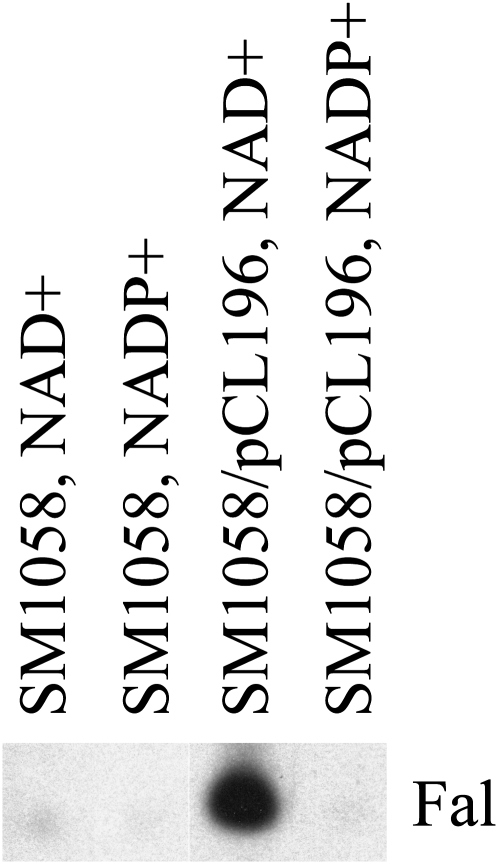Abstract
In Arabidopsis (Arabidopsis thaliana), farnesylcysteine is oxidized to farnesal and cysteine by a membrane-associated thioether oxidase called farnesylcysteine lyase. Farnesol and farnesyl phosphate kinases have also been reported in plant membranes. Together, these observations suggest the existence of enzymes that catalyze the interconversion of farnesal and farnesol. In this report, Arabidopsis membranes are shown to possess farnesol dehydrogenase activity. In addition, a gene on chromosome 4 of the Arabidopsis genome (At4g33360), called FLDH, is shown to encode an NAD+-dependent dehydrogenase that oxidizes farnesol more efficiently than other prenyl alcohol substrates. FLDH expression is repressed by abscisic acid (ABA) but is increased in mutants with T-DNA insertions in the FLDH 5′ flanking region. These T-DNA insertion mutants, called fldh-1 and fldh-2, are associated with an ABA-insensitive phenotype, suggesting that FLDH is a negative regulator of ABA signaling.
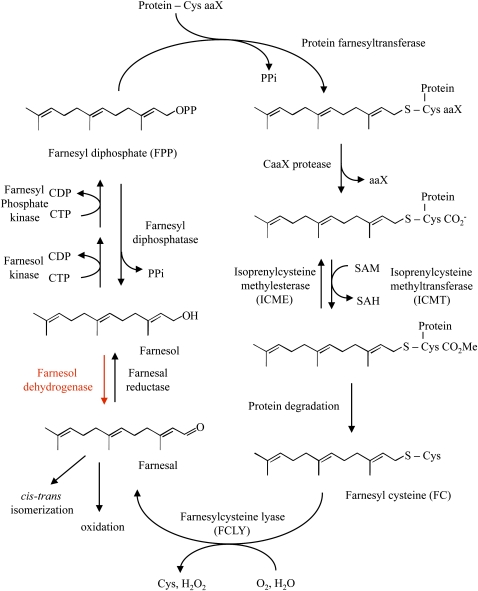
Isoprenylated proteins are modified at the C terminus via cysteinyl thioether linkage to either a 15-carbon farnesyl or a 20-carbon geranylgeranyl group (Clarke, 1992; Zhang and Casey, 1996; Rodríguez-Concepción et al., 1999; Crowell, 2000; Crowell and Huizinga, 2009). These modifications mediate protein-membrane and protein-protein interactions and are necessary for the proper localization and function of hundreds of proteins in eukaryotic cells. In Arabidopsis (Arabidopsis thaliana), the PLURIPETALA (PLP; At3g59380) and ENHANCED RESPONSE TO ABA1 (At5g40280) genes encode the α- and β-subunits of protein farnesyltransferase (PFT), respectively (Cutler et al., 1996; Pei et al., 1998; Running et al., 2004). These subunits form a heterodimeric zinc metalloenzyme that catalyzes the efficient transfer of a farnesyl group from farnesyl diphosphate to protein substrates with a C-terminal CaaX motif, where “C” is Cys, “a” is an aliphatic amino acid, and “X” is usually Met, Gln, Cys, Ala, or Ser (Fig. 1). The PLP and GERANYLGERANYL-TRANSFERASE BETA (At2g39550) genes encode the α- and β-subunits of protein geranylgeranyltransferase type 1 (PGGT1), respectively (Running et al., 2004; Johnson et al., 2005). These subunits form a distinct heterodimeric zinc metalloenzyme that catalyzes the efficient transfer of a geranylgeranyl group from geranylgeranyl diphosphate to protein substrates with a C-terminal CaaL motif, where “C” is Cys, “a” is an aliphatic amino acid, and “L” is Leu. A third protein prenyltransferase, called protein geranylgeranyltransferase type II or RAB geranylgeranyltransferase, catalyzes the dual geranylgeranylation of RAB proteins with a C-terminal XCCXX, XXCXC, XXCCX, XXXCC, XCXXX, or CCXXX motif, where “C” is Cys and “X” is any amino acid. However, RAB proteins must be associated with the RAB ESCORT PROTEIN to be substrates of RAB geranylgeranyltransferase. Plant protein prenylation has received considerable attention in recent years because of the meristem defects of Arabidopsis PFT mutants and the abscisic acid (ABA) hypersensitivity of Arabidopsis PFT and PGGT1 mutants (Cutler et al., 1996; Pei et al., 1998; Running et al., 1998, 2004; Johnson et al., 2005).
Figure 1.
Proposed metabolism of farnesal and farnesol as it relates to protein prenylation. The portion of the cycle shown in red is the subject of this article.
Proteins that are prenylated by either PFT or PGGT1 undergo further processing in the endoplasmic reticulum (Crowell, 2000; Crowell and Huizinga, 2009). First, the aaX portion of the CaaX motif is removed by proteolysis (Fig. 1). This reaction is catalyzed by one of two CaaX endoproteases, which are encoded by the AtSTE24 (At4g01320) and AtFACE-2 (At2g36305) genes (Bracha et al., 2002; Cadiñanos et al., 2003). Second, the prenylated Cys residue at the new C terminus is methylated by one of two isoprenylcysteine methyltransferases (Fig. 1), which are encoded by the AtSTE14A (At5g23320) and AtSTE14B (ICMT; At5g08335) genes (Crowell et al., 1998; Crowell and Kennedy, 2001; Narasimha Chary et al., 2002; Bracha-Drori et al., 2008). A specific isoprenylcysteine methylesterase encoded by the Arabidopsis ICME (At5g15860) gene has also been described, demonstrating the reversibility of isoprenylcysteine methylation (Deem et al., 2006; Huizinga et al., 2008).
Like all proteins, prenylated proteins have a finite half-life. However, unlike other proteins, prenylated proteins release farnesylcysteine (FC) or geranylgeranylcysteine (GGC) upon degradation. Mammals possess a prenylcysteine lyase enzyme that catalyzes the oxidative cleavage of FC and GGC (Zhang et al., 1997; Tschantz et al., 1999; Tschantz et al., 2001; Beigneux et al., 2002; Digits et al., 2002). This FAD-dependent thioether oxidase consumes molecular oxygen and generates hydrogen peroxide, Cys, and a prenyl aldehyde product (i.e. farnesal or geranylgeranial). In Arabidopsis, a similar lyase exists. However, the Arabidopsis enzyme, which is encoded by the FCLY (At5g63910) gene, is specific for FC (Fig. 1; Crowell et al., 2007; Huizinga et al., 2010). GGC is metabolized by a different mechanism.
