Abstract
Membrane compartmentalization and trafficking within and between cells is considered an essential cellular property of higher eukaryotes. We established a high-throughput imaging method suitable for the quantitative detection of membrane compartments at subcellular resolution in intact epidermal tissue. Whole Arabidopsis (Arabidopsis thaliana) cotyledon leaves were subjected to quantitative confocal laser microscopy using automated image acquisition, computational pattern recognition, and quantification of membrane compartments. This revealed that our method is sensitive and reliable to detect distinct endomembrane compartments. We applied quantitative confocal laser microscopy to a transgenic line expressing GFP-2xFYVE as a marker for endosomal compartments during biotic or abiotic stresses, and detected markedly quantitative adaptations in response to changing environments. Using a transgenic line expressing the plasma membrane-resident syntaxin GFP-PEN1, we quantified the pathogen-inducible extracellular accumulation of this fusion protein at fungal entry sites. Our protocol provides a platform to study the quantitative and dynamic changes of endomembrane trafficking, and potential adaptations of this machinery to physiological stress.
A number of recent technological developments in the life sciences allow the establishment of quantitative profiles of biomolecule classes such as gene transcripts, small RNAs, proteins, and metabolites from single cells, tissues, or whole organs. These high-throughput and data-rich technologies are particularly informative when applied in defined genetic contexts. However, most profiling data provide little or no spatial information of the respective molecules. Confocal laser microscopy and the use of fluorescently tagged molecules can overcome this limitation at cellular and subcellular resolution, and reveals their dynamics in vivo. Nipkow disc (spinning disc) imaging technology (Nakano, 2002) in combination with advanced image processing is in principle available to retrieve spatial information of biomolecules at high throughput (Pepperkok and Ellenberg, 2006; Rausch, 2006; Collinet et al., 2010). Few studies have explored the potential of high-throughput imaging at subcellular resolution in a whole organism/tissue context (Crane et al., 2009; Negishi et al., 2009; Wolinski et al., 2009). This is mainly due to difficulties associated with the handling of living organs and/or tissue and the need to develop quantitative image processing tools.
In plants, Arabidopsis (Arabidopsis thaliana) is a model organism for cell biological studies. An increasing number of cell types and cellular structures, including organelles, as well as a variety of membrane compartments can be specifically visualized in living tissue by the use of fluorescence markers. Among the latter are the plasma membrane and a variety of trafficking vesicles such as endosomes, exosomes, and multivesicular bodies (MVBs), where endosomes are sorted and redistributed to their target membranes (Robinson et al., 2008). Vesicle distribution and trafficking pathways are subject to dynamic changes during developmental cell differentiation and in response to environmental signals. For instance, polar gradients of the plant growth hormone auxin are generated by endocytosis and recycling of PINFORMED (PIN) auxin transporters according to environmental or developmental triggers (Friml et al., 2002; Geldner et al., 2003; Dhonukshe et al., 2007). Other plant growth responses depend on the function of the brassinosteroid receptor BRI1, which undergoes constitutive endocytosis (Russinova et al., 2004).
Dynamic processes at the plasma membrane are important for immune reactions upon attack by pathogenic microorganisms. For example, perception of bacterial infection in mammals and plants triggers endosomal translocation of plasma membrane resident Toll-like receptor 4 and the flagellin receptor FLS2, respectively (Husebye et al., 2006; Robatzek et al., 2006), thereby initiating a first line of defense responses. In interactions with fungal pathogens, the Arabidopsis plasma membrane-resident syntaxin PEN1 and vesicle-localized interacting proteins VAMP721/722 become concentrated at incipient fungal entry sites (Collins et al., 2003; Assaad et al., 2004; Bhat et al., 2005; Kwon et al., 2008b), likely reflecting targeted exocytosis and extracellular exosome release at the host cell periphery as part of defense execution pathways (Kwon et al., 2008a; Pajonk et al., 2008; Meyer et al., 2009).
Few numerical data of organelles and endomembrane compartments are available in plants (Pyke and Leech, 1991; Logan et al., 2003; Scott et al., 2006; Ketelaar et al., 2008). It appears that chloroplast numbers exceed mitochondria, and the mean surface of vesicles destined for exocytosis is larger than that of endocytic vesicles. However, such data are difficult to compare since they were obtained from diverse cell types. Moreover, different developmental and environmental cues might influence the number of subcellular structures, as observed for peroxisomes (Hu et al., 2002). In addition, there are almost no quantitative data available describing spatiotemporal changes of membrane compartments in tissue and cell type-specific contexts during development and in response to environmental cues.
Although numerous membrane trafficking components have been identified in plants, the dynamics of the underlying processes in intact cells, the coordination of cellular subfunctions between different endomembrane compartments, as well as their tissue- and cell type-specific differentiation are still poorly understood. In addition, plants have evolved unique protein subfamilies associated with vesicle trafficking, which might reflect plant cell-specific properties such as the absence of an endoplasmic reticulum-Golgi intermediate compartment and the combined activities of the trans-Golgi network and early endosomes (Hwang and Robinson, 2009).
