Learning from the Arabidopsis experience: the next gene search paradigm. Plant Physiol 127: 1354–1360 [PMC free article] [PubMed] [Google Scholar]
- Cheeseman JM. (1988) Mechanisms of salinity tolerance in plants. Plant Physiol 87: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H, Mahé F, Ainouche M. (2010) Transcriptome divergence between the hexaploid salt-marsh sister species Spartina maritima and Spartina alterniflora (Poaceae). Mol Ecol 19: 2050–2063 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirschi KD. (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279: 2922–2926 [DOI] [PubMed] [Google Scholar]
- Chou IT, Gasser CS. (1997) Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol 35: 873–892 [DOI] [PubMed] [Google Scholar]
- Daraselia ND, Tarchevskaya S, Narita JO. (1996) The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol 112: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Haas JS, Bohnert HJ, Cheeseman JM. (2009) Shedding light on an extremophile lifestyle through transcriptomics. New Phytol 183: 764–775 [DOI] [PubMed] [Google Scholar]
- Deng Z, Li Y, Xia R, Wang W, Huang X, Zhang L, Zhang S, Yang C, Zhang Y, Chen M, et al. (2009) Structural analysis of 83-kb genomic DNA from Thellungiella halophila: sequence features and microcolinearity between salt cress and Arabidopsis thaliana. Genomics 94: 324–332 [DOI] [PubMed] [Google Scholar]
- Emory SA, Bouvet P, Belasco JG. (1992) A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev 6: 135–148 [DOI] [PubMed] [Google Scholar]
- Flowers TJ. (2004) Improving crop salt tolerance. J Exp Bot 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. (2008) Salinity tolerance in halophytes. New Phytol 179: 945–963 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Gaut BS, Ross-Ibarra J. (2008) Selection on major components of angiosperm genomes. Science 320: 484–486 [DOI] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Griffith M, Timonin M, Wong AC, Gray GR, Akhter SR, Saldanha M, Rogers MA, Weretilnyk EA, Moffatt B. (2007) Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ 30: 529–538 [DOI] [PubMed] [Google Scholar]
- Guo YQ, Tian ZY, Qin GY, Yan DL, Zhang J, Zhou WZ, Qin P. (2009) Gene expression of halophyte Kosteletzkya virginica seedlings under salt stress at early stage. Genetica 137: 189–199 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK. (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hellendoorn K, Verlaan PW, Pleij CW. (1997) A functional role for the conserved protonatable hairpins in the 5′ untranslated region of turnip yellow mosaic virus RNA. J Virol 71: 8774–8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al. (2004) Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM. (2000) Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol 10: 179–186 [DOI] [PubMed] [Google Scholar]
- Kant S, Bi YM, Weretilnyk E, Barak S, Rothstein SJ. (2008) The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol 147: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S. (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Klaff P, Riesner D, Steger G. (1996) RNA structure and the regulation of gene expression. Plant Mol Biol 32: 89–106 [DOI] [PubMed] [Google Scholar]
- Kore-eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman JC. (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene 341: 83–92 [DOI] [PubMed] [Google Scholar]
- Lieth H, Sucre MG, Herzog B. (2008) Mangroves and Halophytes. Springer, New York [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. (2009) Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher TR, Vassiliadis C. (2005) Phenotypic impacts of repetitive DNA in flowering plants. New Phytol 168: 71–80 [DOI] [PubMed] [Google Scholar]
- Nah G, Pagliarulo CL, Mohr PG, Luo M, Sisneros N, Yu Y, Collura K, Currie J, Goicoechea JL, Wing RA, et al. (2009) Comparative sequence analysis of the SALT OVERLY SENSITIVE1 orthologous region in Thellungiella halophila and Arabidopsis thaliana. Genomics 94: 196–203 [DOI] [PubMed] [Google Scholar]
- Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ. (2010) Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot 61: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane SL, Jr, Al-Shehbaz IA. (2003) Phylogenetic position and generic limits of Arabidopsis (Brassicaceae) based on sequences of nuclear ribosomal DNA. Ann Mo Bot Gard 90: 603–612 [Google Scholar]
- Orsini F, D’Urzo MP, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poroyko V, Spollen WG, Hejlek LG, Hernandez AG, LeNoble ME, Davis G, Nguyen HT, Springer GK, Sharp RE, Bohnert HJ. (2007) Comparing regional transcript profiles from maize primary roots under well-watered and low water potential conditions. J Exp Bot 58: 279–289 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PG, Horton P, Gray JE. (2004) The Arabidopsis cyclophilin gene family. Plant Physiol 134: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez HB, Lemeur R, Van Damme P, Jacobsen SE. (2003) Ecophysiological analysis of drought and salinity stress of quinoa (Chenopodium quinoa Willd.). Food Rev Int 19: 111–119 [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. (2000) PipMaker: a Web server for aligning two genomic DNA sequences. Genome Res 10: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes EA, Sarnow P. (1991) An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol 65: 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR. (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125.1–125.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, Totoki Y, Toyoda A, Sakaki Y, Seki M, Ono H, et al. (2008) Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol 8: 115.1–115.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinocur B, Altman A. (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16: 123–132 [DOI] [PubMed] [Google Scholar]
- Volkov V, Amtmann A. (2006) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J 48: 342–353 [DOI] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A. (2004) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ 27: 1–14 [Google Scholar]
- Wang B, Davenport RJ, Volkov V, Amtmann A. (2006) Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J Exp Bot 57: 1161–1170 [DOI] [PubMed] [Google Scholar]
- Wang Z, Weber JL, Zhong G, Tanksley SD. (1994) Survey of plant short tandem DNA repeats. Theor Appl Genet 88: 1–6 [DOI] [PubMed] [Google Scholar]
- Wang ZI, Li P, Fredricksen M, Gong ZH, Kim CS, Zhang CQ, Bohnert HJ, Zhu J-K, Bressan RA, Hasegawa PM, Zhao YX, Zhang H. (2004) Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci 166: 609–616 [Google Scholar]
- Weigel D, Mott R. (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10: 107.1–107.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Díaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M. (2005) Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol 58: 561–574 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ 25: 131–139 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lai J, Sun S, Li Y, Liu Y, Liang L, Chen M, Xie Q. (2008) Comparison analysis of transcripts from the halophyte Thellungiella halophila. J Integr Plant Biol 50: 1327–1335 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouari N, Ben Saad R, Legavre T, Azaza J, Sabau X, Jaoua M, Masmoudi K, Hassairi A. (2007) Identification and sequencing of ESTs from the halophyte grass Aeluropus littoralis. Gene 404: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.