Abstract
Toc12 is a novel J domain-containing protein identified in pea (Pisum sativum) chloroplasts. It was shown to be an integral outer membrane protein localizing in the intermembrane space of the chloroplast envelope. Furthermore, Toc12 was shown to associate with an intermembrane space Hsp70, suggesting that Toc12 is important for protein translocation across the chloroplast envelope. Toc12 shares a high degree of sequence similarity with Arabidopsis (Arabidopsis thaliana) DnaJ-J8, which has been suggested to be a soluble protein of the chloroplast stroma. Here, we isolated genes encoding DnaJ-J8 from pea and found that Toc12 is a truncated clone of one of the pea DnaJ-J8s. Protein import analyses indicate that Toc12 and DnaJ-J8s possess a cleavable transit peptide and are localized in the stroma. Arabidopsis mutants with T-DNA insertions in the DnaJ-J8 gene show no defect in chloroplast protein import. Implications of these results in the energetics and mechanisms of chloroplast protein import are discussed.
Most chloroplast proteins are encoded by the nuclear genome and synthesized in the cytosol as higher molecular mass precursors with an N-terminal extension known as the transit peptide. Precursor proteins are imported into chloroplasts through a translocon complex located at the chloroplast envelope. Translocon components associated with the outer membrane are called Toc (for translocon of the outer envelope membrane of chloroplast) proteins, and those associated with the inner membrane are called Tic (for translocon of the inner envelope membrane of chloroplast) proteins. Cleavage of the transit peptide from the precursor by a specific stromal processing peptidase during translocation results in the production of the lower molecular mass mature protein. Various translocon components have been assigned functions in the basic steps of the import process (for review, see Inaba and Schnell, 2008; Jarvis, 2008; Li and Chiu, 2010). For example, Toc159 (the no. indicates the calculated molecular mass of the protein) and Toc34 are receptors for the transit peptides, and Toc75 is the protein-translocating channel across the outer membrane. Toc64, on the other hand, has a dual function: it serves as a docking site for the cytosolic Hsp90 through its cytosolic domain and as a scaffold for translocon components located in the intermembrane space through its intermembrane space domain (Qbadou et al., 2007).
Protein import into chloroplasts involves at least two distinct ATP-consuming steps. The first step is called “early import intermediate” or “docking,” in which less than 100 μm ATP is required and precursors are translocated across the outer membrane and come into contact with translocon components in the inner membrane (Olsen et al., 1989; Kouranov and Schnell, 1997; Inaba et al., 2003; Inoue and Akita, 2008). It has been shown that the ATP is used in the intermembrane space (Olsen and Keegstra, 1992), most likely by a yet unidentified intermembrane space Hsp70 called imsHsp70 or Hsp70-IAP (ims for “intermembrane space” and IAP for “import intermediate-associated protein”; Marshall et al., 1990; Schnell et al., 1994; Qbadou et al., 2007). The second ATP-consuming step is the complete translocation of precursors across the two envelope membranes into the stroma. This step requires about 1 mm ATP. The ATP is most likely used by the stromal Hsp93 and chloroplast Hsc70 associated with the translocon to drive protein translocation into the stroma (Nielsen et al., 1997; Shi and Theg, 2010; Su and Li, 2010).
Hsp70 family proteins are involved in many cellular processes, including protein folding, protein translocation across membranes, and regulation of protein degradation. Hsp70 proteins are often recruited to perform a certain function by specifically localized J domain-containing proteins. The J domain-containing proteins interact with Hsp70 when Hsp70 is bound to ATP and stimulate ATP hydrolysis by Hsp70. The specific J domain-containing cochaperone that recruits the stromal chloroplast Hsc70 to the inner envelope membrane to assist in protein translocation has not been identified. The specific J domain-containing cochaperone for imsHsp70 for its function in protein import into chloroplasts is proposed to be a protein named Toc12 (Becker et al., 2004).
Toc12 was identified as a novel J domain-containing protein from pea (Pisum sativum) chloroplasts. It belongs to the type III J domain proteins containing only the J domain without the Gly- and Phe-rich domain (G/F domain) and the zinc-finger domain originally found in Escherichia coli DnaJ. It has been shown that the protein is synthesized at its mature size of 103 amino acids without a cleavable transit peptide. After import, the protein has been shown to anchor in the outer membrane by its N-terminal part, which has been suggested to form a β-barrel-type domain. Its C-terminal part, composed of the J domain, has been shown to localize in the intermembrane space. Toc12 has been shown to associate with imsHsp70. Toc12 and imsHsp70 interact with the intermembrane space domain of Toc64, which in turn associates with another intermembrane space translocon component, Tic22. It is proposed that the Toc12-imsHsp70-Toc64-Tic22 complex mediates protein translocation across the intermembrane space through specific precursor binding and ATP hydrolysis (Becker et al., 2004; Qbadou et al., 2007). However, the existence of imsHsp70 has only been shown on immunoblots by its reactivity to the monoclonal antibody SPA820 raised against human Hsp70. Its encoding gene has never been identified. The Arabidopsis (Arabidopsis thaliana) Hsp70 gene family has 14 members. Only two of them are localized in chloroplasts, and both have been shown to locate in the stroma (Ratnayake et al., 2008; Su and Li, 2008). A recent study has further shown that the major protein recognized by the SPA820 antibody in pea chloroplasts is located in the stroma, indicating that imsHsp70 is most likely a stromal protein (Ratnayake et al., 2008).
Most translocon components were originally identified from pea chloroplasts. While all translocon components identified from pea have easily recognizable Arabidopsis homologs, Toc12 seems to be an exception. The Arabidopsis gene suggested to be the pea TOC12 homolog, At1g80920 (Inoue, 2007; Jarvis, 2008), encodes a protein that is much larger than pea Toc12 and is annotated as J8 (referred to as AtJ8 herein). The entire pea Toc12 has a high sequence similarity to the N-terminal two-thirds of AtJ8. AtJ8 contains an extra C-terminal domain of 60 amino acids that is highly conserved among J8 proteins from other higher plants. However, in contrast to pea Toc12, AtJ8 is predicted to locate in the stroma (Miernyk, 2001; www.arabidopsis.org). Indeed, a fusion protein consisting of the first 80 amino acids of AtJ8 fused at the N terminus of GFP was imported into the chloroplast stroma, and approximately 46 amino acids from the N terminus were processed after import (Lee et al., 2008), indicating that the first 46 amino acids of AtJ8 function as a cleavable stroma-targeting transit peptide. A T-DNA insertion in the AtJ8 gene that causes the truncation of the last three amino acids results in no visible phenotype. However, detailed analyses indicate that the mutant has lower CO2 assimilation and Rubisco activity than the wild type (Chen et al., 2010).
We are interested in identifying J domain-containing proteins interacting with stromal Hsp70. As part of the initial effort, we investigated the suborganellar location of J8 and examined the relationship between Toc12 and J8. We found that, in pea, there are at least two genes encoding J8, which we named PsJ8a and PsJ8b. TOC12 represents part of PsJ8b. Toc12, AtJ8, and the two PsJ8 proteins could be imported into chloroplasts and processed to stromally localized soluble mature proteins. Four alleles of AtJ8 mutants were analyzed, but none of them showed any defect in the import of various chloroplast precursor proteins.