Plant membranes have been shown to contain farnesol kinase, geranylgeraniol kinase, farnesyl phosphate kinase, and geranylgeranyl phosphate kinase activities (Fig. 1; Thai et al., 1999). These membrane-associated kinases differ with respect to nucleotide specificity, suggesting that they are distinct enzymes (i.e. farnesol kinase and geranylgeraniol kinase can use CTP, UTP, or GTP as a phosphoryl donor, whereas farnesyl phosphate kinase and geranylgeranyl phosphate kinase exhibit specificity for CTP as a phosphoryl donor). However, it remains unclear if farnesol kinase is distinct from geranylgeraniol kinase or if farnesyl phosphate kinase is distinct from geranylgeranyl phosphate kinase. Nonetheless, it is clear that these kinases convert farnesol and geranylgeraniol to their monophosphate and diphosphate forms for use in isoprenoid biosynthesis, including sterol biosynthesis and protein prenylation.
Because plants have the metabolic capability to generate farnesal from FC and farnesyl diphosphate from farnesol, we considered the possibility that plant membranes also contain an oxidoreductase capable of catalyzing the reduction of farnesal to farnesol and/or the oxidation of farnesol to farnesal (Fig. 1; Thai et al., 1999; Crowell et al., 2007). To date, the only reports of such an oxidoreductase are from the corpora allata glands of insects, where it participates in juvenile hormone synthesis, and black rot fungus-infected sweet potato (Ipomoea batatas; Baker et al., 1983; Inoue et al., 1984; Sperry and Sen, 2001; Mayoral et al., 2009). Insect farnesol dehydrogenase is an NADP+-dependent oxidoreductase that is encoded by a subfamily of short-chain dehydrogenase/reductase (SDR) genes (Mayoral et al., 2009). Farnesol dehydrogenase from sweet potato is a 90-kD, NADP+-dependent homodimer with broad specificity for prenyl alcohol substrates and is induced by wounding and fungus infection of potato roots (Inoue et al., 1984).
Here, we extended previous work in which [1-3H]FC was shown to be oxidized to [1-3H]farnesal, and [1-3H]farnesal reduced to [1-3H]farnesol, in the presence of Arabidopsis membranes (Crowell et al., 2007). The reduction of [1-3H]farnesal to [1-3H]farnesol was abolished by pretreatment of Arabidopsis membranes with NADase, suggesting that sufficient NAD(P)H is present in Arabidopsis membranes to support the enzymatic reduction of farnesal to farnesol. In this report, we demonstrate the presence of farnesol dehydrogenase activity in Arabidopsis membranes using [1-3H]farnesol as a substrate. Moreover, we identify a gene on chromosome 4 of the Arabidopsis genome (At4g33360), called FLDH, that encodes an NAD+-dependent dehydrogenase with partial specificity for farnesol as a substrate. FLDH expression is repressed by exogenous ABA, and fldh mutants exhibit altered ABA signaling. Taken together, these observations suggest that ABA regulates farnesol metabolism in Arabidopsis, which in turn regulates ABA signaling.
RESULTS
Farnesol Dehydrogenase Activity in Arabidopsis Membranes
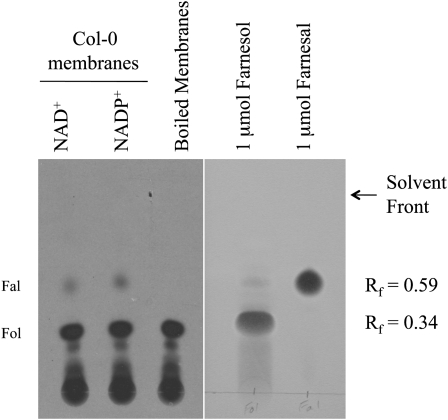
Following the oxidation of FC to farnesal, farnesal is reduced to farnesol, which can be sequentially phosphorylated to farnesyl diphosphate (Fig. 1; Thai et al., 1999; Crowell et al., 2007). We detected the conversion of [1-3H]farnesal to [1-3H]farnesol in the presence of Arabidopsis membranes and showed that this activity is abolished by NADase pretreatment (Crowell et al., 2007). In contrast, NADase does not abolish FC oxidation to farnesal, confirming the reaction order (i.e. FC oxidation to farnesal, followed by farnesal reduction to farnesol). These observations strongly suggest the existence of an NAD(P)H-dependent farnesal reductase/NAD(P)+-dependent farnesol dehydrogenase enzyme in Arabidopsis. To examine this oxidoreductase activity further, and to test the reversibility of the reaction, we used calf intestine alkaline phosphatase to dephosphorylate [1-3H]farnesyl diphosphate and then incubated the reaction mixture at 30°C for 30 min in the presence of either native or boiled Arabidopsis membranes and either 0.1 mm NAD+ or 0.1 mm NADP+. Reactions were resolved by thin-layer chromatography (TLC) and analyzed by fluorography. As shown in Figure 2, alkaline phosphatase treatment of [1-3H]FPP generated significant amounts of [1-3H]farnesol, which was not converted to farnesal in the presence of boiled Arabidopsis membranes. However, in the presence of native Arabidopsis membranes and either NAD+ or NADP+, [1-3H]farnesol was oxidized to [1-3H]farnesal, and both substrate and product comigrated with authentic chemical standards. It is important to note that since oxidation of [1-3H]farnesol to [1-3H]farnesal involved the loss of a hydrogen atom at the 1-position, only 50% of the farnesal product was expected to be radioactive. Furthermore, although oxidation of [1-3H]farnesol was observed in the presence of exogenous NAD+ or NADP+, Arabidopsis membranes contained sufficient cofactor to support oxidation of farnesol (Supplemental Fig. S1; Crowell et al., 2007). Thus, it is not clear from these results if the farnesol dehydrogenase activity, or activities, in Arabidopsis membranes use NAD+, NADP+, or both.
Figure 2.
Oxidation of [1-3H]farnesol to [1-3H]farnesal in the presence of Arabidopsis membranes. [1-3H]Farnesol was generated by calf intestine alkaline phosphatase digestion of [1-3H]farnesyl diphosphate and subsequently incubated with native or boiled Arabidopsis membranes in the presence of 0.1 mm NAD+ or NADP+ for 30 min at 30°C. Reaction products were resolved by silica gel TLC, visualized by fluorography (left), and compared to authentic chemical standards, which were visualized by vanillin staining (right). Retardation factor (Rf) values for farnesal (Rf = 0.59) and farnesol (Rf = 0.34) are indicated. The radioactivity at the origin is [1-3H]farnesyl diphosphate. Fal, Farnesal; Fol, farnesol.
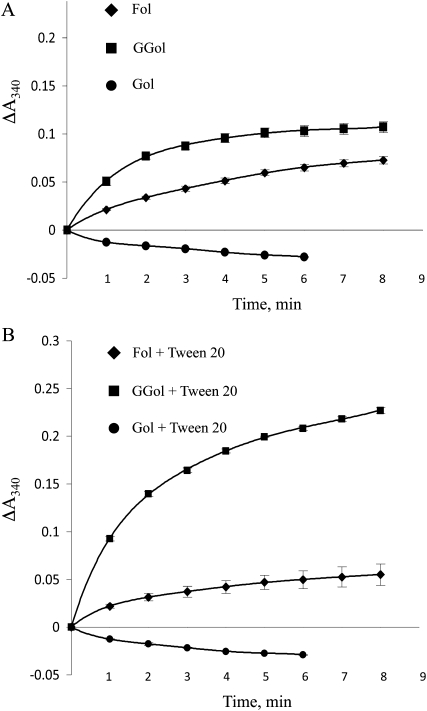
Farnesol dehydrogenase activity in Arabidopsis membranes was analyzed spectrophotometrically at 340 nm (i.e. NADH and NADPH absorb at 340 nm). As shown in Figure 3, reduced cofactor was formed in the presence of 1 mm farnesol and 1 mm geranylgeraniol but not in the presence of 1 mm geraniol (the negative slope in the presence of geraniol suggests oxidation of endogenous NADH or NADPH). These data demonstrate that Arabidopsis farnesol dehydrogenase activity is (1) linear with time for 2 min under these conditions, (2) present in Arabidopsis membranes at a specific activity >10 nmol min−1 mg−1, and (3) specific for biologically relevant prenyl alcohol substrates (farnesol and geranylgeraniol). Similar results were obtained with 0.1 mm NAD+ and 0.1 mm NADP+ as a cofactor.