Here we describe a method using quantitative confocal laser microscopy (QCLM) to reveal stress adjustment of membrane trafficking in living Arabidopsis whole-leaf tissue. We developed endomembrane and plasma membrane microdomain scripts on the basis of an image framework software, which are broadly applicable to trace different membrane compartments. We observed quantitative differences between distinct endomembrane vesicles in leaf epidermal tissue and differences in the quantity of one tested endosomal compartment upon exposure of Arabidopsis plants to biotic or abiotic stresses. We validated the robustness of QCLM by quantifying pathogen-inducible extracellular compartments.
RESULTS
Quantitative High-Throughput Confocal Imaging
To quantify membrane compartments within a cell or tissue area, a set of transgenic Arabidopsis lines expressing fluorescently labeled proteins was subjected to semiautomated confocal imaging. We employed a high-throughput spinning-disc microscope to measure samples in multiwell plates (Opera microscope, “Materials and Methods”). This device was initially developed to analyze monolayer cell cultures derived from animals or humans. Since suspension-cultured plant cells of well-defined cell types are essentially unavailable, and membrane trafficking should be viewed dependent on tissue context, we established whole-tissue imaging in 96-well plates of detached Arabidopsis cotyledon leaves. In contrast to monolayer cell cultures (Collinet et al., 2010), automated imaging of Arabidopsis leaves proved to be challenging due to their natural curvature. To capture images of sectors of the leaf epidermis that were entirely within a focal plane, a consecutive series of optical planes in the z direction were taken (Supplemental Fig. S1; z stack consisted of 21 images for the endomembrane script and 31 images for the plasma membrane microdomain script; step size 1 μm). Individual images were then used to produce a merged pseudo image of the epidermal cell layer, which was further automatically processed by computational tools (Acapella based; “Materials and Methods”). We developed new object-specific algorithms for cell detection along the plasma membrane (“Materials and Methods”; Supplemental Figs. S5 and S6) and adapted spot detection within the existing software to detect endomembrane and plasma membrane microdomain compartments.
An increasing number of fluorescently tagged proteins labeling different endomembrane compartments are available and frequently used in cell biological studies (Marion et al., 2008; Robinson et al., 2008; Geldner et al., 2009). The peptide domain FYVE specifically binds to phosphoinositol 3-phosphate and is enriched in endosomal membranes and MVBs (Gaullier et al., 1998; Voigt et al., 2005). We developed an endomembrane script to detect and quantify endosomal compartments marked by the green fluorescent fusion protein GFP-2xFYVE in leaf epidermal cells of transgenic Arabidopsis lines. Images were taken at 40× magnification, enabling resolution of subcellular structures including endosomes and were then subjected to our customized computational analysis (“Materials and Methods” and Supplemental Data S1). For robust quantification, we focused on endosomal compartments inside cells that were fully separated from cell boundaries. Manual reinspection of the images showed that GFP-2xFYVE compartments were detected with high accuracy (Fig. 1, A, C, and F; 87% of manually detected GFP-2xFYVE-marked endomembrane objects were independently identified by the pattern recognition software, and vice versa, 98.5% of GFP-2xFYVE-marked endomembrane objects detected by the automated object recognition were also found by manual inspection). The quantification of GFP-2xFYVE compartments created large amounts of numerical data, which were displayed in graphic sheets for easy access of the scores (“Materials and Methods”; Supplemental Fig. S2; Supplemental Data S3).
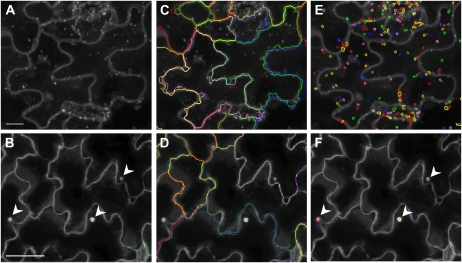
Figure 1.
Pattern recognition of membrane compartments in leaf epidermal tissue at cellular and subcellular resolution. Merged confocal laser microscopic images show Arabidopsis leaf epidermal cells. Top section: GFP-2xFYVE plants were imaged at 40× magnification (scale bar = 20 μm) and images analyzed by the endomembrane script. Bottom section: GFP-PEN1 plants were imaged at 20× magnification (scale bar = 50 μm) and images analyzed by the plasma membrane microdomain script. A and B, Merged pseudo images. C and D, Recognition of epidermal cells is shown by colored lines. E, Recognition of GFP-2xFYVE-labeled endosomal compartments is shown by colored circles. F, Recognition of B. graminis induced GFP-PEN1 accumulation beneath attempted fungal entry sites (indicated by arrowheads) is shown by colored circles. Color coding is random, different colors indicate individual cells and compartments.
