Abstract
SHORT-ROOT (SHR) and SCARECROW (SCR) are required for stem cell maintenance in the Arabidopsis (Arabidopsis thaliana) root meristem, ensuring its indeterminate growth. Mutation of SHR and SCR genes results in disorganization of the quiescent center and loss of stem cell activity, resulting in the cessation of root growth. This paper reports on the role of SHR and SCR in the development of leaves, which, in contrast to the root, have a determinate growth pattern and lack a persistent stem cell niche. Our results demonstrate that inhibition of leaf growth in shr and scr mutants is not a secondary effect of the compromised root development but is caused by an effect on cell division in the leaves: a reduced cell division rate and early exit of the proliferation phase. Consistent with the observed cell division phenotype, the expression of SHR and SCR genes in leaves is closely associated with cell division activity in most cell types. The increased cell cycle duration is due to a prolonged S-phase duration, which is mediated by up-regulation of cell cycle inhibitors known to restrain the activity of the transcription factor, E2Fa. Therefore, we conclude that, in contrast to their specific roles in cortex/endodermis differentiation and stem cell maintenance in the root, SHR and SCR primarily function as general regulators of cell proliferation in leaves.
Stem cells are undifferentiated, totipotent cells that are able to duplicate themselves and to form offspring that differentiates into multiple cell types. They are situated in a microenvironment, the stem cell niche, where extracellular signals maintain stem cell division at low rates and prevent differentiation (Ohlstein et al., 2004; Li and Xie, 2005). In plants, the best studied stem cell niches are within the root and shoot apical meristems. There, stem cells produce somatic daughter cells that go on dividing and expanding, thereby forming the postembryonic tissues and organs that make up the body of the plant. It is the balance between stem cell maintenance within the meristem and differentiation of cells that exit the niche that facilitates indeterminate root and shoot growth.
SHORT-ROOT (SHR) and SCARECROW (SCR) are members of the GRAS family of transcription factors (Pysh et al., 1999; Lee et al., 2008), required for stem cell maintenance in the root apical meristem. Mutation of SHR and SCR genes causes a disorganization of the quiescent center (QC) and loss of stem cell activity, resulting in the depletion of proliferating cells in the root meristem and, consequently, cessation of root growth. Essentially, loss of SHR/SCR function renders root growth determinate. Furthermore, shr and scr mutants lack longitudinal cell divisions that separate the cortex/endodermis initial daughter cells, resulting in only one ground tissue cell layer (Benfey et al., 1993; Scheres et al., 1995; Di Laurenzio et al., 1996; Helariutta et al., 2000; Sabatini et al., 2003; Heidstra et al., 2004). In the shr mutant, this cell layer displays only cortex characteristics, whereas the scr ground tissue layer shows a mixed cortex/endodermis identity. The shr phenotype indicates that SHR is necessary both for the asymmetric division that generates cortex and endodermis and for endodermis cell fate specification (Benfey et al., 1993; Scheres et al., 1995; Di Laurenzio et al., 1996). Expression of SCR in the QC of shr mutants cannot rescue QC function and only partially rescues stem cell maintenance (Sabatini et al., 2003). The observations that shr is epistatic to scr, SCR expression is reduced in shr roots, and SHR binds to the SCR promoter indicate that SCR acts directly downstream of SHR (Helariutta et al., 2000; Levesque et al., 2006).
In shoots, loss of SHR or SCR function affects differentiation of the bundle sheath cell layer in leaves and the endodermis in hypocotyls and inflorescence stems, suggesting that the radial patterning of ground tissue in both root and shoot is regulated by the same molecular mechanism (Fukaki et al., 1998; Wysocka-Diller et al., 2000). Furthermore, several studies reported an overall shoot growth phenotype in the shr and scr mutants. The shoot gravitropism1 mutant, later identified to be allelic to scr, has small leaves, and growth of the shr mutant is severely retarded, resulting in a stunted shoot phenotype (Benfey et al., 1993; Fukaki et al., 1996, 1998). Here, we show that retarded leaf growth in shr and scr mutants is not a secondary effect of the compromised root but is caused by the loss of SHR and SCR function in the leaf tissue. Besides their function in ground tissue specification, SHR and SCR also affect proliferative cell division, driving growth in leaves. This is surprising, as in the root the effect of shr and scr on organ growth appears to be primarily mediated through their effect on stem cell maintenance, not proliferation outside the stem cell niche. Here, we use the Arabidopsis (Arabidopsis thaliana) leaf as a system to specifically study the function of these genes in cell cycle regulation (separate from their role in stem cell specification) and how this relates to the shoot growth phenotype.