RESULTS
Toc12 Is Transcribed from One of the Two Genes Encoding PsJ8
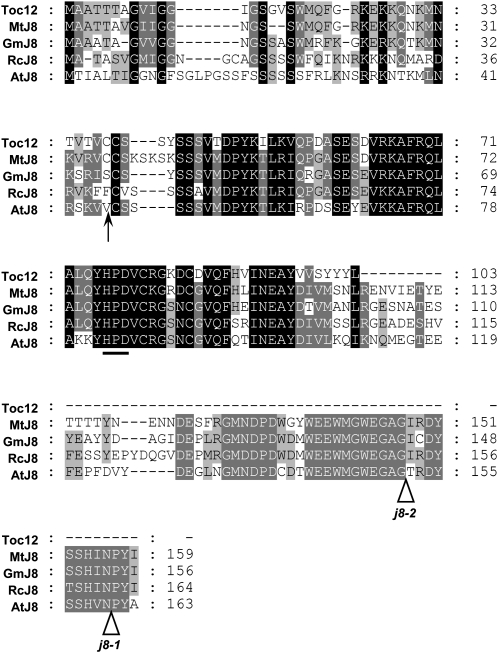
Through multiple sequence alignment, we found that pea Toc12 is extremely similar to the N-terminal two-thirds of J8 homologs from various legume species (Fig. 1). For example, Toc12 is 78.8% identical to the N-terminal two-thirds of Medicago truncatula J8. However, all J8s have an extra C-terminal domain about 60 amino acids in length. Part of this domain is highly conserved from legumes to Arabidopsis (Fig. 1). Instead of this C-terminal domain, Toc12 has a short tail of five amino acids not found in any other species.
Figure 1.
Toc12 is highly similar to the N-terminal two-thirds of J8 from various legume species. Sequence alignment of pea Toc12 and J8 from Medicago (MtJ8), soybean (GmJ8), castor bean (RcJ8), and Arabidopsis (AtJ8) is shown. The predicted transit peptide processing site of AtJ8 is indicated with an arrow. The conserved tripeptide HPD in the J domain is underlined. T-DNA insertion sites of Arabidopsis j8-1 and j8-2 mutants are indicated with white arrowheads.
To isolate J8-encoding genes from pea, we took advantage of the conservation of the C-terminal domain. In J8s of Medicago, castor bean (Ricinus communis), and soybean (Glycine max), the last 18 bases before the stop codon have identical DNA sequences (Supplemental Fig. S1). Using this conserved stretch of sequence to design a reverse primer, part of the 5′ untranslated region (UTR) and the first 11 bases of pea TOC12 open reading frame to design a forward primer (Supplemental Table S1; primers PsJ8-R2 and PsJ8-F1, respectively), and first-strand cDNA reverse transcribed from total pea RNA as templates, we obtained an approximately 450-bp cDNA fragment by PCR amplification. Sequencing revealed that the PCR product was actually a mixture of two different cDNAs (Supplemental Fig. S2), which we designated as PsJ8a and PsJ8b. Clone-specific primers were then designed to isolate their corresponding genomic clones and to perform 3′ RACE to identify the authentic cDNA sequence encoding the C-terminal end and 3′ UTR.
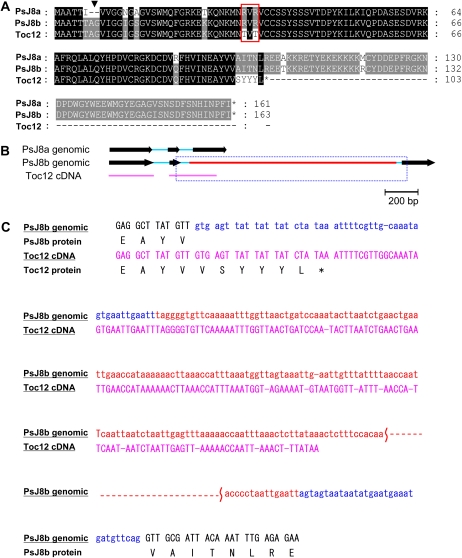
Sequence comparison revealed that PsJ8a has nucleotide differences from PsJ8b throughout the cDNA sequence and resulted in 12 amino acid changes, including a two-amino acid insertion near the N terminus of PsJ8b (the Ala and Gly of the seventh and eighth residues in J8b; Fig. 2A, arrowhead). Most strikingly, however, PsJ8b contains in the second intron a 1,229-bp insertion that is absent in PsJ8a (Fig. 2B, red line).
Figure 2.
Deduced amino acid sequences and genomic structures of the two PsJ8 genes. A, Toc12 is part of PsJ8b. Sequence alignment of PsJ8a, PsJ8b, and Toc12 is shown. The additional Ala and Gly residues present in PsJ8b are indicated with the arrowhead. Thr residues at positions 34 and 36 of Toc12 substituted with Arg in PsJ8a and PsJ8b are boxed in red. Stop codons are marked with asterisks. B, TOC12 is a partially spliced or alternatively spliced RNA of PsJ8b. Exons and introns are shown as black arrows and blue lines, respectively. A unique 1,229-bp insertion in the second intron of PsJ8b is shown as a red line. TOC12 cDNA is represented by the pink line. Sequences of the beginning and end of the boxed region are shown in C. C, Detailed sequences of the beginning and end of the boxed region marked in B. Nucleotide sequences (same color code as in B) and deduced amino acid sequences of PsJ8b and TOC12 are shown.
Toc12 is identical to the N-terminal two-thirds of PsJ8b in both protein (Fig. 2A) and cDNA (Supplemental Fig. S2) sequences except in two places. Residues 34 and 36 are both Thr in Toc12 but Arg in PsJ8a and PsJ8b (Fig. 2A, red box). The last six amino acids and the 3′ UTR of Toc12 are encoded by the sequence corresponding to the beginning of the second intron of PsJ8b, including part of the 1,229-bp insertion unique to PsJ8b (Fig. 2, B and C). This result suggests that TOC12 RNA arises from partially or alternatively spliced PsJ8b. The difference of Arg versus Thr at residues 34 and 36 may be attributed to the different pea varieties used. Using a primer located immediately after the stop codon of Toc12 in the second intron, we could indeed amplify a cDNA with the C-terminal sequence of Toc12, but residues 34 and 36 were still Arg. Using site-directed mutagenesis, we created a cDNA clone encoding the exact protein sequence of the published Toc12 by changing residues 34 and 36 to Thr.