To study and quantify pathogen-induced dynamic changes at the plasma membrane of leaf epidermal cells, we monitored fluorescently tagged syntaxin, GFP-PEN1 expressed in transgenic Arabidopsis plants (Collins et al., 2003). This plasma membrane-resident SNARE domain-containing protein becomes concentrated at incipient fungal entry sites at the periphery of epidermal cells, which is thought to reflect targeted exocytosis as well as exosome formation at pathogen contact sites (Assaad et al., 2004; Kwon et al., 2008b; Pajonk et al., 2008; Meyer et al., 2009). We established a customized script for the recognition of fungus-induced GFP-PEN1 microdomains (designated plasma membrane microdomain script). Leaves were infected with conidiospores of the powdery mildew fungus Blumeria graminis f. sp. hordei (B. graminis) and were imaged at 20× magnification. Automated detection of GFP-PEN1 microdomains was robust because 92% of manually detected accumulation sites were independently identified by the pattern recognition software, and vice versa, 87% of accumulation sites detected by the automated object recognition were also found by manual inspection (“Materials and Methods”; Fig. 1, B, D, and F; Supplemental Fig. S2; Supplemental Data S2). The endomembrane and plasma membrane microdomain scripts recorded a total of 21 and 16 output parameters, respectively (Supplemental Tables S1 and S2). These included the area and fluorescence intensity of the detected compartments.
Quantification of Endomembrane Compartments
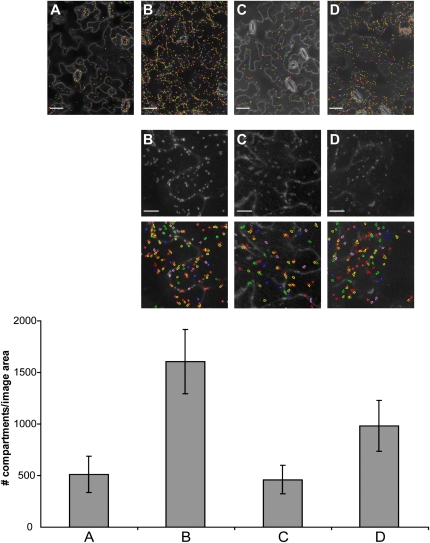
Application of the endomembrane script to quantify GFP-2xFYVE endosomal compartments in leaf epidermal cells revealed average values of 510 ± 177 (n = 70) compartments/image area, and 14 ± 9 (n = 50) compartments/cell (average number of cells/image area: 27 ± 4 [n = 10]). We tested whether the endomembrane script was more broadly applicable to recognize other fluorescently tagged subcellular compartments. Toward this end, we selected transgenic Arabidopsis lines expressing yellow fluorescence protein (YFP)-tagged proteins that reside in different secretory and endocytic compartments, including the early endosome/trans-Golgi network (YFP-VTI12), the post-Golgi/endosome (YFP-Rab C1), and the late endosome/prevacuolar compartment (YFP-RabF2b/ARA7; Sanderfoot et al., 2000; Rutherford and Moore, 2002; Geldner et al., 2009). All fusion constructs are under the control of strong constitutively active promoters (see “Materials and Methods”). Merged pseudo images were produced for all subcellular marker lines (Fig. 2). Although plasma membranes were almost unlabeled in lines expressing YFP-RabF2b/ARA7 and YFP-VTI12, and despite little differences in size and signal contrast compared to GFP-2xFYVE endosomes, YFP fluorescent compartments were correctly recognized and quantified (Fig. 2). We detected marked differences in the quantity of these endomembrane compartments, ranging from 1,606 ± 310 (n = 19) compartments/image area for YFP-VTI12, 461 ± 139 (n = 73) for YFP-Rab C1, and 984 ± 248 (n = 23) for YFP-RabF2b/ARA7 (Fig. 2). This demonstrates that our method can distinguish different endosomal compartments, and may indicate that these occur at different numbers in cotyledon epidermal cells.
Figure 2.
Detection and quantification of different endomembrane compartments in leaf epidermal tissue. Top section: Merged confocal microscopy images were taken from leaf epidermal cells of Arabidopsis lines expressing the indicated subcellular markers. Images were analyzed with the endomembrane script; the recognition of endomembrane compartments is shown by colored circles (scale bar = 50 μm). Middle section: High-magnification sections of confocal images (scale bar = 20 μm) and, below each image, corresponding endomembrane script-based recognition of endomembrane compartments (colored circles). Bottom section: Quantification of endomembrane compartments. Bars represent average numbers of endomembrane compartments ± sd (n = 19–70) per image area. A, GFP-2xFYVE. B, YFP-VTI12. C, YFP-Rab C1. D, YFP-RabF2b/ARA7. Similar results were obtained in three independent experiments.
