Abstract
Xyloglucan is the main hemicellulose in the primary cell walls of most seed plants and is thought to play a role in regulating the separation of cellulose microfibrils during growth. Xylose side chains block the degradation of the backbone, and α-xylosidase activity is necessary to remove them. Two Arabidopsis (Arabidopsis thaliana) mutant lines with insertions in the α-xylosidase gene AtXYL1 were characterized in this work. Both lines showed a reduction to undetectable levels of α-xylosidase activity against xyloglucan oligosaccharides. This reduction resulted in the accumulation of XXXG and XXLG in the liquid growth medium of Atxyl1 seedlings. The presence of XXLG suggests that it is a poor substrate for xyloglucan β-galactosidase. In addition, the polymeric xyloglucan of Atxyl1 lines was found to be enriched in XXLG subunits, with a concomitant decrease in XXFG and XLFG. This change can be explained by extensive exoglycosidase activity at the nonreducing ends of xyloglucan chains. These enzymes could thus have a larger role than previously thought in the metabolism of xyloglucan. Finally, Atxyl1 lines showed a reduced ability to control the anisotropic growth pattern of different organs, pointing to the importance of xyloglucan in this process. The promoter of AtXYL1 was shown to direct expression to many different organs and cell types undergoing cell wall modifications, including trichomes, vasculature, stomata, and elongating anther filaments.
The primary wall that surrounds the growing cells of plants has to be able to extend in response to turgor pressure. This process needs to be tightly regulated to avoid a mechanical failure of the wall. The direction of expansion also needs to be controlled so that different cell types can develop their particular morphology. In addition, the growth of the different tissues in an organ has to be tightly coordinated so that it can achieve its final shape (Baskin, 2005). The mechanical behavior of the expanding cell wall has been likened to a fiber-reinforced composite, with crystalline cellulose microfibrils embedded in an amorphous matrix of hemicellulose and pectin. How this works at the molecular level is still the subject of much research and speculation (Geitmann and Ortega, 2009).
Xyloglucan is the main hemicellulose in the primary cell walls of gymnosperms and most angiosperm families and is present in all extant groups of land plants, although with some differences in structure (Peña et al., 2008; Scheller and Ulvskov, 2010). All these xyloglucans have a backbone of (1→4)-linked β-d-glucopyranosyl residues, many of which are substituted with α-d-xylopyranosyl residues at O6. In many vascular plants, including Arabidopsis (Arabidopsis thaliana), every fourth glucosyl residue of the xyloglucan backbone is unsubstituted (Vincken et al., 1997). In the standard nomenclature for xyloglucan structures, these residues are represented by G, while X, L, and F indicate Glc residues substituted, respectively, with α-d-Xylp, β-d-Galp-(1→2)-α-d-Xylp, and α-l-Fucp-(1→2)-β-d-Galp-(1→2)-α-d-Xylp side chains (Fry et al., 1993). Conventionally, the reducing end of the molecule is positioned to the right. Treatment of Arabidopsis xyloglucan with an endoglucanase that attacks unsubstituted residues results in oligosaccharide mixtures that include XXG, GXXG, XXXG, XXLG, XLXG, XLLG, XXFG, and XLFG, with some of the Gal residues O-acetylated (Madson et al., 2003; Obel et al., 2009).
Although the detailed arrangement and possible connections of the different components of primary cell walls are still unclear, xyloglucan chains are long enough to attach simultaneously to neighboring microfibrils and thus could generate resistance to cell wall extension (Obel et al., 2007). There is also considerable evidence for the covalent linkage of xyloglucan to the pectic polysaccharide rhamnogalacturonan I (Popper and Fry, 2008). The attachment of xyloglucan to cellulose microfibrils is based on hydrogen bonds, and it might be controlled by expansin proteins (Cosgrove, 2005). Xyloglucan connections between microfibrils could also be broken or created by enzymes in the xyloglucan transglycosylase/hydrolase (XTH) family (Nishitani and Vissenberg, 2007). These enzymes cleave xyloglucan chains in front of unsubstituted Glc residues and stay covalently bound to this residue, forming an enzyme-donor complex (Johansson et al., 2004). They can later attach the Glc residue to the nonreducing end of another xyloglucan molecule, acting as xyloglucan endotransglucosylases (XETs). A group of XTHs can also use water as an acceptor, acting as xyloglucan endohydrolases, but they seem to be a minority (Baumann et al., 2007; Eklöf and Brumer, 2010). It is unclear at the moment if endoglucanases from other families are involved in xyloglucan metabolism (Lopez-Casado et al., 2008).
The importance of xyloglucan as a regulator of cell wall extension has been thrown into doubt by the identification of an Arabidopsis mutant that has no detectable xyloglucan but still manages to develop normally (Cavalier et al., 2008). Apart from being slightly smaller, this mutant has defective root hairs, but it seems clear that Arabidopsis must have alternative ways of regulating the separation of cellulose microfibrils. It is interesting nonetheless that microfibrils seem to be more irregularly spaced in the xyloglucan-deficient mutant (Anderson et al., 2010).
The end result of endoglucanase activity on xyloglucan is the release of oligosaccharides with an unsubstituted Glc at the reducing end. Specific exoglycosidase activities are then necessary to release each type of residue (Iglesias et al., 2006). α-Xylosidase activities in both pea (Pisum sativum) and Tropaeolum majus can only remove unsubstituted Xyl residues from the nonreducing end of the molecule (O’Neill et al., 1989; Fanutti et al., 1991). A β-glucosidase is then required to remove the unsubstituted Glc before α-xylosidase can act again (Crombie et al., 1998). β-Galactosidase and α-fucosidase activities are also required for the complete disassembly of the different Arabidopsis oligosaccharides (Edwards et al., 1988; Léonard et al., 2008). There is currently no information on the enzymes that might be involved in xyloglucan deacetylation or on how the presence of acetyl residues affects exoglycosidases.
The Arabidopsis gene AtXYL1 (At1g68560) was identified as coding for an α-xylosidase activity against xyloglucan oligosaccharides by the similarity of its product to purified cabbage (Brassica oleracea var capitata) α-xylosidase (Sampedro et al., 2001). The identification was confirmed through heterologous expression in yeast. According to the Carbohydrate Active Enzymes database (http://www.cazy.org/), AtXYL1 is a member of glycosyde hydrolase family 31, which includes mainly α-glucosidases and α-xylosidases(Cantarel et al., 2009). A reduction of up to 70% of α-xylosidase activity was reported in antisense lines where AtXYL1 was silenced, but this reduction did not cause changes in morphology (Monroe et al., 2003). This article presents the characterization of two independent insertional mutants in AtXYL1 that have no detectable α-xylosidase activity and show remarkable changes in xyloglucan composition along with alterations in the growth pattern.