Figure 3.
Arabidopsis farnesol dehydrogenase is specific for biologically relevant prenyl alcohol substrates. A, Farnesol dehydrogenase reactions were performed in the presence of 1 mm farnesol, geranylgeraniol, or geraniol, and reduced cofactor was detected spectrophotometrically at 340 nm as a function of time. Reduced cofactor production was linear for 2 min and was detected in the presence of farnesol or geranylgeraniol as an isoprenoid substrate. Farnesol dehydrogenase activity was determined to be 10 nmol min−1 mg−1. B, Farnesol dehydrogenase assays were performed as in A in the presence of 0.1% Tween 20. These data are representative of three independent experiments. The se of the mean is shown. Fol, Farnesol; GGol, geranylgeraniol; Gol, geraniol.
Because farnesol and geranylgeraniol are hydrophobic molecules and might not be homogeneously mixed into the reactions described above, we performed an identical set of farnesol dehydrogenase reactions in the presence of 0.1% Tween 20. As shown in Figure 3, 0.1% Tween 20 enhanced the oxidation of geranylgeraniol, suggesting increased dispersion and use of geranylgeraniol, but slightly inhibited the oxidation of farnesol. Because our interest is in the metabolism of farnesal and farnesol, no further reactions were performed in the presence of detergent.
Identification of an Arabidopsis Farnesol Dehydrogenase Gene
To date, farnesol dehydrogenase activity has only been described in insect corpora allata glands and black rot fungus-infected potato (Baker et al., 1983; Inoue et al., 1984; Sperry and Sen, 2001; Mayoral et al., 2009). Moreover, the only gene known to encode a protein with farnesol dehydrogenase activity belongs to the short-chain dehydrogenase gene family from mosquito (AaSDR-1; Mayoral et al., 2009). A search for Arabidopsis genes encoding proteins with significant amino acid sequence similarity to the protein encoded by the mosquito AaSDR-1 gene revealed a single gene on chromosome 5 (At5g04900), called AtNOL1, with weak similarity (E = 3e−20). However, the orthologous NOL gene from rice (Oryza sativa) encodes a chlorophyll b reductase that is involved in the degradation of chlorophyll b and light-harvesting complex II (Kusaba et al., 2007). Because this enzyme reduces chlorophyll b to 7-hydroxymethyl chlorophyll a, it is unlikely to be a bona fide farnesol dehydrogenase.
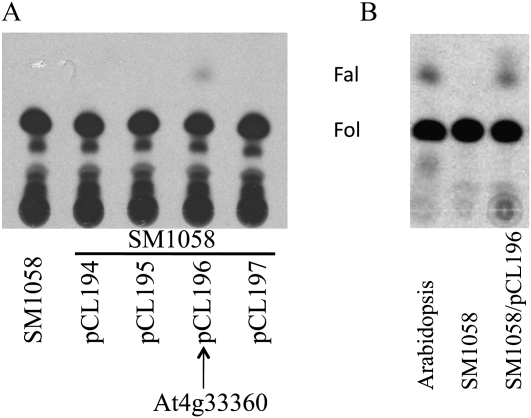
To identify a putative farnesol dehydrogenase gene from Arabidopsis, we searched for genes encoding alcohol dehydrogenases and related oxidoreductases [i.e. with a Rossmann-fold motif or InterPro motif for NAD(P)H binding] that were predicted or known to be membrane localized. This resulted in a large number of candidate genes. We then searched for genes predicted to encode terpenoid metabolic enzymes (search terms included terpene, terpenoid, and isoprenoid) and considered the intersection of this group of genes with the group of membrane-localized oxidoreductases described above. This strategy resulted in a manageable number of candidate genes (Table I), including one member of the Arabidopsis SDR gene family (At3g61220). To determine which gene(s) in this group might encode farnesol dehydrogenase, we amplified the coding sequences of At5g16990, At5g16960, At4g33360, and At3g61220 by reverse transcription (RT)-PCR and inserted the resulting DNA fragments into the pYES2.1/V5-His-TOPO vector (At2g43420 was not included because it was considered to be less likely to encode a farnesol dehydrogenase). After confirming the orientations and DNA sequences of the four coding regions, the resulting plasmids, called pCL194 (At5g16990), pCL195 (At5g16960), pCL196 (At4g33360), and pCL197 (At3g61220), were introduced into Saccharomyces cerevisiae strain SM1058 (MATa leu2, ura3, trp1, his4, can1), and recombinant yeast cells were selected on CSM-ura agar medium. Transformed and untransformed yeast were then grown at 30°C to log phase in medium containing 2% Glc and shifted into medium containing 2% Gal for an additional 14 h. Cells were lysed and membranes assayed for farnesol dehydrogenase activity as described above. As shown in Figure 4, membranes from control yeast cells or recombinant yeast cells harboring pCL194, pCL195, or pCL197 exhibited no farnesol dehydrogenase activity. However, membranes from recombinant yeast cells harboring pCL196, which contained the At4g33360 coding sequence, converted [1-3H]farnesol to [1-3H]farnesal. To our knowledge, this is the first demonstration of a gene that encodes a plant farnesol dehydrogenase and has been submitted to The Arabidopsis Information Resource with the gene class symbol FLDH. Interestingly, the protein product of the FLDH gene exhibited only 12% amino acid sequence identity with the protein product of the AaSDR-1 gene from mosquito.
Table I. Candidate farnesol dehydrogenase genes from Arabidopsis.
SUBA, Subcellular Location Database for Arabidopsis Proteins.
| Arabidopsis Genome Initiative Name | Annotation | Biochemical Function | Structural Features | Location (Proteomic Analyses): SUBA |
| At4g33360 | Terpene-cyclase/mutase-related; similar to cinnamyl (allyl) alcohol dehydrogenase | Terpenoid metabolic function | InterPro NAD(P)+-binding | Endoplasmic reticulum, plasma membrane, vacuole |
| At3g61220 | SDR | (−)-Menthol dehydrogenase activity | Interpro NAD(P)+-binding Interpro SDR | Plasma membrane |
| At5g16990, At5g16960, etc. | 2-Alkenal reductase; similar to allyl alcohol dehydrogenase | Alkanal + NAD(P)+ → alk-2-enal + NAD(P)H | Rossman-fold: NAD(P)+-binding | Plasma membrane |
| At2g43420 | Similar to 3-β-hydroxysteroid dehydrogenase | Steroid biosynthesis | Rossman-fold: NAD(P)+-binding | Plasma membrane |
Figure 4.
At4g33360 (FLDH) encodes an enzyme with farnesol dehydrogenase activity. A, Membranes from SM1058 yeast cells with or without pCL194 (At5g16990), pCL195 (At5g16960), pCL196 (At4g33360), or pCL197 (At3g61220) expression constructs were assayed for farnesol dehydrogenase activity using dephosphorylated [1-3H]FPP as a substrate. B, Membranes from Arabidopsis seedlings, SM1058 cells, and SM1058/pCL196 cells were assayed for farnesol dehydrogenase activity using [1-3H]farnesol from American Radiolabeled Chemicals, which was purified by preparative TLC. Fal, Farnesal; Fol, farnesol.
Because alkaline phosphatase treatment of [1-3H]farnesyl diphosphate resulted in partial dephosphorylation, the reaction observed in the presence of membranes from SM1058 cells harboring the pCL196 plasmid was not well defined. Accordingly, we performed farnesol dehydrogenase reactions in the presence of TLC-purified [1-3H]farnesol. As shown in Figure 4B, incubation of purified [1-3H]farnesol with Arabidopsis membranes or membranes from SM1058 cells transformed with the pCL196 plasmid resulted in oxidation of [1-3H]farnesol to [1-3H]farnesal. However, no farnesol dehydrogenase activity was observed in the presence of membranes from control SM1058 cells.