Quantitative Differences of Endosomal Compartments in Response to Stress and in Different Genetic Backgrounds
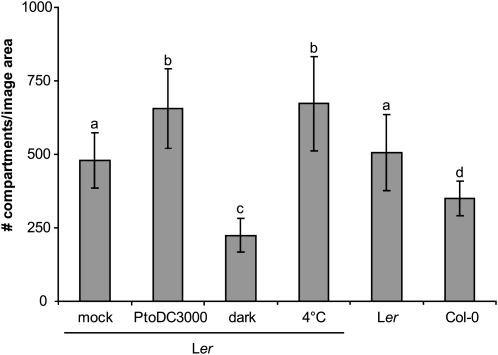
It is not known whether plant cells can adjust the number of individual endomembrane compartments in response to stress cues. We therefore applied our endomembrane script to quantify GFP-2xFYVE compartments upon exposure of the plants to pathogen challenge, or prolonged darkness (72 h), or cold stress (4°C for 42 h). Spray inoculation with virulent Pseudomonas syringae pv tomato DC3000 (PtoDC3000) bacteria resulted within 24 h in an increase of GFP-2xFYVE compartments: While mock-treated plants displayed 479 ± 94 (n = 61) compartments/image area, which is comparable to untreated plants, 656 ± 135 (n = 65) compartments/image area were recorded in infected plants (Fig. 3). Similarly, elevated amounts of 672 ± 161 (n = 59) GFP-2xFYVE compartments/image area were detected upon cold treatment (Fig. 3). By contrast, epidermal cells of dark-incubated plants had markedly decreased the number of these compartments to 224 ± 57 (n = 60). These data demonstrate that cotyledon epidermal cells have the capacity to differentially adjust GFP-2xFYVE compartment numbers to individual environmental stresses. The experiments described above were obtained from transgenic lines expressing GFP-2xFYVE in Arabidopsis accession Landsberg erecta (Ler). Next we subjected a transgenic line expressing YFP-2xFYVE in Arabidopsis accession Columbia-0 (Col-0) for comparative quantitative analysis. A total of 324 ± 91 (n = 76) compartments/image area were counted in the Col-0 background, which is lower than in the Ler background (Fig. 3). Our method thus allows recording of quantitative differences depending on genetic backgrounds (ecotype/transgenic line).
Figure 3.
Quantification of GFP-2xFYVE endosomal compartments in response to stress treatments. Confocal images of leaf epidermal cells were obtained from transgenic lines expressing fluorescently labeled FYVE compartments, recognized, and quantified by the endomembrane script. GFP-2xFYVE plants were untreated (Ler), treated 24 h before imaging with 10 mm MgCl2 (mock), or 0.5 × 108 colony forming units/mL virulent bacteria (PtoDC3000 spray inoculation), incubated for 72 h in the dark, or exposed to 42 h of cold. YFP-2xFYVE (Col-0) plants were included for comparison. Bars represent average numbers of FYVE-labeled compartments ± sd (n = 50–70) per image area. Similar results were obtained in three independent experiments. Letters indicate significant differences P < 0.001.
Quantification of Pathogen-Induced Dynamics at the Plasma Membrane
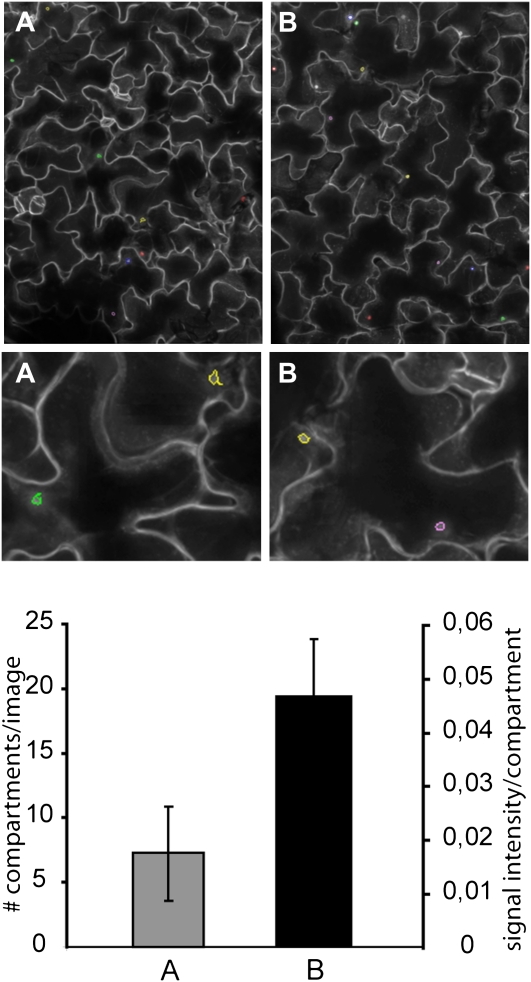
In an independent approach we applied QCLM by using the plasma membrane microdomain script. Plasma membrane-resident PEN1 accumulates within 10 to 15 h as concentric ring underneath incipient B. graminis entry sites (Assaad et al., 2004; Kwon et al., 2008b; Pajonk et al., 2008; Meyer et al., 2009). A transgenic line expressing functional GFP-PEN1 in the pen1-1 background in ecotype Col-0 (Collins et al., 2003) was inoculated with B. graminis conidiospores (approximately 10 conidiospores/image area) and after 24 h detached cotyledons were subjected to QCLM (Fig. 1F; Supplemental Fig. S2, C and D). Leaf epidermal cells displayed an average number of 111 ± 38 (n = 300) GFP-PEN1 accumulation sites/image area (Fig. 4). Additional data recorded were the area of the accumulation sites (45.67 ± 2.45; n = 300), the GFP signal intensity (0.046 ± 0.010; n = 300), and the number of encased fungal haustoria (5.48 ± 3.62; n = 300), a postinvasive fungal feeding structure (O’Connell and Panstruga, 2006). This illustrates that QCLM is not only suited to record mobile endomembrane compartments but can also be applied to quantify local dynamic changes at the plasma membrane.