RESULTS
Rosette Growth Is Reduced in shr and scr Mutants
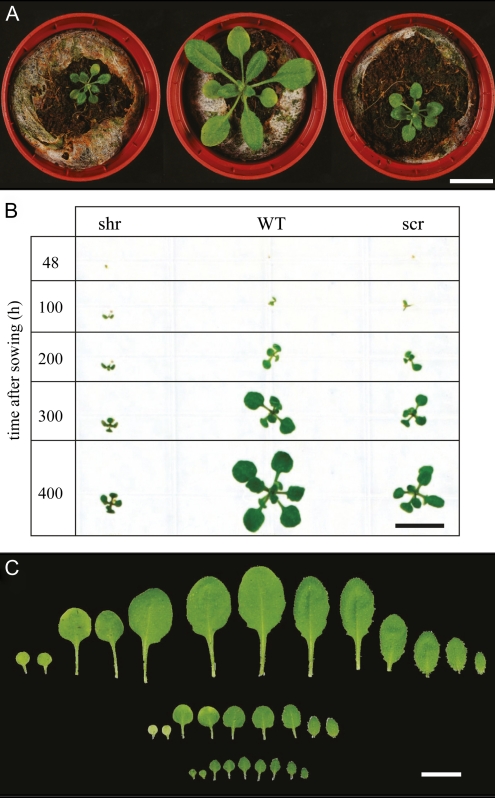
Besides their characteristically strong reduction in primary root growth (Benfey et al., 1993; Di Laurenzio et al., 1996), shr and scr mutants display a severely dwarfed shoot phenotype (Fig. 1A). Following the development of shr and scr plants in vitro shows that germination is not affected, the seedlings being essentially the same size as the wild type 100 h after sowing. Thereafter, however, the rate of growth of shr and scr plants is severely inhibited (Fig. 1B). At 18 d after sowing (DAS), the projected rosette area was reduced 6.1-fold in shr plants and 2.6-fold in scr plants. The observed reduced rosette size is due to fewer leaves being present and leaves at any position being smaller (Fig. 1C; Supplemental Fig. S1). The largest difference occurs under our experimental conditions in leaf 5 and 7 for scr and shr, with reductions of 73% and 95% in leaf area, respectively. Thus, besides a reduction in individual leaf growth, these results indicate that, in contrast to the complete termination in the root apical meristem (Benfey et al., 1993; Di Laurenzio et al., 1996; Helariutta et al., 2000; Sabatini et al., 2003), the activity of the shoot apical meristem is partially inhibited in scr and shr mutant lines.
Figure 1.
Rosette growth of shr and scr mutants. A, Rosettes of shr-6 (left), wild-type (middle), and scr-3 (right) plants at 20 DAS. Bar = 1 cm. B, Representative images for shr-6, scr-3, and wild-type (WT) plants 48, 100, 200, 300, and 400 h after sowing. Bar = 1 cm. C, Rosette leaves of soil-grown wild-type (top), scr-3 (middle), and shr-6 (bottom) plants harvested before bolting, from oldest (left) to youngest (right), including the two cotyledons. Bar = 1 cm.