Characterization of the FLDH-Encoded Farnesol Dehydrogenase
To determine whether the FLDH-encoded enzyme was NAD+ or NADP+ dependent, farnesol dehydrogenase reactions were performed in the presence of membranes from control and recombinant yeast cells harboring the pCL196 plasmid. As shown in Figure 5, very little purified [1-3H]farnesol was oxidized to [1-3H]farnesal in the presence of control membranes. However, in the presence of membranes from recombinant yeast cells expressing FLDH, [1-3H]farnesol was oxidized to [1-3H]farnesal in the presence of NAD+. No oxidation was observed in the presence of NADP+. These results indicate that, unlike the farnesol dehydrogenase detected in insect corpora allata glands and black rot fungus-infected sweet potato, the FLDH-encoded farnesol dehydrogenase is specific for NAD+ (endogenous NAD+ present in Arabidopsis membranes most likely accounts for the results obtained in Fig. 2 with NADP+, but an alternative farnesol dehydrogenase is also possible).
Figure 5.
The FLDH-encoded farnesol dehydrogenase is NAD+ dependent. [1-3H]farnesol from American Radiolabeled Chemicals was used to analyze farnesol dehydrogenase activity in membranes of SM1058 and SM1058/pCL196 (At4g33360, FLDH) cells in the presence of NAD+ or NADP+ as cofactor. Fal, Farnesal.
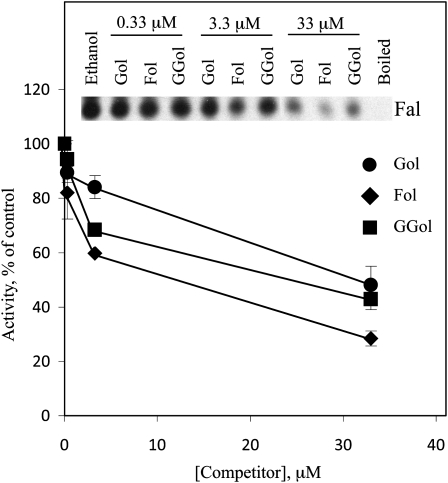
The farnesol dehydrogenase detected in black rot fungus-infected sweet potato exhibited broad specificity for prenyl alcohol substrates. To determine whether the FLDH-encoded farnesol dehydrogenase also exhibited broad substrate specificity, we performed farnesol dehydrogenase assays with membranes from SM1058/pCL196 cells in the presence of unlabeled farnesol, geranylgeraniol, or geraniol as competitors. As shown in Figure 6, unlabeled farnesol was a more effective competitor than geraniol or geranylgeraniol, suggesting that farnesol has the highest affinity for the active site of the FLDH-encoded enzyme. However, geraniol and geranylgeraniol were competitive, indicating that the farnesol dehydrogenase encoded by the FLDH gene exhibits broad specificity for prenyl alcohol substrates.
Figure 6.
The FLDH-encoded farnesol dehydrogenase exhibits partial specificity for farnesol in competition assays. [1-3H]farnesol from American Radiolabeled Chemicals was used to analyze farnesol dehydrogenase activity in membranes from SM1058/pCL196 cells in the presence of NAD+ and unlabeled farnesol, geranylgeraniol, or geraniol as competitor. Ethanol was included as a solvent control. Each competitor was used at 0.33, 3.3, and 33 μm. These results are representative of two independent experiments. The se of the mean is shown. Fol, Farnesol; GGol, geranylgeraniol; Gol, geraniol.
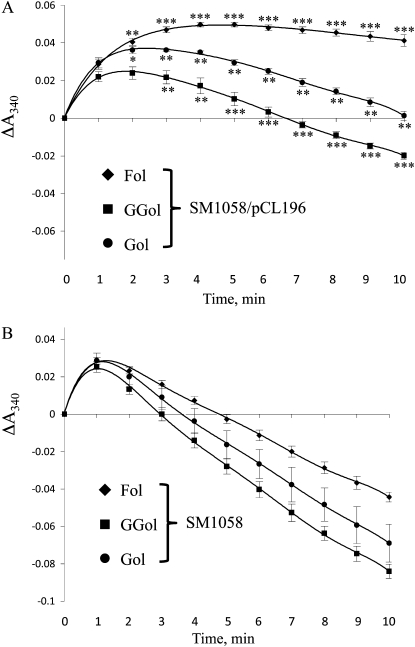
Membranes from control SM1058 cells and recombinant SM1058 cells harboring pCL196 were also analyzed spectrophotometrically at 340 nm. As shown in Figure 7, membranes from control cells, when incubated with 0.1 mm NAD+ and either 1 mm farnesol, geranylgeraniol, or geraniol, exhibited an initial increase in A340, after which absorbance values declined, suggesting oxidation of endogenous NADH and/or NADPH. In contrast, membranes from SM1058/pCL196 cells exhibited less of a decline in absorbance. Consistent with the results shown in Figure 6, which indicate that unlabeled farnesol is more competitive than geranylgeraniol or geraniol in the presence of the FLDH-encoded enzyme, A340 increased and remained elevated in the presence of farnesol. Together, these data demonstrate that FLDH encodes an NAD+-dependent farnesol dehydrogenase enzyme with partial specificity for farnesol. Surprisingly, the FLDH-encoded enzyme does not exhibit appreciable farnesal reductase activity (Supplemental Fig. S2).
Figure 7.
The FLDH-encoded farnesol dehydrogenase exhibits partial specificity for farnesol in spectrophotometric assays. A, Farnesol dehydrogenase reactions were performed in the presence of 1 mm farnesol, geranylgeraniol, or geraniol, and reduced cofactor was detected spectrophotometrically at 340 nm as a function of time. SM1058/pCL196 membranes were used as a source of farnesol dehydrogenase activity. B, Farnesol dehydrogenase assays were performed as in A using SM1058 membranes. These data are representative of two independent experiments. The se of the mean is shown. *, **, and *** represent significant differences compared with the SM1058 control of P < 0.05, P < 0.01, and P < 0.001, respectively, as determined by Student’s t test. Fol, Farnesol; GGol, geranylgeraniol; Gol, geraniol.
ABA Regulation of FLDH Expression
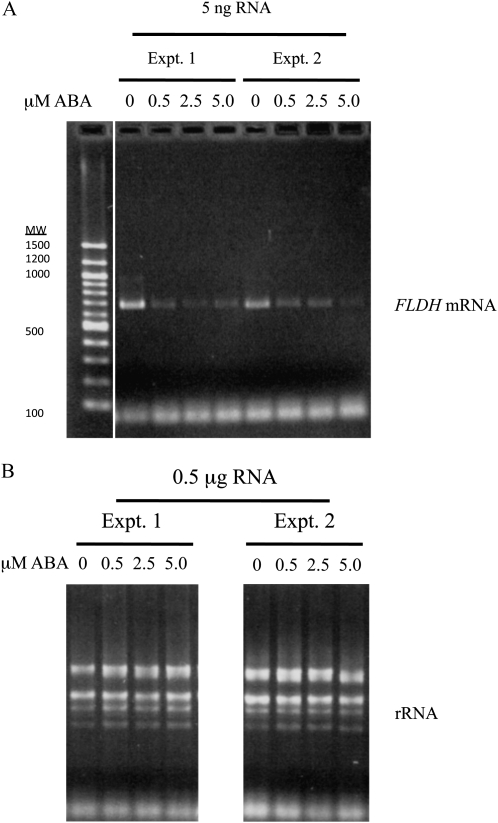
According to microarray data sets visualized using the Bio-Array Resource for Plant Functional Genomics at the University of Toronto, FLDH expression is repressed by ABA, which raises the interesting possibility that ABA regulates farnesol metabolism. As shown in Figure 8, RT-PCR analysis confirmed the repression of FLDH expression by exogenous ABA. To determine whether ABA regulates other genes involved in farnesol metabolism, we also tested the hypothesis that ABA regulates the expression of the FCLY gene. As with FLDH, microarray data sets visualized using the Bio-Array Resource for Plant Functional Genomics indicate that FCLY expression is repressed by ABA. Moreover, RT-PCR analysis confirmed the repression of FCLY expression by ABA (Supplemental Fig. S3). Together, these data suggest that ABA regulates farnesol metabolism at multiple levels in Arabidopsis plants.