Figure 4.
Quantification of pathogen-induced GFP-PEN1 accumulation at the cell periphery. Merged confocal images from the cotyledon leaf epidermis of the GFP-PEN1 line (A and B) 24 h post inoculation with B. graminis conidiospores. GFP-PEN1 accumulation at attempted fungal entry sites were recognized and quantified by the plasma membrane microdomain script. Top section: entire image area (scale bar = 200 μm). Middle section: sector of image area (scale bar = 50 μm). Bottom section: quantification and fluorescence signal intensity of GFP-PEN1 compartments, respectively. Bars represent average ± sd. A, GFP-PEN1 line: 7.27 ± 3.58 GFP-PEN1 compartments/image area (n = 16 areas). B, GFP-PEN1 line: 0.046 ± 0.010 signal intensity of GFP-PEN1 accumulation (n = 300 plants).
DISCUSSION
In an effort to better understand membrane trafficking in physiological contexts and intact live tissues, we developed a procedure for high-throughput confocal microscopy that enables the quantification of constitutive or pathogen-induced subcellular compartments. This allowed us to monitor mobile subclasses of the plant endomembrane system and an immobile pathogen-inducible compartment. Semiautomated sample handling in multiwell plates minimized normally laborious microscopy time. Spinning disc scanning enables capture of single images within 40 ms, and thus within 1 s, images of 10 z sections of 1 μm distance were acquired, which provided computational assembly of pseudo images (endomembrane script). This was critical to overcome inherent out-of-focal-plane blurring of axial images caused by the natural curvature of leaf organs and should eliminate the possibility that even endosomes moving at a rate of 1 μm/s are counted multiple times. Five to eight leaf areas per sample were imaged to cope with natural cell size and cell-type variation within epidermal tissue, and this produced statistically robust numerical data. A subsequent customized computational pipeline allowed the rapid analysis of thousands of leaf images. Inspection of a 96-well plate required only 30 to 80 min, which demonstrates the suitability of QCLM for high-throughput applications.
We developed algorithms to detect and quantify objects in microscopy images on the basis of an image framework software (Acapella based). Both scripts permitted pattern recognition of fluorescently labeled plasma membranes and round-shaped membrane compartments. Multiple parameters of the detected structures were recorded in parallel, which included their abundance, size, and fluorescence signal intensity (Supplemental Tables S1 and S2). These cellular traits are essentially impossible to score by eye using conventional confocal microscopy, and emphasize the benefit of automated image processing. It is however possible to analyze images obtained by conventional confocal microscopy using our scripts (Supplemental Fig. S3). Our current endomembrane and plasma membrane microdomain scripts do not separately calculate output parameters for different cell types, such cell type-specific calculations can be implemented in future versions of our scripts.
The usefulness of quantitative image analysis based on pattern recognition was described for budding yeast (Saccharomyces cerevisiae; Negishi et al., 2009; Wolinski et al., 2009), immortal human cell lines (Pelkmans et al., 2005; Moffat et al., 2006; Collinet et al., 2010), and zebrafish (Danio rerio) embryos (Gehrig et al., 2009). These methods are suitable for imaging single cells or whole organisms at subcellular or cellular resolution. Recently, a microfluidic system was developed for subcellular image analysis of live nematodes (Chung et al., 2008). Our QCLM procedure uses instead multiwell plate assisted confocal microscopy, which allows imaging of intact live organs/tissues at subcellular resolution.
The endomembrane script was optimized to recognize GFP-2xFYVE structures, a marker of late endosomes and MVBs. This script detected equally well YFP-Rab2b/ARA7-tagged late endosomes, YFP-RabC1-marked post-Golgi endosomes, and YFP-VTI12-labeled early endosomes of the trans-Golgi network, thus illustrating its broad application potential to monitor specific endosome classes. Since the spinning-disc microscope permits parallel image acquisition from different fluorescent proteins, our method can be extended to simultaneously assess and quantify more than one endomembrane compartment from transgenic lines coexpressing different endomembrane marker proteins each fused to a distinctive fluorophore.