RESULTS
Atxyl1 Mutants Are Deficient in α-Xylosidase Activity
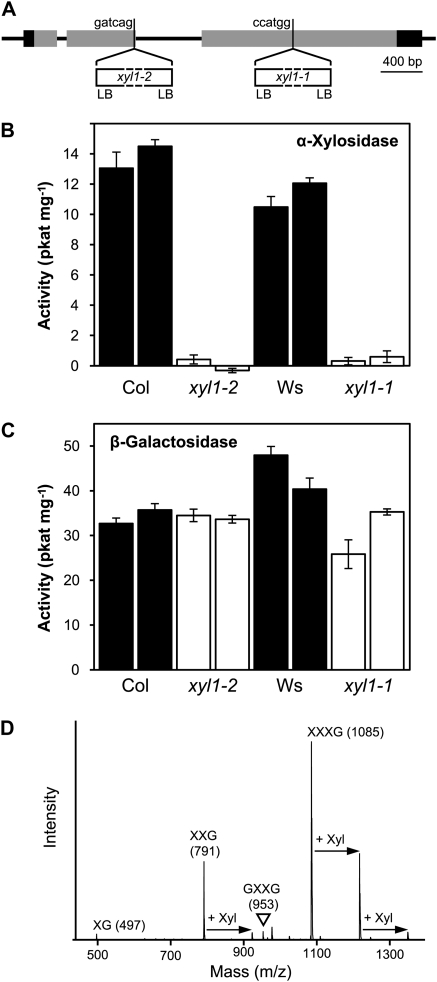
Homozygous lines of two independent insertional mutants in the Atxyl1 gene (At1g68650) were characterized in this work. Atxyl1-1 was found in the collection of the Arabidopsis Knockout Facility at the University of Wisconsin Biotech Center (Krysan et al., 1999). This mutant is in the Wassilewskija genetic background, and the insertion is located within the third exon, as determined by the sequence of the border region (Fig. 1A). The second mutant, Atxyl1-2, has a Columbia background and was obtained from the GABI-KAT collection (Rosso et al., 2003). The insertion is located within the second exon (Fig. 1A). The phenotypes of both insertional lines were compared with that of lines lacking the insertion segregated from the same populations.
Figure 1.
Enzymatic characterization of Atxyl1 insertional mutants. A, Gene model of AtXYL1 showing the location of the T-DNA insertions. Exons are represented by rectangles, with translated regions in gray and untranslated regions in black. The six nucleotides preceding the insertions are shown. In both mutants, left border sequences (LB) are present in both directions. B, α-Xylosidase activity against XXXG in Atxyl1 lines. Two replicate protein extractions were obtained from each wild-type (black bars) and mutant (white bars) line. Error bars show sd. Col, Columbia; Ws, Wassilewskija. C, β-Galactosidase activity against XLLG in Atxyl1 lines. The same protein extracts as in B were used. Error bars show sd. D, MALDI-TOF spectrum showing possible transglycosylation by AtXYL1. The spectrum corresponds to a partial digestion of XXXG with a protein extract from a Columbia wild-type line (16 h, 1.5 mm XXXG, 850 μg mL−1 protein). Peaks corresponding to the [M+Na]+ adduct of the substrate and intermediate products are labeled. Arrows indicate additional peaks whose masses can be explained by transglycosylation of Xyl residues.
In both Atxyl1-1 and Atxyl1-2, α-xylosidase activity against XXXG is reduced below detectable levels (Fig. 1B). On the other hand, β-galactosidase activity against XLLG does not seem to be affected on either of the mutants (Fig. 1C). The Columbia genome includes a close paralog of Atxyl1, annotated as locus At3g45940. In a previous work, we identified this locus as a recently inactivated pseudogene and named it φAtxyl2 (Sampedro et al., 2001). Since Atxyl1-1 is in a Wassilewskija background, the sequence of φAtxyl2 was determined in this ecotype (Supplemental Fig. S1). Inactivating mutations are found throughout the sequence. An 8-bp insertion and an in-frame stop codon in the coding region are shared with Columbia, while an additional 1-bp insertion is not. In Columbia, this locus has been annotated as a gene by assuming two additional introns over conserved regions of the coding sequence, and a third one would be necessary in Wassilewskija. Furthermore, most point mutations between Wassilewskija and Columbia are nonsynonymous. There are four ESTs deposited in GenBank that correspond to locus At3g45940, but these are 454 sequences from ovules of ecotype Landsberg (accession nos. EL981094, EL996745, ES022488, and ES118483). The sequences are very short and do not overlap with the inactivating mutations in Columbia and Wassilewskija.
In addition to α-xylosidase activity, we detected in wild-type plants a possible transglycosylating activity involving the Xyl residues of xyloglucan oligosaccharides. Partial digestions of XXXG with protein extracts from Columbia seedlings similar to those used for quantification were analyzed by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Peaks with the expected mass-to-charge ratio (m/z) of GXXG, XXG, and XG were observed (Fig. 1D). The low level of GXXG suggests that xylosidase is the limiting activity in the degradation of XXXG. In addition to these peaks, a number of unexpected ions were detected in these samples. Particularly prominent is a peak at 1,217, which corresponds to the mass of the [M+Na]+ adduct of XXXG with an additional pentose residue. This peak does not appear when digestions are immediately stopped after adding XXXG. More interestingly, it does not appear when Atxyl1-1 or Atxyl1-2 extracts are used (data not shown). A transglycosylating activity by AtXYL1, competing with hydrolysis and using oligosaccharides as both donors and acceptors of Xyl residues, could explain this peak as well as others at m/z 923 and 1,349 (Fig. 1D). Further evidence of this activity is the fact that partial digestions of XXXG with wild-type protein extracts show levels of free Glc that are usually 25% or more higher than levels of free Xyl (data not shown).
XXXG and XXLG Accumulate in the Growth Medium of α-Xylosidase Mutants
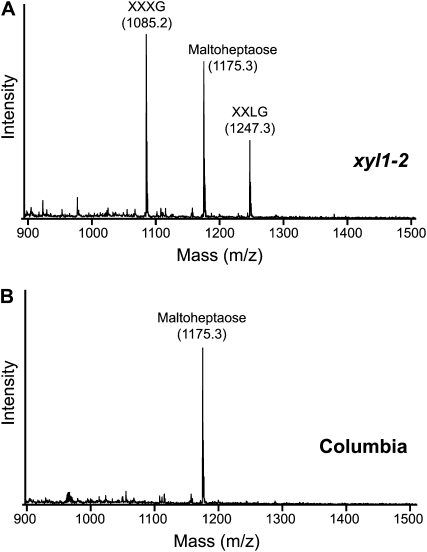
The liquid medium in which seedlings had been grown for 7 d was analyzed through MALDI-TOF MS in both wild-type and mutant lines. Maltoheptaose was added as an internal standard, but the same result was observed when it was not present. In both mutants, but not in the respective wild types, two peaks were observed with the m/z of the [M+Na]+ adduct of XXXG (1,085) and XXLG/XLXG (1,247), as shown for Atxyl1-2 in Figure 2. The identity of the 1,085 peak was confirmed by comparison of its MALDI-TOF/TOF spectrum with that of commercial XXXG (Supplemental Fig. S2, A and B). Both spectra show an almost identical pattern of fragmentation, with most ions corresponding to the breakage of glycosidic bonds.