The Shoot Phenotype Is Not a Consequence of the Aberrant Root Phenotype
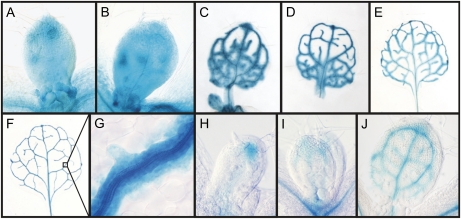
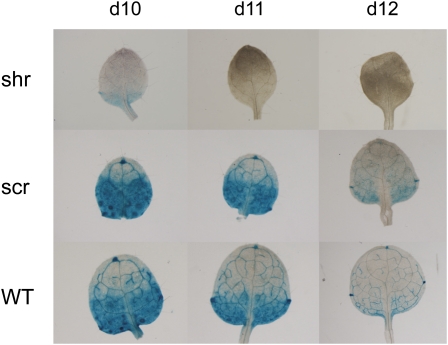
A plausible explanation for the observed shoot phenotype of the shr and scr mutants is that it is an indirect effect of the compromised root development. Therefore, we set out to investigate whether the shr and scr mutant leaf phenotype can be attributed to a direct function of these genes during leaf development. First, SHR expression was analyzed in developing leaves using the SHR:GUS line (Helariutta et al., 2000). Staining of proliferating leaves of 6- and 7-d-old plants for 5 h showed an expression signal throughout the leaf (Fig. 2, A and B). At later stages of leaf development, this global expression pattern changed: from day 10 onward, when cell proliferation is known to stop (Beemster et al., 2005), the pattern gradually became confined to the vascular network (Fig. 2, C–F). In mature leaves of 24-d-old plants, the expression was still high in the vascular bundle. More detailed analysis showed that the GUS signal was present in the entire vascular bundle and bundle sheath cells surrounding it (Fig. 2G). In leaves of 5-, 6-, and 7-d-old plants, incubation for only 2 h revealed that, although the expression occurred throughout the leaf, during the early stages the GUS signal was already higher in developing vascular traces than in the surrounding ground tissue (Fig. 2, H–J). Notably, the data show that the expression was specific for the vascular pattern well before differentiation of the vascular bundles can be observed at the cellular level (data not shown). Thus, analysis of SHR:GUS shows the expression of the SHR gene in proliferating cells, the vascular system, and the surrounding bundle sheath cells. This expression pattern closely resembles that of SCR, which is also expressed in most tissues in young proliferating leaves, whereas during expansion and maturation of the leaf the expression pattern becomes specific to the bundle sheath cells associated with the veins (Wysocka-Diller et al., 2000). Therefore, we conclude that the expression pattern of both genes coincides with general cell proliferation and vascular differentiation during leaf development.
Figure 2.
SHR expression during leaf development. A to F, SHR:GUS expression in leaves 1 and 2 of 6-d-old (A), 7-d-old (B), 10-d-old (C), 12-d-old (D), 16-d-old (E), and 24-d-old (F) plants after 5 h of incubation. G, Expression in the vascular bundle and the bundle sheath cells surrounding it at 24 DAS. H to J, Incubation for 2 h at day 5 (H), day 6 (I), and day 7 (J).
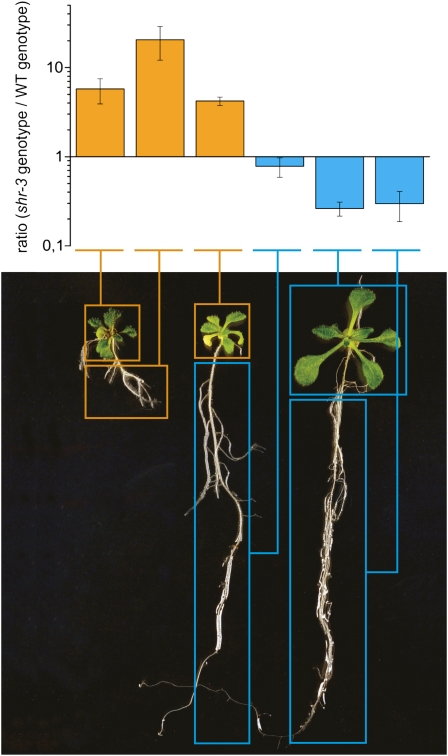
To also obtain genetic evidence that the SHR expression in the leaf directly affects leaf development, we used the shr-3 mutant, which contains the En-1 transposon in the coding sequence of the SHR gene (Helariutta et al., 2000). shr-3 plants were grown on vertical plates and screened for reverted wild-type root phenotypes (indicative of En-1 transposition) at 20 DAS. Approximately 3% of the plants developed a root system that was much longer than normal shr-3 roots, closely resembling a wild-type root system (Fig. 3). The plants with normal root systems could be divided into two groups, those with shoots comparable to shoots of wild-type plants and those with a shr rosette. This second class demonstrates that having normal root development does not rescue the shr shoot phenotype. To investigate whether the reverted shoot and root phenotypes were due to reactivation of the SHR gene, we analyzed DNA from individual shoots and roots, with shr or wild-type phenotypes, respectively. To this end, we performed a quantitative (Q)-PCR using a set of primers that bridge the joint between the SHR coding sequence and the En-1 transposon and therefore only yield a PCR fragment when the transposon is present in the gene (SHR primers). Additionally, we used a second set of primers that flank the insertion site of the complete 8-kb-long En-1 transposon in the SHR gene, which only gives a PCR fragment in the absence of the transposon (i.e. after excision from its original locus [wild-type primers]). The ratio between the relative