In the transgenic line expressing GFP-2xFYVE compartments, their abundance becomes differentially adjusted upon distinct stress treatments. Infection with pathogenic Pseudomonas bacteria resulted in an increase of endosomal compartments. Recent reports provide evidence for pathogen-triggered interference with endosomal trafficking pathways important for virulence (Nomura et al., 2006; Tanaka et al., 2009), but how this affects the abundance of endosomes remains to be demonstrated. We also recorded a reduced abundance of GFP-2xFYVE compartments upon dark incubation. Light was previously shown to induce internalization of the plasma membrane resident receptor kinase phototropin 1, and to affect endosomal trafficking of the PIN2 auxin carrier protein, thereby regulating PIN2 localizing to the plasma membrane or the vacuole (Laxmi et al., 2008; Kaiserli et al., 2009). It might be possible that light accelerates plasma membrane internalization or reduces endosomal recycling and maturation. While cold treatment is known to prevent internalization (Baluska et al., 2002), we revealed elevated numbers of GFP-2xFYVE compartments. This might be also explained by a reduction in endosomal recycling and maturation, or due to the fact that initially cold-treated samples were transferred to room temperature during the imaging process and thereupon an increase of plasma membrane endocytosis was induced.
The interest in high-throughput confocal imaging is evident from a number of recent studies describing forward genetic screens by manual microscopy or on the basis of engineered macroscopic reporter traits (Avila et al., 2003; Logan et al., 2003; Tamura et al., 2005; Teh and Moore, 2007; Boulaflous et al., 2008; Tanaka et al., 2009). Mutants defective in actin organization and vacuole biogenesis, respectively, were discovered with the help of a GFP-targeted vacuolar protein (Avila et al., 2003; Tamura et al., 2005; Nomura et al., 2006). Monitoring intracellular accumulation of a secreted GFP variant allowed the isolation of mutants defective in membrane trafficking to the plasma membrane (Teh and Moore, 2007). Brefeldin A-triggered internalization/accumulation of GFP-PIN1 was used as readout to identify BEN1/MIN7, which encodes an ADP-ribosylation factor-guanine nucleotide exchange factor (Tanaka et al., 2009) shown before to interfere with secretory and endocytic traffic (Nomura et al., 2006). However, these screens were limited because of laborious manual handling and the need to detect clear qualitative differences in the localization of the respective fluorescently labeled proteins. Our method instead enables handling of large sample sets at high-throughput and automated image analysis recording multiparametric quantitative values. This will advance genetic screening at cellular and subcellular resolution, e.g. to identify genetic determinants of membrane trafficking.
MATERIALS AND METHODS
Materials, Growth Conditions, and Treatments
The following Arabidopsis (Arabidopsis thaliana) transgenic plants were used in this study (accession Col-0 if not otherwise indicated): p35S::GFP-PEN1 in pen1-1 (Collins et al., 2003), p35S::GFP-2xFYVE (Ler; Voigt et al., 2005), p35S::YFP-2xFYVE (Vermeer et al., 2006), pUBQ10::3myc-YFP-RabF2b (Geldner et al., 2009), pUBQ10::3myc-YFP-Rab C1 (Geldner et al., 2009), and pUBQ10::3myc-YFP-VTI12 (Geldner et al., 2009). Plants were grown on moist turf substrate (STENDER) containing 10 mg/L Confidor WG 70 pesticide (BAYER) in controlled environments with 12 h light and 60% humidity. Cotyledon leaves of 2-week-old plants were used for microscopy. Blumeria graminis (isolate K1) and PtoDC3000 infection assays were done as previously described (Zipfel et al., 2004; Miklis et al., 2007). Cold treatment was applied by incubation at 4°C for 42 h; dark incubation was done for 72 h.
QCLM
Quantitative confocal high-throughput imaging was performed with the Opera microscope (PERKINELMER CELLULAR TECHNOLOGIES GERMANY GmbH) equipped with three 1.3 MPixel CCD cameras with a Nipkow spinning disc. Excitation of the samples was performed at 488 nm for GFP or YFP. The emission spectrum was taken from 502 to 577 nm. Leaves were prepared in 96-well plates with optical glass bottom (GREINER BIO-ONE GmbH). Sample preparation was supported by a self-made aluminum stamp (400 g) carrying 96 pins (4 mm diameter, mm length) to maximize contact of the cotyledon leaf with the optical glass bottom of the 96-well plate. We adhered on the tip of each pin a 4-mm thick cellular-rubber sheet (ethylene propylene diene rubber, JACOB NETTEKOVEN) with a polyvinyl chloride film (Mactac 8900) on top to minimize leaf tissue damage (Supplemental Fig. S4). Detached cotyledons of 2-week-old Arabidopsis plants were positioned on the pins and the stamp was fitted into optical plates supplied with water.