AtJ8 and PsJ8s Are Localized in the Stroma
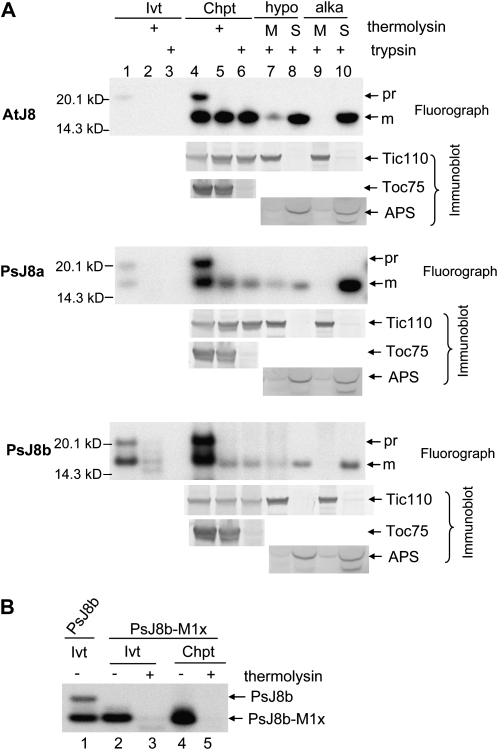
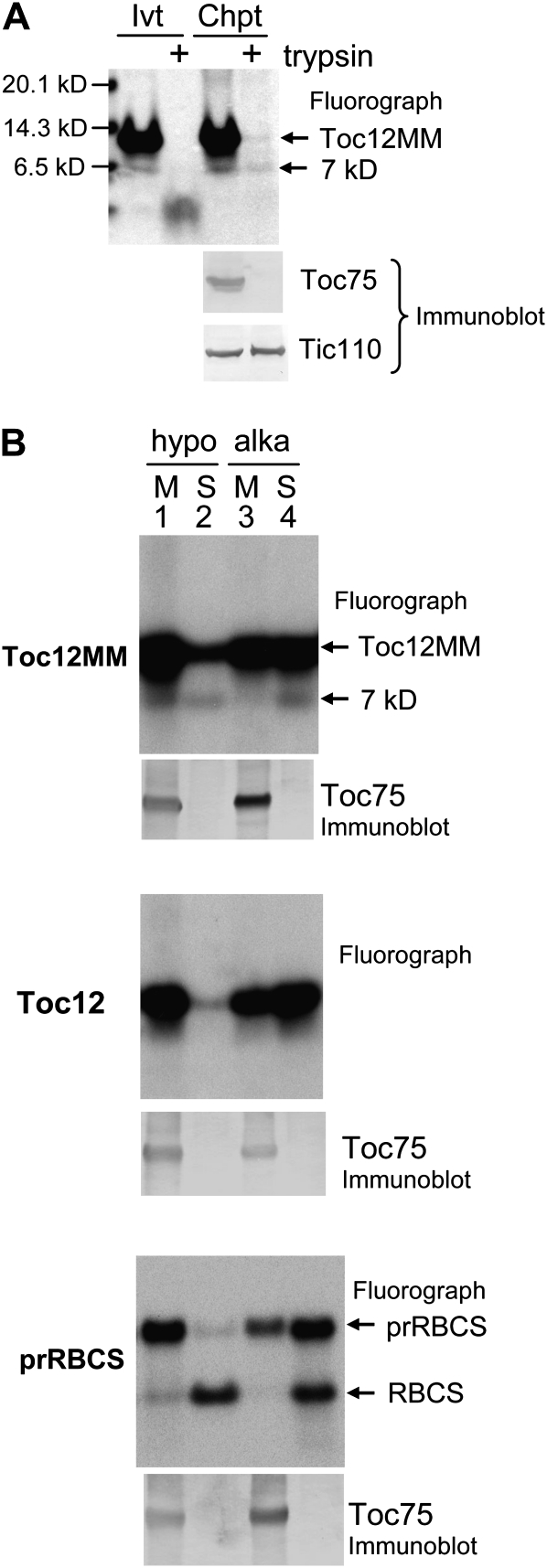
We first investigated the suborganellar location of AtJ8 and the two PsJ8s using in vitro protein import and fractionation. [35S]Met-labeled J8 proteins were incubated with isolated pea chloroplasts under import conditions. AtJ8 was synthesized as an approximately 21-kD protein (Fig. 3A, AtJ8 panel, lane 1). After import into chloroplasts, a smaller protein of approximately 16 kD was produced. Chloroplasts after import were further treated with thermolysin or trypsin. Thermolysin can only degrade proteins exposed on the cytosolic side of the outer envelope membrane, while trypsin can digest proteins in the intermembrane, as it can penetrate the outer but not the inner envelope membrane (Cline et al., 1984; Jackson et al., 1998). The effectiveness of the trypsin treatment was shown by the degradation of the outer membrane protein Toc75 and the resistance of Tic110 (Fig. 3A, lanes 5 and 6), which is an inner membrane protein with a large stromal domain. The 21-kD AtJ8 was degraded by thermolysin and trypsin, but the 16-kD protein produced after import was resistant to both proteases, indicating that the 16-kD protein was inside the inner envelope membrane. Thus, it is likely that the 21-kD in vitro-translated protein was the precursor form and the 16-kD protein was the imported mature form after removal of the transit peptide. A difference of 5 kD agrees well with the ChloroP prediction and the experimental result (Lee et al., 2008) that the first 46 amino acids functions as a cleavable transit peptide.
Figure 3.
AtJ8, PsJ8a, and PsJ8b are synthesized as higher molecular mass precursors with cleavable transit peptides and imported into the chloroplast stroma. A, Import of AtJ8, PsJ8a, and PsJ8b into isolated pea chloroplasts. In vitro-translated, [35S]Met-labeled precursor proteins (Ivt) and chloroplasts after import of the precursor proteins (Chpt) were treated with thermolysin or trypsin. Trypsin-treated chloroplasts were lysed hypotonically (hypo) or treated by alkaline extraction (alka) and then separated into membrane (M) and soluble (S) fractions. Samples were analyzed by SDS-PAGE followed by fluorography (for AtJ8 and PsJ8s) or immunoblotting (for Tic110, Toc75, and APS). pr, Precursor form; m, mature form. The Ivt lanes contained 1% of the in vitro-translated proteins used for the import reactions shown in the Chpt lanes. B, PsJ8b-M1x could not be imported into chloroplasts. In vitro-translated, [35S]Met-PsJ8b-M1x (Ivt; lanes 2 and 3) and chloroplasts after import of PsJ8b-M1x (Chpt; lanes 4 and 5) were treated with thermolysin. All samples were analyzed by SDS-PAGE and fluorography. In vitro-translated PsJ8b (lane 1) was also analyzed for size comparison.
Trypsin-treated chloroplasts were further lysed hypotonically and separated into membrane and soluble fractions. Most of the 16-kD mature AtJ8 was found in the soluble fraction, and a small fraction was associated with the membranes (Fig. 3A, lanes 7 and 8). However, mature AtJ8 was entirely observed in the soluble fraction when chloroplasts were treated by alkaline extraction (Fig. 3A, lanes 9 and 10). These results indicate that the 16-kD mature AtJ8 protein is a soluble stromal protein with a small portion peripherally associated with membranes. Control proteins of the small subunit of ADP-Glc pyrophosphorylase (APS) and Tic110 were localized to the soluble and membrane fractions, respectively.