Figure 2.
Accumulation of xyloglucan oligosaccharides in Atxyl1-2. A, MALDI-TOF spectrum of the concentrated liquid growth medium from 7-d-old Atxyl1-2 seedlings. Maltoheptaose at 50 μm final concentration was added as an internal standard for quantification. The peaks with a m/z of 1,085 and 1,247 were identified as [M+Na]+ adducts of XXXG and XXLG by further fragmentation through MALDI-TOF/TOF MS, as detailed in Supplemental Figure S2. B, MALDI-TOF spectrum of the growth medium from wild-type Columbia seedlings.
To identify the 1,247 peak, its MALDI-TOF/TOF spectrum was compared with that of commercial XLLG (Supplemental Fig. S2, D and E). The spectrum of XLLG shows two prominent ions with m/z of 659 and 773 that can be explained by a single cleavage between the two Gal-containing side chains. In the spectrum of the 1,247 peak, the 659 fragment, diagnostic of XXLG, is much more abundant than the 773 fragment, diagnostic of XLXG (Madson et al., 2003; Tiné et al., 2006). Since a fragment of m/z 773 can also appear from XXLG by double fragmentation, the presence of XLXG in the growth medium is uncertain (Yamagaki et al., 1998). The concentration of XXXG in Atxyl1-2 growth medium was estimated at approximately 8 μm, based on the comparison of the peak area with the internal standard and using a calibration curve of commercial XXXG (Supplemental Fig. S2C). This amounts to 0.08% of the dry weight of the seedlings. Although this value should be considered a rough approximation due to the difficulties of employing MALDI-TOF MS as a quantification method, it nonetheless suggests that we should expect a significant accumulation of oligosaccharides in the apoplast of soil-grown plants. In the growth medium of Atxyl1-1, the concentration of oligosaccharides was approximately 10 times lower (data not shown).
XXLG Subunits Are More Abundant in Xyloglucan from Atxyl1 Lines
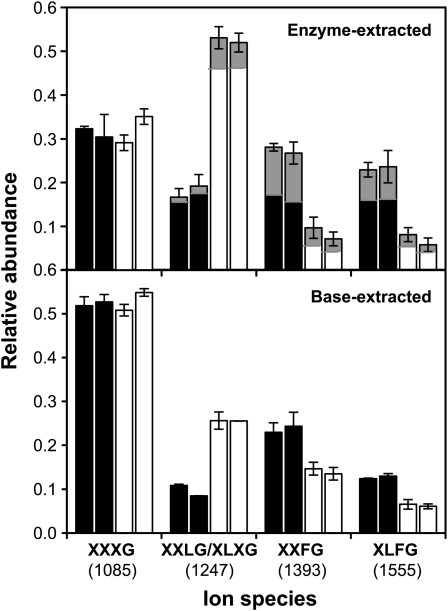
Xyloglucan composition was analyzed by quantifying, on the basis of MALDI-TOF spectra, the relative abundance of the major subunits obtained after endoglucanase digestion. Two separate xyloglucan fractions, enzyme and base extracted, were sequentially obtained from the stems of both Atxyl1-2 and Columbia plants. The effect of the lack of α-xylosidase is particularly evident in the xyloglucan fraction that was directly extracted with an endoglucanase treatment (Fig. 3). The major change observed in the spectra of Atxyl1-2 samples is an increase in the relative area of the peak at m/z 1,247 (XXLG/XLXG subunits) from 18% to 53%. The MALDI-TOF/TOF spectrum of this peak in Atxyl1-2 shows that XLXG is at best a very minor component, suggesting that the increase is due to XXLG subunits (Supplemental Fig. S3).
Figure 3.
Xyloglucan composition in Atxyl1-2 and wild-type plants. Alcohol-insoluble cell wall residues were obtained from elongated stems of wild-type (black bars) or Atxyl1-2 (white bars) plants. Xyloglucan was first extracted with endoglucanase from duplicate samples of each line, and peak areas from two to five MALDI-TOF spectra for each sample were quantified to estimate sd, presented as error bars. Minor peaks (less than 5%) were ignored. The proportion of acetylated subunits is indicated by the gray areas. After enzyme extraction, samples were treated with 24% KOH. Released xyloglucan was digested with endoglucanase and analyzed by MALDI-TOF MS.
Xyloglucan from a second extraction of the cell wall residue with 24% KOH showed a smaller change in composition, with an increase in XXLG/XLXG subunits from 10% to 26% (Fig. 3). In both fractions, the increase in XXLG is associated with a reduction in XXFG and XLFG subunits, with no apparent change in the proportion of XXXG. Subunits with Fuc, therefore, are strongly reduced in Atxyl1-2 plants, from 51% to 15% in enzyme-accessible xyloglucan and from 36% to 20% in the nonaccessible fraction. As for the proportion of acetylated subunits, it can only be estimated in the enzyme-extracted fraction, where it shows a reduction from 20% to 11% in Atxyl1-2. However, for each type of subunit (XXLG, XXFG, or XLFG), the proportion that is acetylated in Atxyl1-2 is almost the same as in the wild type (Fig. 3). Similar changes were observed when comparing enzyme-extracted xyloglucan from stems of Atxyl1-1 and Wassilewskija plants as well as in Atxyl1-2 siliques, although in this last case the change in composition was smaller (data not shown). Other organs were not tested.
Organ Elongation Is Affected in α-Xylosidase-Deficient Plants
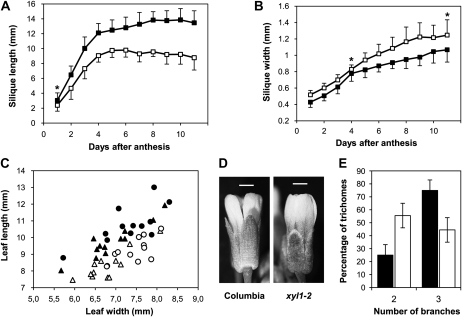
Both Atxyl1-1 and Atxyl1-2 plants are healthy and fertile, and they reach a similar size to wild-type plants. However, a clear alteration of the growth pattern is apparent in the siliques of both mutants, which are significantly shorter and wider than the respective wild types (Fig. 4; Supplemental Fig. S4). At 11 d post anthesis, Atxyl1-2 siliques are 35% shorter and 17% wider than wild-type siliques, with the result that the length-to-width ratio is reduced from 12.6 to 7.0. The development of this phenotype appears to be somewhat different in both mutants. In Atxyl1-2, the reduction in silique length is apparent from very early stages (Fig. 4A), while in Atxyl1-1, the premature cessation of growth seems to be responsible for the final difference in length (Supplemental Fig. S4A). On the other hand, just after anthesis, both mutants already have wider siliques than the respective wild types, and this difference is maintained until maturity (Fig. 4B; Supplemental Fig. S4B). The alteration in growth pattern of the siliques does not seem to affect seed production.