Microscope settings used with the endomembrane script: Images of a consecutive series of 21 planes with a distance of 1 μm were taken with the 40× objective in five predefined areas per leaf. The image area is 443.76 μm × 335.40 μm (1,376 × 1,040 pixel). Thus, in total 105 images were obtained per leaf. Image capture of individual planes was as fast as 40 ms. Measurement and parallel image analysis of one 96-well plate required 30 min. Microscope settings used with the plasma membrane script: Images of a consecutive series of 31 planes with a distance of 1 μm were taken with the 20× objective in eight predefined areas (the image area is 887.52 μm × 670.80 μm) per leaf. Thus, in total 248 images were obtained per leaf. Due to technical reasons images of only four areas per leaf could be taken per run, therefore each plate was measured twice. Per image area about 10 GFP-PEN1 accumulations and per plant around 160 GFP-PEN1 accumulations were analyzed. Measurement of one 96-well plate required 2 × 40 min for the GFP-PEN1 line. Subsequent image analysis was conducted overnight. For setting the threshold levels, 300 plants were analyzed 24 hpi with B. graminis in three independent experiments.
Image Processing and Automated Analysis
Images were analyzed with the Acapella Software (version 2.0, PERKIN ELMER CELLULAR TECHNOLOGIES GERMANY GmbH). To merge the three-dimensional stack of 21 or 31 optical planes, respectively, an image projection was performed, resulting in a two-dimensional pseudo image. Subsequently, the pseudo image was analyzed with either the endomembrane script or the plasma membrane microdomain script (Supplemental Data S1 and S2). To enable image processing of plant tissues, it was necessary to develop a novel algorithm for the detection of the cell outline and from that generating cells within the Acapella Software. Cell detection of animal suspension cells typically proceeds by identifying large centers of brightness that are assumed to represent the nuclei of the cells and from these move outwards to find other consistent local maxima of brightness that represent the cell membrane edges. Due to the different topology of the plant cell, cell images usually do not have prominent nuclei so these cannot be used as a starting point. As a consequence, standard cell finding methods fail when challenged with plant leaf cell images. Nuclei detection algorithms here typically find only the bright centers, which are actually the edges of the cell and divide the edges into many separate objects. Subsequent cytoplasm finding proceeds into the center of the cell, rather than the edges and the objects that are found are artefactual subdivisions of the real cells centered on the brightest spots in the membranes (Supplemental Figs. S5A and S6A). The approach we have taken successfully identifies the plant cell by taking into account the differing intensity source in the image. The first step is to use a line-finding algorithm to trace paths along the mazy outlines of the cell images. Typically this will result in one or a very few objects that seem like fuzzy paths through the bright parts of the image that are called skeletons. The fuzzy line must next be tidied to make a clearer line and this is achieved by a series of skeleton branch trimming and filtering steps. This process is repeated until the line is clean of branches. At this point the line is eroded to allow it to cover the cell membrane completely and it is now straightforward to perform object identification using the accurately defined cell outline as an external guide (Supplemental Figs. S5B and S6B).
We provide an implementation of our plant leaf cell detection algorithm as a proc that can be easily included into the Opera/Acapella imaging and analysis system. The library is available from http://www.ringtail.tsl.ac.uk/sr_acapellacode/LeafDetection.html (provided under the GNU Public License V3). Although our implementation of the leaf cell detection algorithm is made in a closed system, the code uses standard functions available and could easily be translated or implemented in other image analysis systems.
In summary our scripts enable detection of plant leaf cells and subcellular membrane compartments, and each script recorded 21 and 16 output parameters, respectively (Supplemental Tables S1 and S2). For easy access of the processed image analysis, the data files were imported into PERL and displayed in graphic bars (Supplemental Data S3 and S4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Example for z sections that are used to assemble a pseudo image.
Supplemental Figure S2. Quantitative parameters of GFP-2xFYVE and GFP-PEN1 membrane compartments.
Supplemental Figure S3. Pattern recognition of GFP-2xFYVE compartments from images obtained by conventional confocal microscopy.
Supplemental Figure S4. Self-made aluminum stamp used for leaf preparation.
Supplemental Figure S5. Comparative image processing of GFP-2xFYVE leaf samples.
Supplemental Figure S6. Comparative image processing of GFP-PEN1 leaf samples.
Supplemental Table S1. Overview of parameters recorded by the endomembrane script.
Supplemental Table S2. Overview of parameters recorded by the plasma membrane microdomain script.
Supplemental Data S1. Endomembrane script.
Supplemental Data S2. Plasma membrane microdomain script.
Supplemental Data S3. PERL scripts for endomembrane script.
Supplemental Data S4. PERL script for plasma membrane microdomain script.