The two PsJ8s were also translated as 21-kD proteins, but a protein with a size slightly larger than 16 kD was also produced, most likely due to internal initiations from one of the downstream Met residues (Fig. 3A, PsJ8 panels, lane 1). To check whether this smaller protein could be imported into chloroplasts, we abolished translation initiation from the first Met of PsJ8b by mutating the initiation ATG codon into ATT and generated the mutant clone PsJ8b-M1x. In vitro transcription/translation of PsJ8b-M1x indeed produced a protein that is the same size as the internal initiation product synthesized from PsJ8b (Fig. 3B, lanes 1 and 2). Incubation of PsJ8b-M1x with chloroplasts under import conditions resulted in PsJ8b-M1x associating with chloroplasts, but the proteins remained entirely thermolysin sensitive (Fig. 3B, lanes 4 and 5), indicating that PsJ8b-M1x was only sticking to the chloroplast surface and could not be imported.
Import of the two PsJ8s into chloroplasts produced essentially the same results as those from AtJ8 import. For both PsJ8 proteins, a 16-kD mature protein resistant to both thermolysin and trypsin was produced after import. When trypsin-treated chloroplasts were lysed hypotonically, most of the imported 16-kD mature proteins were in the soluble fraction and a small fraction was associated with the membrane fraction (Fig. 3B, lanes 7 and 8). When chloroplasts were lysed by alkaline extraction, all of the imported 16-kD mature PsJ8s were in the soluble fraction (Fig. 3B, lanes 9 and 10). Results in Figure 3 indicate that AtJ8 and the two PsJ8s are synthesized as higher molecular mass precursors with a cleavable transit peptide and that the imported mature proteins are soluble and localized in the stroma.
Toc12 Is Processed to a 7-kD Mature Protein Localized in the Stroma
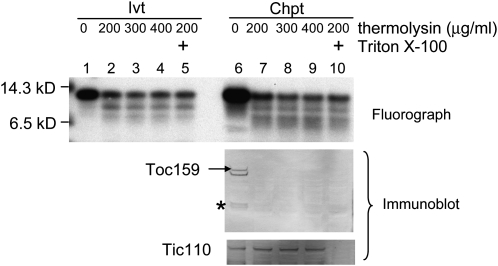
Toc12 is essentially identical to the N-terminal two-thirds of PsJ8b. Therefore, if PsJ8b has a cleavable transit peptide, Toc12 should possess an identical transit peptide, except residues 34 and 36 are Thr in Toc12 but Arg in PsJ8b. Yet, PsJ8b is a soluble protein in the stroma, and Toc12 was shown to be an integral outer membrane protein. The transit peptide region of PsJ8b is the region predicted to be a β-barrel-type membrane anchor in Toc12. To determine whether the sequence differences in PsJ8b and Toc12 have resulted in their different localizations, we reinvestigated the localization of Toc12. [35S]Met-labeled Toc12 was incubated with isolated pea chloroplasts under import conditions. Chloroplasts after import were treated with thermolysin. It was reported that Toc12 was completely thermolysin resistant after import due to its localization in the intermembrane space (Becker et al., 2004). However, we found that about 30% of Toc12 was already thermolysin resistant even before import (Fig. 4, lanes 1–5). After incubation with chloroplasts, the same percentage of Toc12 was thermolysin resistant. The percentage of Toc12 that was thermolysin resistant remained unchanged even after the thermolysin concentration was increased to 400 μg mL−1 or after the addition of 0.1% Triton X-100 in the digestion to permeabilize the chloroplasts (Fig. 4, lanes 6–10). Under these conditions, Toc159, an outer membrane protein with a large cytosolic domain, was degraded by the lowest concentration of thermolysin without Triton X-100 addition. In comparison, Tic110 became accessible to thermolysin only after Triton X-100 addition, confirming the effectiveness of the thermolysin and Triton X-100 treatments (Fig. 4, immunoblot panels). These results suggest that some Toc12 molecules are intrinsically thermolysin resistant, probably due to their tight folding or aggregation and not due to protection by the outer membrane. As a result, thermolysin is not a suitable protease for determining the location of Toc12.
Figure 4.
A fraction of Toc12 is intrinsically resistant to thermolysin digestion. In vitro-translated [35S]Met-Toc12 before import (Ivt) and chloroplasts after import of Toc12 (Chpt) were treated with 200, 300, or 400 μg mL−1 thermolysin or with 200 μg mL−1 thermolysin plus 0.1% Triton X-100. Samples were analyzed by SDS-PAGE followed by fluorography (for Toc12) or immunoblotting (for Toc159 and Tic110). Lanes 1 to 5 (Ivt) contained 3.6% of the in vitro-translated Toc12 used for chloroplast import shown in lanes 6 to 10. Toc159 is easily degraded into various fragments during chloroplast isolation. The full-length Toc159 is indicated by the arrow, and the major 86-kD degradation fragment is indicated with the asterisk.
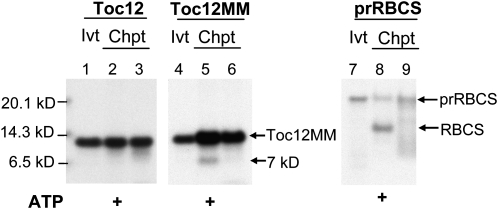
We noticed that if Toc12 has a transit peptide like PsJ8b and the processing of Toc12 occurs at the corresponding site to the one predicted for AtJ8 (Fig. 1, arrow), the mature Toc12 produced after removal of the transit peptide would contain no Met residues and thus would not be visible in autoradiographs. Therefore, we added two Met residues at the C terminus of Toc12 by site-directed mutagenesis and named this protein Toc12MM. When [35S]Met-labeled Toc12MM was incubated with isolated pea chloroplasts under import conditions, a 7-kD protein was produced (Fig. 5, lane 5). This 7-kD protein was produced only when 5 mm ATP was added to the import reaction and was not produced when no ATP was added (Fig. 5, lane 6), suggesting that it was the mature protein produced after import into chloroplasts. However, possibly due to its tight folding and tendency to aggregate, the import efficiency of Toc12MM was very low. Under identical conditions, no band was visible in the same region of the Toc12 import samples either with or without ATP addition (Fig. 5, lanes 2 and 3). In a control reaction using the precursor to the small subunit of ribulose 1,5-bisphosphate carboxylase (prRBCS), mature RBCS was only produced when ATP was added, confirming the effectiveness of ATP addition (Fig. 5, lane 8).
Figure 5.
Toc12MM can be processed into a 7-kD protein after import into chloroplasts. In vitro-translated [35S]Met-Toc12, [35S]Met-Toc12MM, and [35S]Met-prRBCS were incubated with isolated pea chloroplasts in the presence (lanes 2, 5, and 8) or absence (lanes 3, 6, and 9) of 5 mm ATP. In vitro-translated proteins (Ivt) and chloroplasts after the import reactions (Chpt) were analyzed by SDS-PAGE and fluorography. Lanes 1, 4, and 7 represent 0.8%, 0.24%, and 0.24% of the in vitro-translated proteins used for the corresponding import experiments, respectively.