Figure 8.
FLDH is negatively regulated by ABA. A, RT-PCR was performed on 5 ng of total RNA from wild-type (Col-0) seedlings, which were grown for 4 d on 0.5× MS plates containing 1.0% Suc and 0.8% agar and then transferred to identical plates containing 0, 0.5, 2.5, or 5.0 μm cisABA for 16 h. The FLDH RT-PCR product was expected to be 693 bp in length. B, Ribosomal RNA is shown for the RNA samples used in A. Expt., Experiment.
Role of FLDH in ABA Signaling
We identified homozygous T-DNA insertions in the 5′ flanking region of the FLDH gene (the fldh-1 and fldh-2 insertions are located at −25 and −282, respectively). Genomic PCR using an At4g33360 forward primer that anneals in the promoter region upstream of the T-DNA insertions (At4g33360-P) and an At4g33360 reverse primer that anneals in the coding region downstream of the T-DNA insertions (At4g33360-R) generated the expected product from wild-type Arabidopsis (Columbia-0 [Col-0]) DNA but not fldh-1 (SALK_111066) DNA (Supplemental Fig. S4). In contrast, genomic PCR using At4g33360-P or At4g33360-R and a T-DNA left border primer (TDNA SALK-LBb1) produced products from fldh-1 DNA but not wild-type Arabidopsis DNA. These results support the hypothesis that fldh-1 is homozygous (other T2 plants were heterozygous and amplified fragments with At4g33360-P and At4g33360-R; data not shown). Moreover, the appearance of an amplified product with At4g33360-P and TDNA SALK-LBb1, as well as At4g33360-R and TDNA SALK-LBb1, indicates the presence of a double or rearranged T-DNA insertion in fldh-1 (i.e. left border sequences are oriented toward the 5′ and 3′ ends of the FLDH gene). The SALK_060297 (fldh-2) line was identified as a homozygous T-DNA insertion line at the Salk Institute Genomic Analysis Laboratory and confirmed by genomic PCR (data not shown).
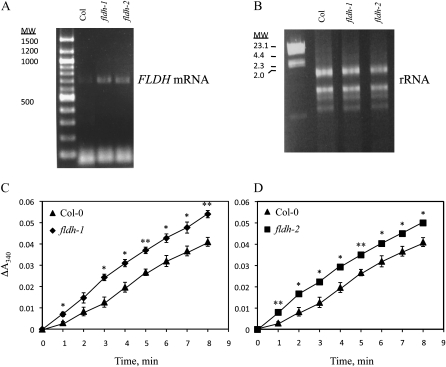
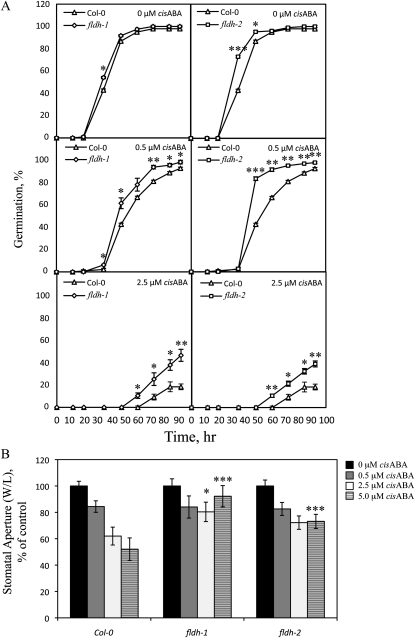
The fldh-1 and fldh-2 mutants described in the preceding paragraph were analyzed for expression of the FLDH gene. As shown in Figure 9, fldh-1 and fldh-2 contained elevated levels of FLDH transcripts, as judged by RT-PCR. These results indicate that both T-DNA insertions disrupt a cis-acting negative regulatory element in the FLDH promoter. Moreover, membranes isolated from both mutants exhibited increased farnesol dehydrogenase activity compared to the wild type (Fig. 9). No developmental phenotypes were observed for either fldh-1 or fldh-2, but, as shown in Figure 10, both mutants exhibited an ABA-insensitive phenotype in seed germination and stomatal closure assays. These results indicate that FLDH negatively regulates ABA signaling in Arabidopsis.
Figure 9.
Biochemical characterization of fldh mutants of Arabidopsis. A, RT-PCR with At4g33360-RT5 and At4g33360-RT3 primers demonstrates that fldh-1and fldh-2 exhibit increased FLDH expression. B, Ethidium bromide-stained RNA used for RT-PCR in A. MW, Molecular weight. C and D, Col-0, fldh-1, and fldh-2 were analyzed spectrophotometrically for farnesol dehydrogenase activity using farnesol and NAD+ as substrates. The se of the mean is indicated for all data points. * and ** represent significant differences compared with the Col-0 control of P < 0.5 and P < 0.01, respectively, as determined by Student’s t test.
Figure 10.
Effects of T-DNA insertions in the 5′ flanking region of the FLDH gene on ABA signaling. A, Seed germination was scored for Col-0, fldh-1, and fldh-2 lines as a function of time in the presence of 0, 0.5, or 2.5 μm cisABA (each data point represents three independent trials, 72 < n < 120). B, Stomatal apertures were measured for Col-0, fldh-1, and fldh-2 lines in the presence of 0, 0.5, 2.5, or 5.0 μm cisABA (83 < n < 166). The se of the mean is indicated for all data points. *, **, and *** represent significant differences compared with the Col-0 control of P < 0.05, P < 0.01, and P < 0.001, respectively, as determined by Student’s t test. W/L, Stomatal pore width divided by length.
DISCUSSION
Previous work from our laboratory demonstrated the oxidation of FC to farnesal and that of Thai et al. (1999) established the sequential phosphorylation of farnesol to farnesyl monophosphate and farnesyl diphosphate in plants (Thai et al., 1999; Crowell et al., 2007). These observations suggested the existence of oxidoreductases capable of catalyzing the interconversion of farnesal and farnesol. Consistent with this hypothesis, farnesal is reduced to farnesol in the presence of Arabidopsis membranes (Crowell et al., 2007). Moreover, reduction of farnesal to farnesol is inhibited by pretreatment of Arabidopsis membranes with NADase, suggesting the involvement of an NAD(P)H-dependent farnesal reductase/NAD(P)+-dependent farnesol dehydrogenase. In this report, farnesol dehydrogenase activity in Arabidopsis membranes is demonstrated directly, and a gene on chromosome 4 of the Arabidopsis genome (FLDH, At4g33360) is shown to encode farnesol dehydrogenase. Expression of FLDH, the protein product of which is an NAD+-dependent farnesol dehydrogenase with partial selectivity for farnesol, is repressed by ABA. In addition, mutants with elevated FLDH expression are less sensitive to ABA than wild-type plants, suggesting that FLDH is a negative regulator of ABA signaling.