Figure 4.
Morphological phenotypes of Atxyl1 lines. A, Silique elongation in Atxyl1-2. Siliques from 12 wild-type (black squares) or Atxyl1-2 (white squares) plants were measured using a scanner. Error bars indicate sd. The asterisk indicates a difference significant with a 95% confidence; all other differences are significant with a 99.5% confidence. B, Silique width in Atxyl1-2. Data are from the same siliques as in A, and symbols are used in the same way. C, Leaf shape in Atxyl1-1 plants. Length and width of the third (triangles) and fourth (circles) leaves were measured in 26-d-old rosettes from wild-type (black symbols) and Atxyl1-1 (white symbols) plants. D, Flowers from Columbia and Atxyl1-2 plants. Bars = 0.5 mm. E, Trichome branching in Atxyl1-1. The number of branches of all trichomes from eight first-pair leaves of wild-type (black bars) and Atxyl1-1 (white bars) plants were counted. Unbranched trichomes, less than 1% in both lines, were ignored. Error bars show sd.
A comparable defect in elongation was observed in other organs. When 26-d-old rosettes from Atxyl1-1 plants were compared with those of the Wassilewskija wild type, a significantly lower ratio of length to width was observed for the five oldest leaves (P < 0.01). In the third and four leaves in particular, width was not affected by the mutation, while length was considerably reduced (Fig. 4C). Similarly, sepals appear shorter in both mutants than in the respective wild types (Fig. 4D). An additional phenotype was observed in the leaf trichomes of Atxyl1-1 plants. In wild-type leaves, 75% of trichomes have three branches and 25% of them have two, while in xylosidase-deficient plants, most trichomes are two branched (Fig. 4E). This phenotype was not observable in Atxyl1-2 plants. However, trichomes with two branches are very rare in Columbia wild-type plants.
The AtXYL1 Promoter Directs Expression to Many Cell Types and Organs Undergoing Cell Wall Modifications
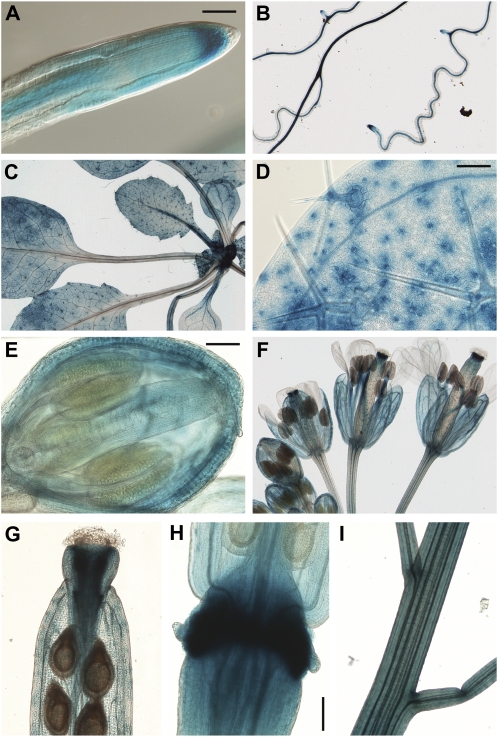
A 3-kb fragment of AtXYL1 promoter was cloned upstream of the GUS gene. Arabidopsis plants transformed with this construct showed widespread expression of the reporter gene (Fig. 5). Expression was detected with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid and was strong enough that incubation for 3 h was usually sufficient. In root tips, in both soil and agar, expression is strongest in the stem cell area extending into the meristematic region (Fig. 5A). In the elongation zone, the highest expression is located in the vascular cylinder, and the same pattern can be observed in older areas of the root (Fig. 5B). Expression in lateral root buds is usually weak before emergence. In growing leaves, there is strong reporter expression in trichomes, guard cells, and vasculature, along with diffuse expression throughout the parenchyma (Fig. 5, C and D). In addition, the youngest leaves show a high level of expression in petioles. In developing flower buds, expression levels are elevated in the sepals and gynoecium (Fig. 5E). As the flower opens, anther filaments show strong reporter expression, while in the gynoecium, expression becomes concentrated in the upper part (Fig. 5F). In petals, expression is located mainly in the vasculature. Developing siliques show some diffuse expression in the valves, but the highest expression is found in the style (Fig. 5G) and in the area of the abscission zone (Fig. 5H), where expression starts before abscission of flower organs but persists until siliques are mature. There is also expression in the pedicels, particularly during silique development, as well as in the younger sections of the stem (Fig. 5I).
Figure 5.
Expression of GUS driven by the AtXYL1 promoter. A, Root tip from a 12-d-old plant grown on soil. Bar = 50 μm. B, Roots from a 12-d-old plant. C, Rosette leaves from a 21-d-old plant. D, First-pair leaf from a 12-d-old plant. Bar = 100 μm. E, Stage 12 flower bud. Bar = 100 μm. F, Flowers at different stages. G, Upper part of an elongated silique. H, Base of a developing silique. Bar = 100 μm. I, Young stem and pedicels.
DISCUSSION
Insertions in AtXYL1 reduce the α-xylosidase activity against XXXG in growing plants to undetectable levels (Fig. 1). Additionally, in both Columbia and Wassilewskija ecotypes, the closest paralog to AtXYL1 is a pseudogene (Supplemental Fig. S1). This locus appears to be transcriptionally active in Landsberg ovules, but it is unclear what significance this has. The next closest paralog in the Arabidopsis genome is AtAGLU1, a putative α-glucosidase whose translation product has been located in the vacuole in several proteomic studies (Carter et al., 2004; Shimaoka et al., 2004). We have previously presented evidence of the cell wall location of Arabidopsis α-xylosidase activity (Sampedro et al., 2001), and numerous studies have identified AtXYL1 in the cell wall proteome (Jamet et al., 2006). It is thus likely that the only cell wall enzyme capable of removing Xyl residues from xyloglucan oligosaccharides is AtXYL1. The specificity of plant α-xylosidases for residues located at the nonreducing end of xyloglucan molecules, which is key to their function, is also present in similar enzymes from prokaryotes that also belong to family 31 of glycosyl hydrolases, and it is likely due to the pocket-shaped active site of this family (Moracci et al., 2000; Lovering et al., 2005).