To identify the location of the 7-kD protein produced after import of Toc12MM, chloroplasts after import were treated with trypsin. As shown in Figure 6A, after import into chloroplasts, the 7-kD protein produced after import was trypsin resistant, indicating that it was inside the inner membrane. When a high amount of in vitro-translated Toc12MM before import was analyzed, an internal initial product very similar in size to the 7-kD mature protein could be observed (Fig. 6A, lane 1). This internal initiation product was completely trypsin sensitive, further supporting the notion that the 7-kD trypsin-resistant mature protein was produced after importing into chloroplasts. The same import samples were analyzed by immunoblotting using antibodies against Toc75 and Tic110. Toc75 was trypsin sensitive and Tic110 was trypsin resistant, indicating that trypsin had indeed penetrated the outer but not the inner membrane.
Figure 6.
The 7-kD protein produced after import of Toc12MM is localized in the stroma. A, In vitro-translated [35S]Met-Toc12MM (Ivt) and chloroplasts after import of Toc12MM (Chpt) were treated with trypsin. Samples were analyzed with SDS-PAGE followed by fluorography (for Toc12MM) or immunoblotting (for Toc75 and Tic110). The Ivt lanes contained 0.4% of the in vitro-translated Toc12MM used for the import reactions shown in the Chpt lanes. B, Fractionation of Toc12MM, Toc12, and prRBCS after import into chloroplasts. Chloroplasts after import of Toc12MM, Toc12, and prRBCS were lysed hypotonically (hypo) or treated by alkaline extraction (alka) and then separated into membrane (M) and soluble (S) fractions. Samples were analyzed by SDS-PAGE followed by fluorography (for imported proteins) or immunoblotting (for Toc75). For Toc12MM and Toc12 import reactions, 5 mm ATP was added. For the prRBCS import reaction, no additional ATP was supplied. Import reactions were performed under light, and the in vitro-translated protein products contained a low amount of ATP from the in vitro translation system.
To further analyze whether Toc12MM and the 7-kD mature protein are integral membrane proteins or soluble proteins, chloroplasts after import of Toc12MM were lysed and separated into membrane and soluble fractions. Import of prRBCS and Toc12 was performed as controls. A lower concentration of ATP was present in the prRBCS reaction in order to produce a higher amount of prRBCS associated with the envelope membranes to serve as an envelope protein control. When chloroplasts were lysed hypotonically, the 7-kD protein produced after import of Toc12MM was present both in the membrane and the soluble fractions. However, when chloroplasts were treated by alkaline extraction, all the 7-kD mature protein was in the soluble fraction. Most Toc12MM and Toc12 fractionated with the membranes when chloroplasts were lysed hypotonically, but more than half of Toc12MM and Toc12 were in the soluble fractionation when chloroplasts were lysed by alkaline extraction (Fig. 6B, lane 4). This membrane association pattern of Toc12MM and Toc12 was similar to that of prRBCS bound to the chloroplast envelope, partially translocated through the translocon (Fig. 6B, prRBCS panel). In comparison, Toc75, a protein with a β-barrel-type membrane anchor, was not affected by the alkaline extraction and was entirely in the membrane fraction. These results suggest that the 7-kD protein produced after import of Toc12MM is a soluble protein with a fraction peripherally associated with membranes. Together with its trypsin resistance, our results indicate that this 7-kD protein is located in the stroma.
It has been shown that a commercially available anti-DnaJ antibody can also recognize Toc12 (Becker et al., 2004). We used the same antibody in order to know the relative position of endogenous Toc12 to PsJ8. We used the antibody on immunoblots of purified outer membrane proteins and total chloroplast soluble and membrane proteins. However, perhaps due to different immunoblotting conditions, we failed to detect specific proteins in these fractions (Supplemental Fig. S4). A few bands were detected after a longer exposure, but they were also detected by the anti-Toc75 antibody and on a control blot probed with the secondary antibody alone.
Mutations in AtJ8 Result in No Import Defect
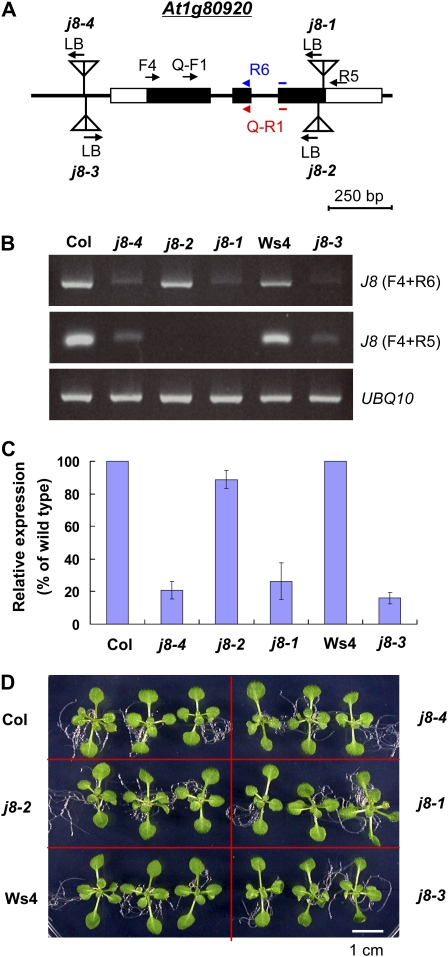
To further investigate the possible function of J8 in chloroplast protein import, we obtained four Arabidopsis mutants with T-DNA insertions in the AtJ8 gene (Fig. 7A). The j8-1 mutant (Salk_024617) is the same allele as reported previously (Chen et al., 2010). The j8-1, j8-2 (Gabi _922G05), and j8-4 (WiscDxLoxHs005M07G) alleles are in the Columbia ecotype, and the j8-3 (Flag_349D04) allele is in the Wassilewskija-4 ecotype. The j8-1 and j8-2 alleles have a T-DNA insertion close to the C terminus of AtJ8 (Figs. 1 and 7A). The j8-3 and j8-4 alleles have a T-DNA insertion in the promoter region 80 and 85 bp upstream of the transcription start site, respectively. Reverse transcription (RT)-PCR using primers on one side of the T-DNA insertion sites (F4 + R6, Fig. 7A) indicated that the j8-1, j8-3, and j8-4 alleles had a very small amount of AtJ8 RNA left, and the j8-2 allele still had a substantial amount of AtJ8 RNA (Fig. 7B, top panel). When using a pair of primers located on two sides of the T-DNA insertion sites of j8-1 and j8-2 (F4 + R5, Fig. 7A), no AtJ8 transcript could be detected in j8-1 or j8-2 (Fig. 7B). Primers were further designed to quantify the amount of AtJ8 RNA in the mutants using real-time quantitative RT-PCR. When using a pair of primers located on one side of the T-DNA insertion sites (Q-F1 + Q-R1, Fig. 7A), the j8-1, j8-3, and j8-4 alleles had about 26%, 21%, and 16% of AtJ8 transcript left, respectively, compared with their corresponding wild types. The j8-2 allele still had about 89% of AtJ8 transcript left (Fig. 7C). However, all four mutants appear indistinguishable from the wild types (Fig. 7D). They also have the same chlorophyll and carotenoid contents as those in the wild type (Supplemental Fig. S3), similar to the reported data for j8-1 (Chen et al., 2010).