The protein product of the FLDH gene has been detected in proteomic analyses of tonoplast proteins (Jaquinod et al., 2007). This is consistent with the tonoplast localization of FC lyase, which catalyzes the oxidation of FC to farnesal and Cys (Jaquinod et al., 2007). However, the FLDH-encoded enzyme has also been detected in proteomic analyses of plasma membrane and endoplasmic reticulum proteins (Alexandersson et al., 2004; Dunkley et al., 2006; Marmagne et al., 2007). It is currently unclear if the latter observations reflect the true localization of the FLDH-encoded farnesol dehydrogenase or if contamination of plasma membrane and endoplasmic reticulum fractions with tonoplast proteins resulted in the mislocalization of the enzyme to these fractions. Whichever it is, experimental confirmation of the intracellular location of the FLDH-encoded farnesol dehydrogenase is necessary to support or refute the hypothesis that FC lyase and farnesol dehydrogenase coexist in the vacuolar membrane for the purpose of FC, farnesal, and farnesol metabolism.
Previously published data indicate that, unlike FC lyase, farnesal reductase activity may not be ubiquitously distributed in Arabidopsis tissues and organs (Crowell et al., 2007). Incubation of FC with membranes isolated from various Arabidopsis tissues and organs resulted in farnesal accumulation in all membranes tested. However, conversion of farnesal to farnesol was restricted to seedlings, flowers, stems, and roots (Crowell et al., 2007). Reduction of farnesal to farnesol was virtually undetectable in leaves, suggesting differential expression of farnesal reductase or decreased availability of reduced nicotinamide cofactors in leaves. Why this might be is uncertain, but it is possible that farnesal is less toxic to the tissues in which farnesal reductase activity is lowest. Alternatively, it is possible that farnesol is more toxic to the tissues in which farnesal reductase activity is lowest (Hemmerlin and Bach, 2000; Hemmerlin et al., 2006). Our data suggest a primary role for FLDH in farnesol oxidation, rather than farnesal reduction. Thus, it is reasonable to suggest that tissues in which FLDH is expressed may be more sensitive to the toxic effects of farnesol. To address this important question, it will be necessary to analyze seedlings, stems, leaves, flowers, and roots of wild-type plants and fldh mutants for farnesol dehydrogenase activity, farnesal content, and farnesol content.
The results shown in Figures 2 and 3 using Arabidopsis membranes as a source of farnesol dehydrogenase activity may represent the activity of a single enzyme or the combined activities of multiple enzymes. To address this question, we identified a farnesol dehydrogenase gene from Arabidopsis to determine if the encoded protein exhibited the same behavior and apparent substrate specificity as the activity detected in Arabidopsis membranes. Because Arabidopsis membranes contain sufficient cofactor to support the interconversion of farnesol and farnesal, it was not possible to determine the cofactor requirement(s) of the enzyme(s) present in Arabidopsis membranes (i.e. activity is observed with or without exogenous cofactor; Crowell et al., 2007). Interestingly, farnesol and geranylgeraniol dehydrogenase activities were detected in Arabidopsis membranes, with the highest activity in the presence of geranylgeraniol, less activity in the presence of farnesol, and no activity in the presence of geraniol (Fig. 3). In contrast, the FLDH-encoded enzyme exhibited the highest activity in the presence of farnesol, less activity in the presence of geraniol, and the least activity in the presence of geranylgeraniol (Fig. 7). Because the substrate profile of the FLDH-encoded farnesol dehydrogenase does not match the substrate profile observed in Arabidopsis membranes (i.e. geranylgeraniol is the preferred substrate when Arabidopsis membranes are used, but farnesol is the preferred substrate of the FLDH-encoded enzyme), it is likely that the activity detected in Arabidopsis membranes represents multiple dehydrogenases, including a geranylgeraniol dehydrogenase and possibly an NADP+-dependent farnesol dehydrogenase (Inoue et al., 1984; Mayoral et al., 2009). In addition, our data suggest that the FLDH-encoded farnesol dehydrogenase catalyzes farnesol oxidation rather than farnesal reduction. Thus, other enzymes must also exist to catalyze farnesal reduction in Arabidopsis.
As stated above, the FLDH-encoded farnesol dehydrogenase was active in the presence of farnesol, geraniol, and geranylgeraniol (in descending order). However, competition assays demonstrated that farnesol was the most potent competitor, followed by geranylgeraniol and geraniol (in descending order). These observations suggest that farnesol has the highest affinity for the active site and highest catalytic turnover rate. In contrast, geranylgeraniol appears to bind to the active site better than geraniol, but with a slower catalytic turnover rate. To confirm or refute these predictions, careful enzymatic analyses with purified enzyme will be necessary to determine precisely how different prenyl alcohols interact with the active site of the FLDH-encoded farnesol dehydrogenase.
ABA regulates the expression of multiple genes involved in farnesol metabolism. For example, the RT-PCR data shown in Figure 8 demonstrate that ABA represses the expression of the FLDH gene. This observation is supported by microarray data visualized using the Bio-Array Resource for Plant Functional Genomics at the University of Toronto. RT-PCR and microarray data also demonstrate that FCLY expression is repressed by ABA (Supplemental Fig. S3). Given that mutants with T-DNA insertions in the FCLY gene exhibit decreased FCLY expression and an enhanced response to ABA, it is reasonable to speculate that ABA repression of FCLY expression also causes an enhanced response to ABA (Crowell et al., 2007). Similarly, the decreased ABA sensitivity of T-DNA insertion mutants with elevated levels of FLDH mRNA and activity suggest that FLDH negatively regulates ABA signaling (Figs. 9 and 10). The mechanism by which FLDH regulates ABA signaling remains unknown, but it is possible that it occurs via modulation of FC lyase activity (i.e. FCLY expression may be elevated in fldh-1 and fldh-2 mutants). Whatever the mechanism, direct or indirect, our data indicate that ABA represses FLDH expression and FLDH expression reduces ABA sensitivity.
CONCLUSION
In this study, our goal was to establish the existence of a farnesol dehydrogenase enzyme in Arabidopsis, characterize the enzyme with respect to isoprenoid and cofactor specificity, identify the corresponding gene, and examine the regulation and function of the gene. From the data shown here, we conclude that Arabidopsis membranes possess farnesol dehydrogenase activity and that the FLDH (At4g33360) gene encodes an NAD+-dependent farnesol dehydrogenase with partial specificity for farnesol as a substrate. Moreover, we conclude that ABA represses the expression of the FLDH gene and that FLDH expression negatively regulates ABA signaling. These findings suggest a regulatory feedback mechanism whereby ABA regulation of FLDH expression increases ABA responsiveness of plant cells.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were sterilized according to the following procedure: 95% ethanol for 5 min, 20% to 50% bleach for 5 to 20 min, followed by five washes in sterile deionized water. Seeds were then suspended in 0.1% agar, stratified on 0.5× Murashige and Skoog (MS) plates containing 1% Suc and 0.8% agar for 3 d at 4°C, and germinated at 22°C under long-day conditions (18 h of white light at 100 μmol m−2 s−1 followed by 6 h of dark) in a vertical orientation. Seedlings were harvested after 4 d for extraction of membranes or isolation of total RNA or transferred to soil and grown under the same conditions. Plants were fertilized with a standard mixture of macro- and micronutrients from below.
Preparation of Arabidopsis Seedling Membranes
Arabidopsis seedlings were pulverized after 4 d of growth at 4°C in a buffer containing 50 mm HEPES, pH 7.4, 500 mm mannitol, 5 mm EDTA, 5 mm dithiothreitol (DTT), and Complete protease inhibitors (Roche Diagnostics). Seedling extracts were then filtered through four layers of cheesecloth and centrifuged for 10 min at 8,000g, and extract supernatants were centrifuged for 60 min at 100,000g. Membrane pellets were resuspended in a buffer containing 2.5 mm HEPES, pH 7.0, 250 mm mannitol, and 1 mm DTT, and aliquots were stored at −80°C in the presence of 15% glycerol.
Farnesol Dehydrogenase Assays
Farnesol dehydrogenase assays were performed in the presence of Arabidopsis or yeast membranes (100 μg membrane protein), [1-3H]farnesol, 20 mm Tris-HCl, pH 7.5, and 0.1 or 0.2 mm NAD+ or NADP+ at 30°C for 30 min. Reactions were spotted onto a plastic-backed silica gel plate, developed using hexane:tetrahydrofuran (3:1) as a mobile phase, and analyzed by fluorography using En3hance fluorographic reagent (Perkin-Elmer) and Kodak X-OMAT film (Eastman Kodak).