We have found evidence that, in addition to hydrolysis of α-Xyl residues, AtXYL1 may be able to catalyze their transglycosylation using xyloglucan oligosaccharides as both donors and acceptors (Fig. 1D). Family 31 enzymes, such as AtXYL1, use a retaining mechanism forming a glycosyl-enzyme intermediate that can potentially transfer the sugar residue to an acceptor other than water (Henrissat and Davies, 1997; Trincone and Giordano, 2006). Family 31 includes several transferases and also an archaeal xyloglucan-specific α-xylosidase with significant transglycosylase activity (Trincone et al., 2001). This enzyme can transfer Xyl residues to different OH groups in a terminal Glc. A similar mechanism for AtXYL1 could explain the mass of the putative transglycosylation products, but a detailed characterization would be necessary to confirm this hypothesis (Fig. 1D).
The absence of α-xylosidase activity results in the accumulation of two specific oligosaccharides, XXXG and XXLG, in the liquid growth medium of mutant seedlings (Fig. 2; Supplemental Fig. S2). A similar accumulation is expected in the apoplast of soil-grown plants. Since β-galactosidase activity against xyloglucan does not seem to be affected in Atxyl1 lines (Fig. 1C), the presence of XXLG in the medium suggests that it is not a good substrate for this activity. A xyloglucan-specific β-galactosidase purified from Copaifera langsdorfii had no activity on XXLG (de Alcântara et al., 1999), and a similar enzyme from T. majus had a much higher activity on XLXG than XXLG (Crombie et al., 1998; de Alcântara et al., 1999). Although these enzymes are involved in xyloglucan mobilization in germinating seeds, our results are consistent with similar substrate specificity for the primary wall β-galactosidase activity in Arabidopsis.
Change in Xyloglucan Composition
Xyloglucan composition in plants without detectable α-xylosidase activity is different from that of wild-type plants, particularly in the enzyme-accessible fraction, as would be expected if caused directly by changes in the metabolism of xyloglucan within the wall (Fig. 3). In Atxyl1 lines, xyloglucan oligosaccharides in the apoplast can only be attacked by galactosidase and fucosidase activities, presumably leading to an accumulation of XXXG and XXLG, as seen in the growth medium (Fig. 2). XET activities could then use these oligosaccharides as acceptors, reincorporating them onto polymeric xyloglucan. Since this mechanism would only add one modified subunit per chain at the reducing end, it is clearly insufficient to explain the magnitude of the alteration observed in Atxyl1 lines.
An alternative mechanism to explain the loss of Gal and Fuc involves direct action of cell wall exoglycosidases on xyloglucan chains. A β-galactosidase from Tropaeolum can efficiently remove Gal residues from midchain regions of polymeric xyloglucan (Edwards et al., 1988; Reid et al., 1988). However, β-galactosidases from Copaifera and Hymenaea courbaril have weak activity on the polymer and much higher activity on xyloglucan oligosaccharides (de Alcântara et al., 1999, 2006). A similar preference for oligosaccharides was reported for an Arabidopsis α-fucosidase (Léonard et al., 2008). This preference could indicate that these enzymes attack the polymer mainly at the nonreducing ends. There is also some evidence that α-xylosidase could attack the nonreducing ends of polymeric xyloglucan. A Tropaeolum α-xylosidase can remove Xyl from oligosaccharides with a backbone of at least eight Glc residues (Fanutti et al., 1991, 1996). Additionally, the presence of up to 17% Xyl-deficient subunits, such as GXXG, exclusively in the enzyme-accessible fraction of pea xyloglucan has been attributed to α-xylosidase activity at the nonreducing ends of the polymer (Guillén et al., 1995; Pauly et al., 2001).
If all four exoglycosidases can attack the nonreducing ends of xyloglucan chains, our results are easier to explain. In Atxyl1 lines, only α-fucosidase and β-galactosidase would be able to act; thus, terminal XXFG and XLFG subunits would become XXLG. XXXG subunits would be left unmodified, and their proportion at nonreducing ends would only increase slightly due to β-galactosidase attack on rare XLXG subunits (Madson et al., 2003). Although this process would affect a single xyloglucan subunit per chain, endotransglycosylation reactions can repeatedly generate new nonreducing ends while placing previous ones at midchain positions. This would lead to the progressive increase in XXLG subunits at the expense of XXFG and XLFG, as observed in Atxyl1 lines (Fig. 3). This hypothesis also requires that in wild-type plants, α-fucosidase and β-galactosidase are followed by α-xylosidase and β-glucosidase activities, so that entire subunits would be removed without altering the composition. This would result in the trimming of the nonreducing ends of xyloglucan chains located in the enzyme-accessible domain.
The level of xyloglucan acetylation is also reduced in the enzyme-accessible fraction of Atxyl1-2 (Fig. 3). If most of the XXLG in Atxyl1-2 comes from XXFG and XLFG, we would expect the proportion of acetylated XXLG (12%) to be closer to that of XXFG (43%) and XLFG (33%). This suggests that some acetyl groups are being removed together with Fuc and Gal.
Altered Growth Pattern
Both Atxyl1 mutants show a reduction in the elongation of several organs. It is most noticeable in siliques, where a decrease in length is accompanied by an increase in width (Fig. 4; Supplemental Fig. S4). Leaf elongation is also significantly reduced in Atxyl1-1 plants, while leaf width is less affected or not at all (Fig. 4C). A similar phenotype is apparent in the sepals (Fig. 4D). This morphological phenotype points to a specific defect in the regulation of anisotropic expansion. As discussed above, lack of α-xylosidase activity is expected to result in a high concentration of xyloglucan oligosaccharides in the apoplast. In addition, the alteration of xyloglucan composition observed in Atxyl1 lines suggests active trimming of the nonreducing ends of polymeric xyloglucan in wild-type plants, a process that is blocked in Atxyl1 plants. These two developments could potentially interfere with the action of XET activities in the wall.
Xyloglucan oligosaccharides can increase the elongation rate of pea segments, especially if they have been treated with auxin (McDougall and Fry, 1990; Takeda et al., 2002). This is most likely caused by XET activities that employ oligosaccharides as acceptors of transglycosylation, breaking down xyloglucan chains and weakening the wall (Fry et al., 1993; Takeda et al., 2002). The elongation defect in Atxyl1 lines cannot be directly explained by this mechanism, although it is possible that excessive weakening triggers a strengthening response.