Figure 7.
Characterizations of four Arabidopsis j8 mutants. A, Schematic representation of T-DNA insertion sites of the j8 mutants. Black and white boxes represent the translated and untranslated exon regions, respectively. LB, Left border of the T-DNA. Positions of primers used for RT-PCR shown in B, and real-time quantitative RT-PCR shown in C, are indicated by arrows. Primers R6 and Q-R1 span the junction between the second and the third exons. B, RT-PCR analyses of AtJ8 transcripts in the j8 mutants. The primers used are indicated in parentheses on the right. The parental ecotype of the j8-1, j8-2, and j8-4 alleles is Columbia (Col). The j8-3 allele is in the Wassilewskija-4 (Ws4) ecotype. The amount of UBQ10 transcripts was analyzed as a control. C, Real-time quantitative RT-PCR analyses of the AtJ8 transcript levels in the j8 mutants. The AtJ8 transcript level was first normalized to the level of UBQ10 in each sample, and the expression level in the wild type was then set as 100%. Primers for AtJ8 detection are Q-F1 and Q-R1 as shown in A. Data are means ± sd; n = 2. D, Phenotype of the j8 mutants. Arabidopsis seedlings were grown on MS medium under 16 h of light/8 h of dark for 14 d. [See online article for color version of this figure.]
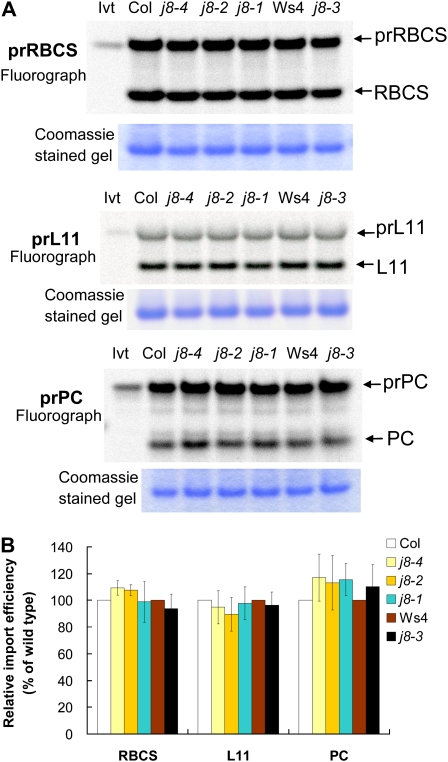
Chloroplasts were isolated from 14-d-old seedlings of the four j8 mutants and their corresponding wild types. They were incubated with chloroplast precursor proteins under import conditions. Three precursors were tested: prRBCS, the precursor to the nucleus-encoded chloroplast 50S ribosomal subunit L11 (prL11), and the precursor to the thylakoid lumen protein plastocyanin (prPC). As shown in Figure 8, for all three precursors tested, the import efficiency of the four mutants was similar to that of their corresponding wild types.
Figure 8.
The Arabidopsis j8 mutants show no detectable defect in chloroplast protein import. A, In vitro-translated [35S]Met-prRBCS, [35S]Met-prL11, and [35S]Met-prPC were imported into chloroplasts isolated from the j8 mutants and their corresponding wild types. Samples were analyzed by SDS-PAGE. The gel was first stained with Coomassie Brilliant Blue and then dried for fluorography. The amount of endogenous RBCS as revealed by Coomassie Brilliant Blue staining is shown below the fluorograph of the same gel. Col, Columbia; Ivt, in vitro-translated precursor protein; Ws4, Wassilewskija-4. B, Quantification of imported mature proteins shown in A. The amount of imported mature protein was first normalized to the amount of endogenous RBCS of the same sample. The import efficiency of the wild type was then set as 100%. Data are means ± sd; n = 3 to 6. [See online article for color version of this figure.]
DISCUSSION
We show here that the pea TOC12 gene is part of the PsJ8b gene. Furthermore, all four proteins, AtJ8, PsJ8a, PsJ8b, and Toc12, are synthesized as higher molecular mass precursors with cleavable transit peptides. After import into chloroplasts, the processed mature proteins are localized in the stroma. A fraction of the imported mature proteins was associated with membranes but was extracted to the soluble fraction when chloroplasts were treated with alkaline extraction. This tendency to associate with membranes is similar to the other two stromal J proteins investigated, Arabidopsis J11 (Chen et al., 2010) and pea PCJ1 (Schlicher and Soll, 1997).
Several experiments were performed to investigate the localization of Toc12 when it was first identified (Becker et al., 2004). In vitro-translated, [35S]Met-labeled Toc12 was imported into isolated chloroplasts and shown to be thermolysin resistant after import, suggesting that Toc12 was located inside the outer membrane. However, the in vitro-translated, [35S]Met-labeled Toc12 before import was not tested for its thermolysin resistance. We show here that a fraction of Toc12 was intrinsically thermolysin resistant even before import. The amount of Toc12 that was thermolysin resistant did not increase after import. The fraction of Toc12 that was thermolysin resistant remained resistant even in the presence of Triton X-100. Therefore, thermolysin resistance cannot be used as an indication that Toc12 was inside the outer membrane. In the original report, a fusion protein of Toc12 fused at the N terminus of GFP (Toc12-GFP) was transiently expressed in tobacco (Nicotiana tabacum) protoplasts. No microscopy data were provided for the location of Toc12-GFP. Chloroplasts isolated from the transfected protoplasts were analyzed by immunoblots, and Toc12-GFP was shown to be trypsin sensitive, suggesting that Toc12-GFP was outside the inner membrane. However, in the same experiment, Tic110 was also trypsin sensitive. Although there are still some debates about the topology of Tic110, all models agree that the large majority of the Tic110 polypeptide is localized in the stroma (Jackson et al., 1998; Balsera et al., 2009). Therefore, it is possible that in the original experiment, trypsin was not properly quenched and had gained access into the stroma. Nonetheless, Toc12 was identified by electrospray ionization tandem mass spectrometry from isolated pea chloroplast outer membrane proteins. Although only one peptide was identified, the peptide was located close to the C terminus of Toc12 and contains two amino acids unique to Toc12. An antibody generated against the entire length of Toc12, including the transit peptide region, recognized a single protein around 14 kD on immunoblots when isolated pea chloroplast outer membrane vesicles were analyzed (Becker et al., 2004). Therefore, it is possible that TOC12 RNA is an alternatively spliced form of PsJ8b, leading to the generation of Toc12 protein in pea. If this is the case, it seems to be unique to pea. No EST corresponding to a similar alternatively spliced form of J8 has been found in Arabidopsis or in other species. Whole genome tiling arrays of Arabidopsis also indicate that the second intron is not transcribed (http://signal.salk.edu/cgi-bin/atta; Yamada et al., 2003). During the revision of this manuscript, a review article was published (Schwenkert et al., 2010) indicating that a thorough analysis of a pea EST database revealed that the pea Toc12 should be the same length as that of all the J8 proteins from other species (Fig. 2 in Schwenkert et al., 2010). The revised Toc12 assembled from the pea EST database is essentially the same as our PsJ8b, which we show here is a stromally localized soluble protein (Fig. 3A).