[1-3H]Farnesol was generated by calf intestine alkaline phosphatase treatment of [1-3H]farnesyl diphosphate (26.2 Ci mmol−1; Perkin-Elmer) according to the manufacturer’s instructions (New England Biolabs). Alternatively, [1-3H]farnesol was purchased from American Radiolabeled Chemicals (60 Ci mmol−1) and purified by preparative TLC using a plastic-backed silica gel plate and hexane:tetrahydrofuran (3:1) as a mobile phase (the [1-3H]farnesol from American Radiolabeled Chemicals was not pure and produced a TLC radiofluorogram identical to that of dephosphorylated [1-3H]farnesyl diphosphate). [1-3H]Farnesol was eluted from excised TLC spots with hexane, dried under nitrogen gas, dissolved in ethanol, and used in farnesol dehydrogenase assays as described above.
Spectrophotometric assays were performed as described above except that unlabeled farnesol, geranylgeraniol, or geraniol was used at a concentration of 1 mm. Reactions were started with cofactor, transferred to a quartz cuvette, and absorbance was monitored at 340 nm for 10 min. Specific activity was calculated using Beer’s Law and an extinction coefficient for NADH of 6.22 cm−1 mM−1.
Expression of Recombinant Arabidopsis Farnesol Dehydrogenase Activity in Yeast
The coding sequences of the At5g16990, At5g16960, At4g33360, and At3g61220 genes were amplified using the Platinum Quantitative RT-PCR Thermoscript One-Step System (Invitrogen/Life Technologies) and the following primers: At5g16990-5, 5′-GGGGGATCCATGACGACGAACAAGCAGGTCATATTC-3′; At5g16990-3, 5′-GGGGGATCCTCACTCACGAGCAATAACAACAACTTGT-3′; At5g16960-5, 5′-GGGGGATCCATGGCGACAACGATCAACAAGCAAGTC-3′; At5g16960-3, 5′-GGGGGATCCTTATGATGGCGAAACCACGACAAGTTGT-3′; At4g33360-5, 5′-GGGGGATCCATGGGCCCAAAGATGCCCAACACAGAA-3′; At4g33360-3, 5′-GGGGGATCCTCAGTAGTGAATGACGCCCAGACTCTTC-3′; At3g61220-5, 5′-GGGGGATCCATGGCAGAGGAAACTCCAAGATATGCTG-3′; At3g61220-3, 5′-GGGGGATCCTCAGAATTCTGAAACTTGCTTGCGACTAAAG-3′. The resulting fragments were inserted into the pYES2.1/V5-His-TOPO vector and sequences and orientations confirmed by DNA sequence analysis. The resulting plasmids, called pCL194, pCL195, pCL196, and pCL197, respectively, were introduced into Saccharomyces cerevisiae strain SM1058 (MATa leu2, ura3, trp1, his4, can1). For yeast transformations, cultures were grown at 30°C overnight in 2 mL of YPAD (1% w/v yeast extract, 2% w/v bacto-peptone, 2% w/v Glc, and 100 μg/mL adenine) and diluted into 100 mL of fresh, prewarmed YPAD. After 90 min at 30°C, the cells were sedimented for 5 min at 3,000 rpm, washed twice in 10 mL of sterile water, washed once in 10 mL of LiAc/TE buffer (0.1 m lithium acetate, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA), and resuspended in LiAc/TE buffer to a concentration of 2 × 109 cells mL−1 (1 × 107 cells mL−1 corresponds to A660 of approximately 0.7). The cells were then incubated without agitation for 15 min at 30°C, and 50-μL aliquots were dispensed into 1.5-mL microfuge tubes. The following additions were made to individual aliquots of cells: 5 μL of 10 μg/μL of salmon sperm DNA, 1 μg of pCL194, pCL195, pCL196, or pCL197 DNA, and 300 μL of 40% w/v polyethylene glycol in LiAc/TE buffer. After incubation without agitation at 30°C for 30 min, the cells were heat shocked at 42°C for 20 min, sedimented for 15 s in a microcentrifuge, and resuspended in 0.5 mL of CSM-ura medium (0.08% w/v CSM-ura, pH 5.8, 0.17% w/v yeast nitrogen base, 0.5% w/v ammonium sulfate, and 2.0% w/v Glc). Transformed yeast cells were selected on CSM-ura agar plates. Transformed and untransformed yeast were then grown at 30°C to log phase in liquid CSM medium containing 2% Glc in the presence or absence of uracil (CSM-ura was used to maintain selective pressure on cells containing At5g16990, At5g16960, At4g33360, and At3g61220 expression constructs). Cells were shifted into CSM medium containing 2% Gal and incubated at 30°C for an additional 14 h prior to harvest. Cells were then lysed in a buffer containing 100 mm Tris-HCl, pH 7.5, 1 mm DTT, 20% v/v glycerol, and Complete protease inhibitors (Roche Diagnostics) by vigorous vortexing in the presence of glass beads, and membranes were prepared by ultracentrifugation at 100,000g for 1 h. Membranes were then assayed for farnesol dehydrogenase activity as described above.
RNA Isolation and RT-PCR
Wild-type Col-0 seeds were surface sterilized and plated on sterile Whatman filters, which were overlaid on 0.5× MS plates containing 1.0% Suc and 0.8% agar. After 3 d of stratification at 4°C, seedlings were germinated in a vertical orientation at 22°C under long-day conditions (18 h of white light at 100 μmol m−2 s−1 followed by 6 h of dark) and grown for an additional 4 d. Filters and seedlings were then transferred onto identical plates containing 0, 0.5, 2.5, or 5.0 μm ABA for 16 h (DMSO was used as a solvent control), and total RNA was isolated using TRIzol Reagent according to the manufacturer’s instructions (Invitrogen/Life Technologies). RT-PCR was then performed to analyze FLDH transcript levels using 5 ng of input RNA, 5 pmol of forward primer, 5 pmol of reverse primer, and the Platinum Quantitative RT-PCR Thermoscript One-Step System (Invitrogen/Life Technologies) in a total reaction volume of 25 μL. The FLDH forward and reverse primers were as follows: At4g33360-RT5, 5′-GTAACGGATTACCGTTCTCTAACGG-3′, and At4g33360-RT3, 5′-TGGAAGCTTTCCTGTAACCCGAGAG-3′. RT-PCR conditions included a 30-min reverse transcription step at 50°C, followed by a 2-min presoak at 95°C, and 40 cycles of the following PCR program: 95°C, 30 s; 55°C, 30 s; 68°C, 90 s. A postsoak was performed at 68°C for 4 min to ensure complete product synthesis. RT-PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Analysis of T-DNA Insertion Mutants
Genomic DNA was isolated from wild-type Col-0 and fldh seedlings using Plant DNAzol according to the manufacturer’s instructions (Invitrogen/Life Technologies). Genomic analysis of wild-type and fldh mutant lines was then performed by PCR using 0.2 ng of genomic DNA, 5 pmol of forward primer, 5 pmol of reverse primer, and Ex-Taq polymerase (TaKaRa Bio) in a total reaction volume of 25 μL. PCR conditions varied, but generally consisted of a 5-min hot start presoak at 95°C and 40 cycles of the following PCR program: 95°C, 30 s; 55°C, 30 s; 72°C, 1 min. A postsoak was performed at 72°C for 7 min to ensure complete product synthesis. Two different PCR analyses were performed. The first used two gene-specific primers: At4g33360-P, 5′-TCTGATGGATACAGAGGAGAGGTG-3′, and At4g33360-R, 5′-CATTCTTCAGTCCACCAACGTTGAC-3′. The second PCR analysis used a T-DNA-specific primer and one of these two gene-specific primers. The T-DNA-specific primer was TDNA SALK-LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′.