signals for these primer pairs was used to determine the genotype with respect to shr-3. As expected, in plants with a shr phenotype in both root and shoot, the expression ratio between the signal from SHR and wild-type primers was significantly higher than 1 in root and shoot samples, consistent with a shr-3 genotype. In contrast, in plants showing a completely reverted phenotype, both shoot and root samples gave a signal ratio below 1, consistent with the absence of the En-1 transposon in the SHR gene. Finally, in plants with a dwarfed shoot but a root with a wild-type phenotype, the expression ratio in the shoot was higher than 1, corresponding to a shr genotype, whereas in the root it was below 1, indicating the restoration to the wild-type genotype (Fig. 3). The reduction of the expression ratio in the plants with only a reversion in the roots was smaller than in fully reverted plants. This can be explained by the presence of the transposon in a part of the root because the reversion events that give rise to root-only rescue probably occurred later in development than whole plant reversions. Nevertheless, these results suggest that translocation of the En-1 transposon in shr-3 plants independently reverts the dwarfed shoot and short root phenotype to the wild type, indicating that SHR expression in the shoot is needed for normal rosette growth, irrespective of the genotype and the associated phenotype of the root.
Figure 3.
The effect of SHR mutation in the shoot is independent of its effect on root growth. Revertant transposon mutants show genetic independence of the root and shoot phenotypes in shr-3 mutants. The genotype of reverted shoot and root phenotypes (left, normal shr-3 plant; middle, reverted root, shr-3 rosette; right, reverted root and shoot) was analyzed in individual roots and shoots. The ratio between DNA containing the shr-3 genotype and the wild-type (WT) genotype was measured via Q-RT-PCR. Error bars represent se (n ≥ 3).
To confirm physiologically that the shr and scr shoot phenotypes are due to their function in the leaf, rather than a consequence of their effect on root development, we compared their effect on leaf growth with that of plants in which the root growth was physically restricted to the same extent. For this, the roots of wild-type plants growing in vitro were cut at 2 cm below the hypocotyl every 3 d after germination, roughly the size that the roots of shr plants reach. At 24 DAS, leaves 1 and 2 of untreated controls and derooted plants were harvested and compared with shr and scr leaves. Removing the roots of wild-type plants did reduce the size of the first leaves by 33% compared with control plants. The leaf area in scr and shr, however, was reduced by 55% and 80%, respectively (Supplemental Fig. S2A). In plants from which the roots were cut, the reduced leaf area was related to a 17% reduction in cell size in the abaxial epidermis compared with control plants. In contrast, the cell size in the shr and scr mutants was not significantly different from the wild type (Supplemental Fig. S2B). In the absence of an effect on cell size, the decrease in leaf area in scr and shr was due to a reduced number of epidermal cells per leaf by 52% and 79%, respectively, whereas cutting the roots reduced the number of cells by only 18% (Supplemental Fig. S2C). Thus, cutting the roots of wild-type plants decreases the final leaf area by reducing cell number and cell size in approximately equal amounts, whereas in shr and scr leaves, the reduction in leaf size is entirely due to a decrease in cell number. Moreover, despite the comparable reduction of the root system and the potential induction of additional wounding stress, the effect of cutting the roots on leaf growth was only small compared with the phenotypes in shr and scr. In the light of these observations, it is unlikely that the shoot phenotype in the mutants is due to an indirect effect of the compromised root growth. Together, the expression patterns of SHR and SCR and the genetic and physiological experiments provide evidence that the rosette phenotype observed in the shr and scr mutants is due to the loss of SHR/SCR function in the shoot itself.