J domain-containing proteins are important cochaperones for the Hsp70 family proteins. The J domain interacts with Hsp70 when Hsp70 is bound to ATP and stimulates ATP hydrolysis by Hsp70. Specific J domain proteins often recruit Hsp70 to perform specific functions. It has recently been shown that chloroplast stromal Hsp70s are important for protein import into chloroplasts (Shi and Theg, 2010; Su and Li, 2010). Because J8 is localized in the stroma, we investigated whether J8 is involved in protein import by analyzing the j8 mutant phenotype. We analyzed four T-DNA insertion mutant alleles. Two of the alleles, j8-3 and j8-4, exhibited a significant decrease (approximately 80%) in AtJ8 transcript. The other two alleles, j8-1 and j8-2, do not possess any full-length AtJ8 transcript. However, all four mutants did not show any defect in importing various chloroplast precursor proteins, suggesting that J8 is unlikely to be involved in protein import. A recent report indicated that the j8-1 mutant has a lower CO2 assimilation and a reduced Rubisco activity (Chen et al., 2010), suggesting that J8 may be a cochaperone for the activity of Hsp70 in the folding and assembly of enzymes in the carbon fixation reaction. However, since the mutants we analyzed might still produce a low level of truncated J8 proteins, we cannot exclude the possibility that the highly conserved C-terminal region is not required for the activity of J8 in protein import.
It has been generally assumed that the ATP required for the binding/early-import-intermediate step is consumed by imsHsp70. Now that imsHsp70 and its cochaperone Toc12 have been shown to be most likely localized in the stroma, the only currently known intermembrane space components of the translocon are Tic22 and the intermembrane space domain of Toc64. Neither of these two components possesses any ATPase activity. Therefore, the proteins that use the ATP in the binding step remain unknown. It needs to be pointed out that, although it has been generally assumed that an ATPase like an Hsp70 facilitates the formation of the early import intermediates, it has been shown that GTP can also support the formation of early import intermediates without being converted to ATP (Olsen and Keegstra, 1992). When supplied with 2.5 mm GTP as the sole energy source at room temperature, the transit peptide plus a part of the mature region of precursors can be transported across the outer membrane. Interestingly, this transport is inhibited by ATPγS (Inoue and Akita, 2008). Therefore, it is possible that the translocon component that supports the formation of early import intermediates may be a novel intermembrane space NTPase with a broad nucleotide specificity that can use ATP or GTP, but with a preference for ATP. This inference makes the Hsp70 family proteins unlikely candidates as the component driving transport across the outer membrane. However, it is also possible that multiple NTPases, including the Hsp70 family proteins, are involved in the formation of early import intermediates. In summary, the linkage between the Toc and Tic complexes and the process of precursor translocation across the outer membrane and the intermembrane space remain to be elucidated.
MATERIALS AND METHODS
Isolation of J8 and Toc12 cDNA and Genomic Clones
First-strand cDNAs were synthesized using the Moloney murine leukemia virus reverse transcriptase (Invitrogen) with RNA isolated from pea (Pisum sativum ‘Little Marvel’) or Arabidopsis (Arabidopsis thaliana) leaves. Primers used to amplify the PsJ8 cDNA and genomic DNA as well as the AtJ8 cDNA are listed in Supplemental Tables S1 and S2. PCR-amplified fragments were subcloned into vector pGEM-T or pSP72 (Promega). Site-directed mutagenesis to create Toc12 and Toc12MM was performed on plasmids containing the coding sequence for PsJ8b using the QuikChange site-directed mutagenesis kit (Stratagene) and primers listed in Supplemental Table S1. 3′ RACE was performed according to the manufacturer’s protocol of the 5′/3′ RACE Kit (Roche).
Chloroplast Protein Import and Postimport Treatments
Arabidopsis seedlings were grown on 0.3% Gelrite-solidified Murashige and Skoog (MS) medium containing Gamborg’s B5 vitamins and 2% Suc. Plants were grown in growth chambers under a 16-h photoperiod at 22°C with a light intensity of about 80 μmol m−2 s−1. For growing pea seedlings (De Bruyn Seed Store), imbibed seeds were grown on vermiculite under a 12-h photoperiod at 20°C with a light intensity of about 150 μmol m−2 s−1. Chloroplasts were isolated from 9-d-old pea seedlings or 14-d-old Arabidopsis seedlings as described (Perry et al., 1991), except that the grinding buffer for Arabidopsis chloroplast isolation was modified to 50 mm HEPES-KOH (pH 8.0), 330 mm sorbitol, 2 mm EDTA, and 0.5% bovine serum albumin. Synthesis of [35S]Met-labeled precursors through in vitro transcription and in vitro translation using wheat germ lysate (Promega), protein import into chloroplasts, and treatment of chloroplasts with thermolysin after import were performed as described (Perry et al., 1991). Reticulocyte lysates cannot be used for translating Toc12 because Toc12 migrated at the same position as hemoglobin that is sometimes produced in reticulocyte lysates. In vitro-translated Toc12 tends to aggregate and therefore was centrifuged at 20,000g for 15 min to remove large aggregates before use for further experiments. For import under ATP-depleted conditions, ATP was removed from the in vitro-translated precursors by gel filtration as described (Perry et al., 1991) and import was performed under a green safelight. Hypotonic lysis of chloroplasts after import was performed by resuspending reisolated intact chloroplasts in 25 mm HEPES-KOH (pH 8.0) and 4 mm MgCl2. Alkaline extraction was performed by resuspending reisolated intact chloroplasts after import in 0.1 m Na2CO3 (pH 11.5) and incubating for 30 min at 4°C. Hypotonically lysed and alkaline-extracted samples were then separated into membranes and soluble fractions by ultracentrifugation at 100,000g for 45 min. Soluble fractions were precipitated in 10% TCA, washed with ice-cold acetone, and dissolved in SDS-PAGE sample buffer. Trypsin treatment of chloroplasts after import was performed as described (Jackson et al., 1998). Immunoblotting was performed as described (Su and Li, 2010) using specific primary antibodies and an alkaline phosphatase-conjugated secondary antibody and visualized by the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate colorimetric system, except for Supplemental Figure S4, which was visualized using the horseradish peroxidase-conjugated secondary antibodies and the Immobilon Western Chemiluminescent HRP system (Millipore), visualized with the UVP BioSpectrum 600 Image System (Ultra Violet Products). Antibodies against Arabidopsis Tic110 and Toc159 and pea Toc75 were generated as described (Tu et al., 2004). The antibody against Escherichia coli DnaJ (ADI-SPA-410) was purchased from Stressgen.