An alternative explanation is suggested by the observation that polymeric xyloglucan added to pea segments reduces their elongation, presumably by participating in endotransglycosylation reactions and therefore increasing the number of connections between microfibrils (Takeda et al., 2002). A similar mechanism could explain the reduced root elongation caused by the addition of two purified XTHs with XET activity (Maris et al., 2009). If trimming of nonreducing ends by exoglycosidases is blocked in Atxyl1 lines, this could facilitate a similar strengthening of the wall and explain the reduced elongation of siliques, leaves, and sepals (Fig. 4). Interestingly, in pea and Tropaeolum epicotyls, approximately 15% of XET activity appears to be covalently linked to xyloglucan, as part of enzyme-donor complexes, due to a lack of appropriate acceptors (Sulová et al., 2001). It is possible that a function of exoglycosidases, such as α-xylosidase, is to limit the formation of linkages among microfibrils by eliminating the loose ends of xyloglucan chains.
Besides reduced elongation, Atxyl1 siliques show an excessive width (Fig. 4B). Addition of polymeric xyloglucan to growing pea segments causes a faster reorientation of microtubules from transversal to longitudinal orientation (Takeda et al., 2002). It would be interesting to investigate if lack of α-xylosidase activity, by increasing microfibril connections, has a similar effect. The transverse orientation of microtubules during elongation helps to maintain a similar orientation for cellulose microfibrils, which appears to constrain increases in cell width (Geitmann and Ortega, 2009). Besides microtubule orientation, other factors that affect organ shape need to be considered, such as tensions between different tissues of the silique or the fact that the reduced growth of the sepals could limit the elongation of the gynoecium.
Xyloglucan from Atxyl1 lines has a reduced proportion of Fuc and Gal, and this could affect its interaction with cellulose. However, mur3 mutants have a comparable xyloglucan composition, with 55% XXXG and 45% XLXG, and they show no elongation defects (Madson et al., 2003). Finally, it is worth mentioning that no morphological phenotype was observed in AtXYL1 antisense lines, even though activity was reduced up to 70%, suggesting that a small amount of α-xylosidase is enough to achieve a superficially normal growth pattern (Monroe et al., 2003). This result suggests that silencing may not be a good approach to study the role of xyloglucan exoglycosidases, even without gene redundancy.
Regulation of α-Xylosidase Activity
Expression of AtXYL1, as revealed by a promoter-reporter fusion (Fig. 5), appears to be widespread, as would be expected for the only α-xylosidase active against xyloglucan. The ubiquity of reporter expression under the control of the AtXYL1 promoter at the organ level is also reflected in published genome-wide microarray analyses (Schmid et al., 2005; Jakoby et al., 2008). Cell type-specific studies confirm the expression of AtXYL1 in guard cells and trichomes as well as a lack of expression in pollen (Pina et al., 2005; Jakoby et al., 2008; Yang et al., 2008). The expression pattern revealed by the promoter-reporter fusion is remarkably similar to that of MUR3, a galactosyltransferase involved in xyloglucan synthesis (Madson et al., 2003). Both promoters show high levels of expression in root stem cells and vascular cylinder, aerial vasculature, trichomes, sepals, and the tip and base of developing siliques. It is possible that these are sites of accelerated xyloglucan turnover with high levels of synthesis and degradation.
The pattern of expression of AtXYL1 revealed by our construct needs to be confirmed by other methods, but some of the locations of high expression can be linked to modifications in the cell wall where xyloglucan metabolism is likely to be involved. It has been proposed, for instance, that dividing root cells in maize (Zea mays) recycle a large amount of cell wall material through endocytosis, including xyloglucan molecules (Baluška et al., 2005). Similarly xyloglucan remodeling by AtXTH28 seems critical for the correct elongation of anther filaments during anthesis (Kurasawa et al., 2009). As for the role of xyloglucan metabolism during vasculature development, AtXTH27 is expressed during secondary wall deposition in xylem and is required for the correct expansion of leaf tracheary elements (Matsui et al., 2005). Additionally, xyloglucan hydrolase activity has been detected in xylem and fibers of Arabidopsis stems using fluorogenic substrates (Ibatullin et al., 2009).
In addition to transcriptional regulation, several posttranscriptional modifications of AtXYL1 could be involved in modulating α-xylosidase activity. In proteomic studies, AtXYL1 sequences are usually obtained from several discrete spots that differ in size, pI, or both (Kwon et al., 2005; Albenne et al., 2009). Some of this heterogeneity can be explained by differences in glycosylation or by an N-terminal propeptide that seems to be lost within the wall (Sampedro et al., 2001; Albenne et al., 2009). More surprising is the fact that some forms of AtXYL1 appear to be phosphorylated and that dephosphorylation by cell wall phosphatases could lead to inactivation (Kwon et al., 2005; Kaida et al., 2010).
α-Xylosidase activity is likely to be the limiting step in the degradation of the xyloglucan backbone in Arabidopsis, since β-glucosidase activity seems to be higher in the apoplast (Iglesias et al., 2006). If α-xylosidase, as we have proposed, can degrade a large amount of polymeric xyloglucan and therefore limit the possible connections between microfibrils, the need for a tight regulation is understandable. Even if xyloglucan is not the only cross-linking polymer that regulates cell wall extension, it is likely to be essential for the precise control of this process in both time and space. We have shown that preventing its normal degradation has a significant effect on the elongation of several organs. Further investigation of α-xylosidase mutants, as well as those affected in other exoglycosidases, offers a new avenue to explore the various and poorly understood roles of xyloglucan in the cell walls.
MATERIALS AND METHODS
Plant Material and Growth Conditions
To identify Arabidopsis (Arabidopsis thaliana) Atxyl1-1 in the T-DNA collection at the University of Wisconsin Biotech Center (Krysan et al., 1999), several rounds of screening were carried out with primers MUT1 and JL-202 (Supplemental Table S1). Atxyl1-2 was ordered from the GABI-KAT collection, where its line identifier is 749G08 (Rosso et al., 2003). Homozygous and wild-type plants for the T-DNA insertions were selected from segregating populations of both mutants. Absence and presence of the insertions were determined by PCR with the primer pairs detailed in Supplemental Table S1. Wild-type and mutant plants were grown on the same tray in 16-h days at 22°C/18°C light/dark temperature and 60 μmol m−2 s−1 light intensity.
Activity Assays
Approximately 1.5 g of 11-d-old rosettes was homogenized in liquid N2. Proteins were extracted for 1 h at 4°C in 9 mL of 50 mm Na-acetate buffer (pH 4.5), 1 m NaCl, 1.5% polyvinylpolypyrrolidone, and 0.1% (v/v) Protease Inhibitor Cocktail for plant cell and tissue extracts (Sigma). After centrifugation, the supernatant was concentrated using Amicon Ultra 30K (Millipore) and washed twice with 10 mL of 20 mm Na-acetate buffer (pH 5.0). Protease inhibitor cocktail at 0.1% (v/v) was added to the second wash. Protein concentration was quantified with Coomassie Plus (Thermo Scientific).