Total RNA was isolated from seedlings of wild-type and fldh plants using TRIzol Reagent according to the manufacturer’s instructions (Invitrogen/Life Technologies). RT-PCR was then performed to analyze FLDH transcript levels in wild-type and fldh plants as described above.
Seed Germination Assays
Seeds used for germination assays were harvested from control and experimental plants, which were grown together under identical conditions. Seeds were surface sterilized, suspended in sterile 0.1% agar, and placed on 0.5× MS plates containing 1% Suc and 0.8% agar in the dark at 22°C. Seeds from control and experimental plants were sown on the same plates, and germination (radical emergence) was scored in the presence of various concentrations of exogenous ABA under a dissecting microscope.
Stomatal Closure Assays
Rosette leaves were excised and incubated for 2 h in the presence of various concentrations of ABA or an equivalent volume of DMSO in 10 mL of water. Epidermal peels were then prepared by peeling away the leaf surface with Scotch tape. Epidermal peels were stained with toluidine blue, mounted on a microscope slide, and visualized with a Leica DMRB microscope interfaced to a SPOT digital camera. Data are recorded as the average width per length of individual apertures (83 < n < 166) relative to the 0 μm ABA sample for each line. Excision and incubation of leaves in the presence of various concentrations of ABA was performed in random order by J.B. Epidermal peels, photography, and measurement of stomatal apertures were performed by A.H.F. without knowledge of sample identities.
Statistical Methods
Data are presented as the mean plus or minus the se of the mean. Statistically significant differences were determined by Student’s t test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_119490.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Arabidopsis membranes contain sufficient cofactor to support farnesol dehydrogenase activity.
Supplemental Figure S2. The FLDH-encoded farnesol dehydrogenase is not a farnesal reductase.
Supplemental Figure S3. FCLY is negatively regulated by ABA.
Supplemental Figure S4. Molecular characterization of fldh mutants of Arabidopsis
Supplementary Material
References
- Alexandersson E, Saalbach G, Larsson C, Kjellbom P. (2004) Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol 45: 1543–1556 [DOI] [PubMed] [Google Scholar]
- Baker FC, Mauchamp B, Tsai LW, Schooley DA. (1983) Farnesol and farnesal dehydrogenase(s) in corpora allata of the tobacco hornworm moth, Manduca sexta. J Lipid Res 24: 1586–1594 [PubMed] [Google Scholar]
- Beigneux A, Withycombe SK, Digits JA, Tschantz WR, Weinbaum CA, Griffey SM, Bergo M, Casey PJ, Young SG. (2002) Prenylcysteine lyase deficiency in mice results in the accumulation of farnesylcysteine and geranylgeranylcysteine in brain and liver. J Biol Chem 277: 38358–38363 [DOI] [PubMed] [Google Scholar]
- Bracha K, Lavy M, Yalovsky S. (2002) The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J Biol Chem 277: 29856–29864 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Lubetzky TC, Yalovsky S. (2008) Functional analysis of Arabidopsis postprenylation CaaX processing enzymes and their function in subcellular protein targeting. Plant Physiol 148: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadiñanos J, Varela I, Mandel DA, Schmidt WK, Díaz-Perales A, López-Otín C, Freije JM. (2003) AtFACE-2, a functional prenylated protein protease from Arabidopsis thaliana related to mammalian Ras-converting enzymes. J Biol Chem 278: 42091–42097 [DOI] [PubMed] [Google Scholar]
- Clarke S. (1992) Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem 61: 355–386 [DOI] [PubMed] [Google Scholar]
- Crowell DN. (2000) Functional implications of protein isoprenylation in plants. Prog Lipid Res 39: 393–408 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Huizinga DH. (2009) Protein isoprenylation: the fat of the matter. Trends Plant Sci 14: 163–170 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Huizinga DH, Deem AK, Trobaugh C, Denton R, Sen SE. (2007) Arabidopsis thaliana plants possess a specific farnesylcysteine lyase that is involved in detoxification and recycling of farnesylcysteine. Plant J 50: 839–847 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Kennedy M. (2001) Identification and functional expression in yeast of a prenylcysteine alpha-carboxyl methyltransferase gene from Arabidopsis thaliana. Plant Mol Biol 45: 469–476 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Sen SE, Randall SK. (1998) Prenylcysteine alpha-carboxyl methyltransferase in suspension-cultured tobacco cells. Plant Physiol 118: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Deem AK, Bultema RL, Crowell DN. (2006) Prenylcysteine methylesterase in Arabidopsis thaliana. Gene 380: 159–166 [DOI] [PubMed] [Google Scholar]
- Digits JA, Pyun HJ, Coates RM, Casey PJ. (2002) Stereospecificity and kinetic mechanism of human prenylcysteine lyase, an unusual thioether oxidase. J Biol Chem 277: 41086–41093 [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A, Bach TJ. (2000) Farnesol-induced cell death and stimulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase activity in tobacco cv bright yellow-2 cells. Plant Physiol 123: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A, Reents R, Mutterer J, Feldtrauer JF, Waldmann H, Bach TJ. (2006) Monitoring farnesol-induced toxicity in tobacco BY-2 cells with a fluorescent analog. Arch Biochem Biophys 448: 93–103 [DOI] [PubMed] [Google Scholar]
- Huizinga DH, Denton R, Koehler KG, Tomasello A, Wood L, Sen SE, Crowell DN. (2010) Farnesylcysteine lyase is involved in negative regulation of abscisic acid signaling in Arabidopsis. Mol Plant 3: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga DH, Omosegbon O, Omery B, Crowell DN. (2008) Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis. Plant Cell 20: 2714–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Tsuji H, Uritani I. (1984) Characterization and activity change of farnesol dehydrogenase in black rot fungus-infected sweet potato. Agric Biol Chem 48: 733–738 [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Chary SN, Chernoff EA, Zeng Q, Running MP, Crowell DN. (2005) Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol 139: 722–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G. (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Mayoral JG, Nouzova M, Navare A, Noriega FG. (2009) NADP+-dependent farnesol dehydrogenase, a corpora allata enzyme involved in juvenile hormone synthesis. Proc Natl Acad Sci USA 106: 21091–21096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha Chary S, Bultema RL, Packard CE, Crowell DN. (2002) Prenylcysteine alpha-carboxyl methyltransferase expression and function in Arabidopsis thaliana. Plant J 32: 735–747 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Gruissem W. (1999) Protein prenylation in plants: old friends and new targets. Plant Mol Biol 39: 865–870 [DOI] [PubMed] [Google Scholar]
- Running MP, Fletcher JC, Meyerowitz EM. (1998) The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development 125: 2545–2553 [DOI] [PubMed] [Google Scholar]
- Running MP, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S. (2004) Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc Natl Acad Sci USA 101: 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry AE, Sen SE. (2001) Farnesol oxidation in insects: evidence that the biosynthesis of insect juvenile hormone is mediated by a specific alcohol oxidase. Insect Biochem Mol Biol 31: 171–178 [DOI] [PubMed] [Google Scholar]
- Thai L, Rush JS, Maul JE, Devarenne T, Rodgers DL, Chappell J, Waechter CJ. (1999) Farnesol is utilized for isoprenoid biosynthesis in plant cells via farnesyl pyrophosphate formed by successive monophosphorylation reactions. Proc Natl Acad Sci USA 96: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschantz WR, Digits JA, Pyun HJ, Coates RM, Casey PJ. (2001) Lysosomal prenylcysteine lyase is a FAD-dependent thioether oxidase. J Biol Chem 276: 2321–2324 [DOI] [PubMed] [Google Scholar]
- Tschantz WR, Zhang L, Casey PJ. (1999) Cloning, expression, and cellular localization of a human prenylcysteine lyase. J Biol Chem 274: 35802–35808 [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65: 241–269 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tschantz WR, Casey PJ. (1997) Isolation and characterization of a prenylcysteine lyase from bovine brain. J Biol Chem 272: 23354–23359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.