Isolation of T-DNA-Tagged Arabidopsis j8 Mutants
Arabidopsis mutants with T-DNA insertion in the AtJ8 genomic region were obtained from various sources. The j8-1 (Salk_024617; Alonso et al., 2003) and j8-4 (WiscDxLoxHs005M07G; Nishal et al., 2005) mutants were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). The j8-2 mutant (Gabi _922G05; Rosso et al., 2003) was obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). The j8-3 mutant (Flag_349D04; Samson et al., 2002) was obtained from the French National Institute for Agricultural Research (http://www.inra.fr/). Primers for the identification of T-DNA insertions in the AtJ8 genomic region are as follows: AtJ8geno-F2, AtJ8geno-R2, and various left border primers. Left border primers used are Salk-LBa1 for j8-1, Gabi-LB for j8-2, Flag-LB4 for j8-3, and Wisc-L4 for j8-4. Primer sequences are listed in Supplemental Table S1. T-DNA insertion sites were confirmed by sequencing.
RT-PCR, Quantitative RT-PCR, and Chlorophyll Content Analyses of j8 Mutants
Total RNA was isolated using TRIzol reagent (Invitrogen) from Arabidopsis leaves. First-strand cDNAs were synthesized using total RNA, Moloney murine leukemia virus reverse transcriptase (Invitrogen), and a poly(dT) primer. For RT-PCR (Fig. 7B), cycle numbers for amplification were 20 for UBQ10 and 25 for AtJ8. Real-time quantitative RT-PCR was performed with the LightCycler System (Roche) and the LightCycler-FastStart DNA Master SYBR Green I kit (Roche). For each PCR, 10 to 50 ng of cDNA and 0.5 μm primer pairs were used. The initial denaturing step of 10 min was followed by 40 cycles of 95°C for 10 s, 60°C for 5 s, and 72°C for 1 s per 25 bp of the expected product. A standard curve based on serial dilutions of the wild-type cDNA for each primer pair was included. The absence of nonspecific products was checked by melting curve analysis. Quantification was performed using the LightCycler Relative Quantification software version 1.0. Normalization was done using the transcript level of UBQ10. Primers used to amplify each transcript are listed in Supplemental Tables S1 and S2. Chlorophyll and carotenoid contents were determined by the method of Lichtenthaler (1987).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY357119 (pea Toc12), BT051272 (J8 of Medicago truncatula), XM_002512691 (J8 of castor bean [Ricinus communis]), BT094680 (J8 of soybean [Glycine max]), NP_178207 or At1g80920 (AtJ8 of Arabidopsis), HM565932 (PsJ8a cDNA of pea), HM565933 (PsJ8b cDNA), HM565934 (PsJ8a genomic DNA), and HM565935 (PsJ8b genomic DNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of J8 cDNA sequences from Medicago (MtJ8), soybean (GmJ8), and castor bean (RcJ8).
Supplemental Figure S2. Sequence alignment of PsJ8a and PsJ8b cDNA and TOC12.
Supplemental Figure S3. Pigment contents of Arabidopsis j8 mutants.
Supplemental Figure S4. The antibody against E. coli DnaJ failed to detect specific proteins in various fractions from pea chloroplasts.
Supplemental Table S1. Sequences of primers used in this study.
Supplemental Table S2. Primers used for the amplification of PsJ8s and Toc12 cDNA and genomic DNA and site-directed mutagenesis.
Supplementary Material
Acknowledgments
We thank Dr. Jychian Chen for the anti-APS antibody, Dr. Heiko Kuhn for English editing, Dr. Yi-shin Su for the purified pea chloroplast outer membrane vesicles, and Drs. Bo Liu and Yuh-Ru Lee for helping us purchase the anti-DnaJ antibody.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Balsera M, Goetze TA, Kovács-Bogdán E, Schürmann P, Wagner R, Buchanan BB, Soll J, Bölter B. (2009) Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J Biol Chem 284: 2603–2616 [DOI] [PubMed] [Google Scholar]
- Becker T, Hritz J, Vogel M, Caliebe A, Bukau B, Soll J, Schleiff E. (2004) Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol Biol Cell 15: 5130–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Holmström M, Raksajit W, Suorsa M, Piippo M, Aro EM. (2010) Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K. (1984) Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol 75: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ. (2003) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem 278: 38617–38627 [DOI] [PubMed] [Google Scholar]
- Inaba T, Schnell DJ. (2008) Protein trafficking to plastids: one theme, many variations. Biochem J 413: 15–28 [DOI] [PubMed] [Google Scholar]
- Inoue H, Akita M. (2008) Three sets of translocation intermediates are formed during the early stage of protein import into chloroplasts. J Biol Chem 283: 7491–7502 [DOI] [PubMed] [Google Scholar]
- Inoue K. (2007) The chloroplast outer envelope membrane: the edge of light and excitement. J Integr Plant Biol 49: 1100–1111 [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem 273: 16583–16588 [DOI] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. (1997) Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol 139: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kim JK, Lee S, Choi S, Kim S, Hwang I. (2008) Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-m, Chiu CC. (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Marshall JS, DeRocher AE, Keegstra K, Vierling E. (1990) Identification of heat shock protein hsp70 homologues in chloroplasts. Proc Natl Acad Sci USA 87: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. (2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishal B, Tantikanjana T, Sundaresan V. (2005) An inducible targeted tagging system for localized saturation mutagenesis in Arabidopsis. Plant Physiol 137: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ, Keegstra K. (1992) The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J Biol Chem 267: 433–439 [PubMed] [Google Scholar]
- Olsen LJ, Theg SM, Selman BR, Keegstra K. (1989) ATP is required for the binding of precursor proteins to chloroplasts. J Biol Chem 264: 6724–6729 [PubMed] [Google Scholar]
- Perry SE, Li H-m, Keegstra K. (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Bionda T, Reger K, Ruprecht M, Soll J, Schleiff E. (2007) Toc64: a preprotein-receptor at the outer membrane with bipartite function. J Mol Biol 367: 1330–1346 [DOI] [PubMed] [Google Scholar]
- Ratnayake RM, Inoue H, Nonami H, Akita M. (2008) Alternative processing of Arabidopsis Hsp70 precursors during protein import into chloroplasts. Biosci Biotechnol Biochem 72: 2926–2935 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicher T, Soll J. (1997) Chloroplastic isoforms of DnaJ and GrpE in pea. Plant Mol Biol 33: 181–185 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. (1994) Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Schwenkert S, Soll J, Bölter B. (2010) Protein import into chloroplasts: how chaperones feature into the game. Biochim Biophys Acta (in press) [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li H-m. (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li H-m. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Chen LJ, Smith MD, Su YS, Schnell DJ, Li H-m. (2004) Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.