For each assay, 40 μL of protein extract (500–750 μg mL−1) was mixed with 20 μL of either 4.5 mm XXXG or 4.5 mm XLLG (Megazyme). After a 16-h incubation at 37°C, samples were boiled for 5 min. Released Xyl was quantified with the d-Xylose assay kit (Megazyme) and free Gal with the d-Galactose assay kit (Megazyme).
Liquid Culture
Approximately 90 mg of sterile seeds was germinated and grown for 7 d in 25-mL flasks containing 10 mL of medium (0.5× Murashige and Skoog medium with 0.5 g L−1 MES at pH 5.7) in 16-h days at 24°C/20°C light/dark temperature and 25 μmol m−2 s−1 light intensity.
Growth medium was collected, and 500 μL was dialyzed for 24 h against water using a Biodialyser with 500-D membranes (Sigma). After freeze drying, the samples were resuspended in 50 μL of 20 mm Na-acetate buffer (pH 5.0) and analyzed through MALDI-TOF MS.
Mass Spectrometry
Samples were mixed with an equal volume of 2,5-dihydroxybenzoic acid solution (10 mg mL−1 in 70% acetonitrile). For analyses of liquid medium, maltoheptaose was added to 50 μm final concentration. MALDI-TOF spectra were obtained with an UltraFlex mass spectrometer (Bruker Daltonics) equipped with a Smartbeam pulsed UV laser (337 nm) and operating in positive-ion mode. Ions were accelerated to a kinetic energy of 25 kV. Each spectrum was obtained by summing 450 shots in reflectron mode. For MALDI-TOF/TOF spectra, precursor ions were accelerated to 8 kV and selected in a timed ion gate. Fragment ions generated by laser-induced decomposition of the precursors were further accelerated by 19 kV in the LIFT cell.
Xyloglucan Composition Analysis
After removing the top 6 cm, 12 mature green stems (approximately 35 cm) were homogenized in liquid nitrogen. The powder was boiled in 8 mL of ethanol for 10 min, washed six times with ethanol and twice with diethyl ether, and then dried. Xyloglucan extractions were then performed in duplicate for each sample. Five milligrams of cell wall residue was resuspended in 400 μL of 10 mm pyridine-acetate buffer at pH 4.5 and then washed four times. Accessible xyloglucan was extracted with 4 units of endocellulase from Trichoderma longibranchiatum (Megazyme) in an overnight digestion at 37°C. Undigested material was washed twice with water and then extracted for 24 h with 900 μL of 24% KOH and 0.1% NaBH4. After centrifugation, the supernatant was neutralized with 300 μL of acetic acid. Extracted cell wall material was filtered through a 10K centrifugal filter (VWR), repeatedly washed with 10 mm pyridine-acetate buffer at pH 4.5, and digested overnight with 4 units of endocellulase. Reducing sugars were determined with the p-hydroxybenzoic acid hydrazide assay (Lever, 1972). Samples were finally dried and resuspended in 20 mm Na-acetate buffer (pH 5.0) to a concentration of 3 mm reducing sugars.
DNA Cloning
AtXYL2 was amplified from Wassilewskija genomic DNA using PrimeSTAR HS DNA Polymerase (Takara) and primers XYL2L1 and XYL2R1 (Supplemental Table S1). The PCR product was cloned using the Zero Blunt TOPO PCR Cloning Kit for Sequencing (Invitrogen). Two independent clones were sequenced, and only confirmed differences from Columbia were considered.
A 3-kb fragment of the AtXYL1 promoter starting 40 bp from the ATG was amplified with primers PRXYL1L1 and PRXYL1R1. The fragment was introduced into vector pDONR/Zeo through BP recombination (Invitrogen). Through LR recombination, it was subsequently cloned upstream of the GUS gene in vector pMDC162 (Curtis and Grossniklaus, 2003).
Generation and Characterization of Transgenic Plants
The promoter:GUS construct was introduced into Agrobacterium tumefaciens EHA105 by electroporation. Arabidopsis Columbia plants were transformed through floral dipping (Clough and Bent, 1998). A total of 16 transgenic plants were recovered in T1, most with similar expression patterns. Three lines were selected, and T2 plants were analyzed at different stages. GUS activity was detected by incubation in a solution of 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 100 mm Na-phosphate buffer (pH 7.0), 10 mm EDTA, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, and 0.1% Triton X-100.
The sequence of AtXYL2 in Wassilewskija can be found in GenBank (accession no. GQ397971).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequences of φAtxyl2 in Wassilewskija and Columbia.
Supplemental Figure S2. MALDI-TOF/TOF spectra of oligosaccharides in Atxyl1-2 growth medium.
Supplemental Figure S3. MALDI-TOF/TOF spectrum of the peak at m/z 1,247 in Atxyl1-2 stems.
Supplemental Figure S4. Silique development in Atxyl1-1.
Supplemental Table S1. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Elene Ruth Valdivia for revising the manuscript.
References
- Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E. (2009) Plant cell wall proteomics: mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2: 977–989 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F, Liners F, Hlavacka A, Schlicht M, Van Cutsem P, McCurdy DW, Menzel D. (2005) Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225: 141–155 [DOI] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklöf JM, Michel G, Kallas AM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Crombie HJ, Chengappa S, Hellyer A, Reid JSG. (1998) A xyloglucan oligosaccharide-active, transglycosylating beta-D-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L) seedlings: purification, properties and characterization of a cDNA clone. Plant J 15: 27–38 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alcântara PHN, Dietrich SMC, Buckeridge MS. (1999) Xyloglucan mobilisation and purification of a (XLLG/XLXG) specific β-galactosidase from cotyledons of Copaifera langsdorffii. Plant Physiol Biochem 37: 653–663 [Google Scholar]
- de Alcântara PHN, Martim L, Silva CO, Dietrich SMC, Buckeridge MS. (2006) Purification of a beta-galactosidase from cotyledons of Hymenaea courbaril L. (Leguminosae): enzyme properties and biological function. Plant Physiol Biochem 44: 619–627 [DOI] [PubMed] [Google Scholar]
- Edwards M, Bowman YJ, Dea IC, Reid JS. (1988) A beta-D-galactosidase from nasturtium (Tropaeolum majus L.) cotyledons: purification, properties, and demonstration that xyloglucan is the natural substrate. J Biol Chem 263: 4333–4337 [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. (1991) A xyloglucan-oligosaccharide-specific α-D-xylosidase or exo-oligoxyloglucan-α-xylohydrolase from germinated nasturtium (Tropaeolum majus L.) seeds. Planta 184: 137–147 [DOI] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. (1996) Substrate subsite recognition of the xyloglucan endo-transglycosylase or xyloglucan-specific endo-(1→4)-β-D-glucanase from the cotyledons of germinated nasturtium (Tropaeolum majus L.) seeds. Planta 200: 221–228 [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, Mort AJ, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Geitmann A, Ortega JKE. (2009) Mechanics and modeling of plant cell growth. Trends Plant Sci 14: 467–478 [DOI] [PubMed] [Google Scholar]