Supplementary Material
Acknowledgments
We thank M. Beck, M. Beckers, S. Boztepe, M. Hallstein, M. Kwaaitaal, G. Mancel, E. Schmelzer, and Perkin Elmer (formerly Evotec Technologies, Hamburg, Germany); in particular A. Kirsch and O. Ollikainen for technical help; and J. Samaj (University of Bonn), T. Munnik (University of Amsterdam), H. Thordal-Christensen (University of Copenhagen), and N. Geldner (University of Lausanne) for providing materials.
References
- Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila EL, Zouhar J, Agee AE, Carter DG, Chary SN, Raikhel NV. (2003) Tools to study plant organelle biogenesis: point mutation lines with disrupted vacuoles and high-speed confocal screening of green fluorescent protein-tagged organelles. Plant Physiol 133: 1673–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Hlavacka A, Samaj J, Palme K, Robinson DG, Matoh T, McCurdy DW, Menzel D, Volkmann D. (2002) F-actin-dependent endocytosis of cell wall pectins in meristematic root cells: insights from brefeldin A-induced compartments. Plant Physiol 130: 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulaflous A, Faso C, Brandizzi F. (2008) Deciphering the Golgi apparatus: from imaging to genes. Traffic 9: 1613–1617 [DOI] [PubMed] [Google Scholar]
- Chung K, Crane MM, Lu H. (2008) Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods 5: 637–643 [DOI] [PubMed] [Google Scholar]
- Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, et al. (2010) Systems survey of endocytosis by multiparametric image analysis. Nature 464: 243–249 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Crane MM, Chung K, Lu H. (2009) Computer-enhanced high-throughput genetic screens of C. elegans in a microfluidic system. Lab Chip 9: 38–40 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. (1998) FYVE fingers bind PtdIns(3)P. Nature 394: 432–433 [DOI] [PubMed] [Google Scholar]
- Gehrig J, Reischl M, Kalmár E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Müller F. (2009) Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat Methods 6: 911–916 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multi-color marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JP, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Husebye H, Halaas O, Stenmark H, Tunheim G, Sandanger O, Bogen B, Brech A, Latz E, Espevik T. (2006) Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J 25: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Robinson DG. (2009) Transport vesicle formation in plant cells. Curr Opin Plant Biol 12: 660–669 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. (2009) Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell 21: 3226–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Galway ME, Mulder BM, Emons AM. (2008) Rates of exocytosis and endocytosis in Arabidopsis root hairs and pollen tubes. J Microsc 231: 265–273 [DOI] [PubMed] [Google Scholar]
- Kwon C, Bednarek P, Schulze-Lefert P. (2008a) Secretory pathways in plant immune responses. Plant Physiol 147: 1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. (2008b) Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R. (2008) Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Scott I, Tobin AK. (2003) The genetic control of plant mitochondrial morphology and dynamics. Plant J 36: 500–509 [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD. (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P. (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. (2007) Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Nakano A. (2002) Spinning-disk confocal microscopy—a cutting-edge tool for imaging of membrane traffic. Cell Struct Funct 27: 349–355 [DOI] [PubMed] [Google Scholar]
- Negishi T, Nogami S, Ohya Y. (2009) Multidimensional quantification of subcellular morphology of Saccharomyces cerevisiae using CalMorph, the high-throughput image-processing program. J Biotechnol 141: 109–117 [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Panstruga R. (2006) Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol 171: 699–718 [DOI] [PubMed] [Google Scholar]
- Pajonk S, Kwon C, Clemens N, Panstruga R, Schulze-Lefert P. (2008) Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J Biol Chem 283: 26974–26984 [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436: 78–86 [DOI] [PubMed] [Google Scholar]
- Pepperkok R, Ellenberg J. (2006) High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol 7: 690–696 [DOI] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. (1991) Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol 96: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch O. (2006) High content cellular screening. Curr Opin Chem Biol 10: 316–320 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Jiang LW, Schumacher K. (2008) The endosomal system of plants: charting new and familiar territories. Plant Physiol 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Moore I. (2002) The Arabidopsis Rab GTPase family: another enigma variation. Curr Opin Plant Biol 5: 518–528 [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. (2000) The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol 124: 1558–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Tobin AK, Logan DC. (2006) BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J Exp Bot 57: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. (2005) KATAMARI1/MURUS3 Is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kitakura S, De Rycke R, De Groodt R, Friml J. (2009) Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr Biol 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Teh OK, Moore I. (2007) An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TWJ, Jr, Munnik T. (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Voigt B, Timmers ACJ, Samaj J, Hlavacka A, Ueda T, Preuss M, Nielsen E, Mathur J, Emans N, Stenmark H, et al. (2005) Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur J Cell Biol 84: 609–621 [DOI] [PubMed] [Google Scholar]
- Wolinski H, Petrovic U, Mattiazzi M, Petschnigg J, Heise B, Natter K, Kohlwein SD. (2009) Imaging-based live cell yeast screen identifies novel factors involved in peroxisome assembly. J Proteome Res 8: 20–27 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.