- Guillén R, York WS, Pauly M, An J, Impallomeni G, Albersheim P, Darvill AG. (1995) Metabolism of xyloglucan generates xylose-deficient oligosaccharide subunits of this polysaccharide in etiolated peas. Carbohydr Res 277: 291–311 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7: 637–644 [DOI] [PubMed] [Google Scholar]
- Ibatullin FM, Banasiak A, Baumann MJ, Greffe L, Takahashi J, Mellerowicz EJ, Brumer H. (2009) A real-time fluorogenic assay for the visualization of glycoside hydrolase activity in planta. Plant Physiol 151: 1741–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Abelenda JA, Rodiño M, Sampedro J, Revilla G, Zarra I. (2006) Apoplastic glycosidases active against xyloglucan oligosaccharides of Arabidopsis thaliana. Plant Cell Physiol 47: 55–63 [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148: 1583–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF. (2006) Cell wall proteins: a new insight through proteomics. Trends Plant Sci 11: 33–39 [DOI] [PubMed] [Google Scholar]
- Johansson P, Brumer H, III, Baumann MJ, Kallas AM, Henriksson H, Denman SE, Teeri TT, Jones TA. (2004) Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Serada S, Norioka N, Norioka S, Neumetzler L, Pauly M, Sampedro J, Zarra I, Hayashi T, Kaneko TS. (2010) Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol 153: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa K, Matsui A, Yokoyama R, Kuriyama T, Yoshizumi T, Matsui M, Suwabe K, Watanabe M, Nishitani K. (2009) The AtXTH28 gene, a xyloglucan endotransglucosylase/hydrolase, is involved in automatic self-pollination in Arabidopsis thaliana. Plant Cell Physiol 50: 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HK, Yokoyama R, Nishitani K. (2005) A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol 46: 843–857 [DOI] [PubMed] [Google Scholar]
- Léonard R, Pabst M, Bondili JS, Chambat G, Veit C, Strasser R, Altmann F. (2008) Identification of an Arabidopsis gene encoding a GH95 alpha1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry 69: 1983–1988 [DOI] [PubMed] [Google Scholar]
- Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47: 273–279 [DOI] [PubMed] [Google Scholar]
- Lopez-Casado G, Urbanowicz BR, Damasceno CM, Rose JK. (2008) Plant glycosyl hydrolases and biofuels: a natural marriage. Curr Opin Plant Biol 11: 329–337 [DOI] [PubMed] [Google Scholar]
- Lovering AL, Lee SS, Kim YW, Withers SG, Strynadka NCJ. (2005) Mechanistic and structural analysis of a family 31 α-glycosidase and its glycosyl-enzyme intermediate. J Biol Chem 280: 2105–2115 [DOI] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K. (2009) Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J Exp Bot 60: 3959–3972 [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K. (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42: 525–534 [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC. (1990) Xyloglucan oligosaccharides promote growth and activate cellulase: evidence for a role of cellulase in cell expansion. Plant Physiol 93: 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Garcia-Cazarin ML, Poliquin KA, Aivano SK. (2003) Antisense Arabidopsis plants indicate that an apoplastic alpha-xylosidase and alpha-glucosidase are encoded by the same gene. Plant Physiol Biochem 41: 877–885 [Google Scholar]
- Moracci M, Cobucci Ponzano B, Trincone A, Fusco S, De Rosa M, van Der Oost J, Sensen CW, Charlebois RL, Rossi M. (2000) Identification and molecular characterization of the first alpha-xylosidase from an archaeon. J Biol Chem 275: 22082–22089 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Vissenberg K. (2007) Roles of the XTH protein family in the expanding cell. Verbelen JP, Vissenberg K, , The Expanding Cell. Springer-Verlag, Berlin, pp 89–116 [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M. (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Obel N, Neumetzler L, Pauly M. (2007) Hemicelluloses and cell expansion. Verbelen JP, Vissenberg K, , The Expanding Cell. Springer-Verlag, Berlin, pp 57–88 [Google Scholar]
- O’Neill RA, Albersheim P, Darvill AG. (1989) Purification and characterization of a xyloglucan oligosaccharide-specific xylosidase from pea seedlings. J Biol Chem 264: 20430–20437 [PubMed] [Google Scholar]
- Pauly M, Qin Q, Greene H, Albersheim P, Darvill A, York WS. (2001) Changes in the structure of xyloglucan during cell elongation. Planta 212: 842–850 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Darvill AG, Eberhard S, York WS, O’Neill MA. (2008) Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 18: 891–904 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. (2008) Xyloglucan-pectin linkages are formed intra-protoplasmically, contribute to wall-assembly, and remain stable in the cell wall. Planta 227: 781–794 [DOI] [PubMed] [Google Scholar]
- Reid JSG, Edwards M, Dea ICM. (1988) Enzymatic modification of natural seed gums. Phillips GO, Wedlock DJ, Williams PA, , Gums and Stabilisers for the Food Industry. IRL Press, Oxford, pp 391–398 [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Sieiro C, Revilla G, González-Villa T, Zarra I. (2001) Cloning and expression pattern of a gene encoding an alpha-xylosidase active against xyloglucan oligosaccharides from Arabidopsis. Plant Physiol 126: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T. (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45: 672–683 [DOI] [PubMed] [Google Scholar]
- Sulová Z, Baran R, Farkaš V. (2001) Release of complexed xyloglucan endotransglycosylase (XET) from plant cell walls by a transglycosylation reaction with xyloglucan-derived oligosaccharides. Plant Physiol Biochem 39: 927–932 [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA 99: 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiné MAS, Silva CO, de Lima DU, Carpita NC, Buckeridge MS. (2006) Fine structure of a mixed-oligomer storage xyloglucan from seeds of Hymenaea courbaril. Carbohydr Polym 66: 444–454 [Google Scholar]
- Trincone A, Cobucci-Ponzano B, Di Lauro B, Rossi M, Mitsuishi Y, Moracci M. (2001) Enzymatic synthesis and hydrolysis of xylogluco-oligosaccharides using the first archaeal alpha-xylosidase from Sulfolobus solfataricus. Extremophiles 5: 277–282 [DOI] [PubMed] [Google Scholar]
- Trincone A, Giordano A. (2006) Glycosyl hydrolases and glycosyltransferases in the synthesis of oligosaccharides. Curr Org Chem 10: 1163–1193 [Google Scholar]
- Vincken JP, York WS, Beldman G, Voragen AGJ. (1997) Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol 114: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagaki T, Mitsuishi Y, Nakanishi H. (1998) Determination of structural isomers of xyloglucan octasaccharides using post-source decay fragment analysis in MALDI-TOF mass spectrometry. Tetrahedron Lett 39: 4051–4054 [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.