Kinematic Analysis of Leaf Growth
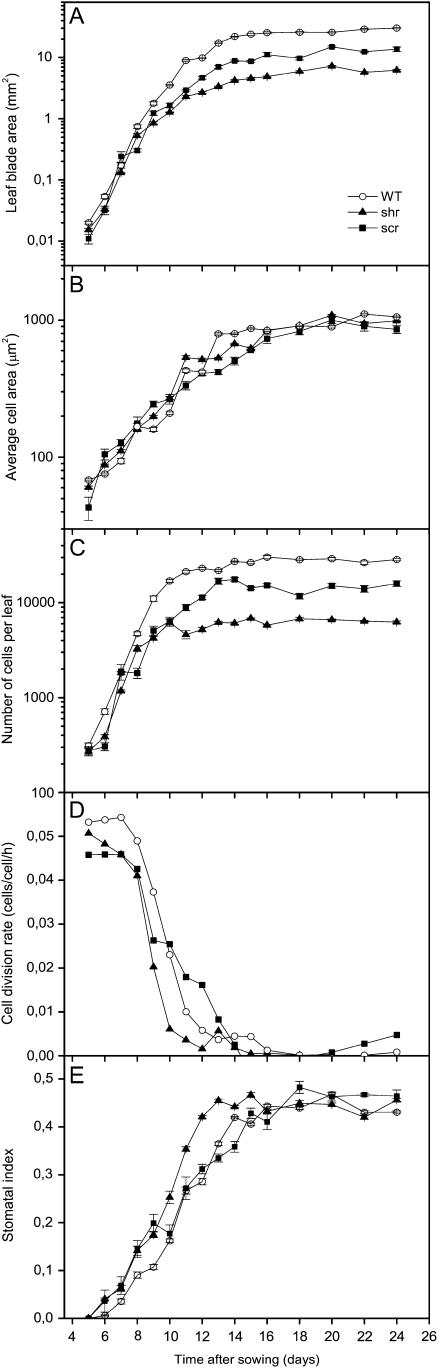
The global expression of SHR and SCR in proliferating leaf tissues, together with the reduced cell number in mature shr and scr leaves, suggest a function of SHR and SCR in the regulation of proliferative cell divisions in the leaf. To investigate this possibility, a kinematic growth analysis of the first leaf pair was performed starting at 5 DAS, when the leaves are actively proliferating, until maturity, at 24 DAS.
Measurements of leaf blade area during leaf development showed that leaf area increased exponentially until day 11 in wild-type plants, after which expansion rates gradually decreased until the mature size was reached at approximately 18 DAS (Fig. 4A). Until day 8 or 9, there is very little difference in leaf area between the wild type and mutants, but at that time the growth rate of scr and especially shr declines more rapidly than in the wild type, resulting in a 2- and 5-fold reduction in mature leaf blade area. To understand whether the differences in leaf growth were related to cell division or cell expansion, we obtained cellular data of the abaxial leaf epidermis during development. The average epidermal cell size in scr and shr mutants was comparable to the wild type throughout leaf development (Fig. 4B), suggesting that differences in the number of cells per leaf are responsible for the smaller leaf size. Similar to the leaf blade area, the number of cells first increases exponentially, followed by a slower increase until the final number of cells per leaf is reached (Fig. 4C). Again, the rate of increase in shr and scr decreased earlier and more rapidly than in wild-type plants, resulting in a 4.6- and 1.8-fold reduction in cell number per leaf compared with wild-type plants at 24 DAS. This indicates that the duration of the proliferation phase, the period in which cell division occurs, is strongly reduced by shr mutation. Indeed, there is a rapid drop in the calculated rates of cell division that occurs between day 9 and 11 in the wild type and between day 8 and 10 in shr. In scr, the initial decline also starts already at day 8 but continues until day 13 (Fig. 4D). Cell division rates were also reduced in the mutants during the first 3 d of the kinematic analysis, when all leaves were still fully proliferative: average cell division rates were 0.046, 0.048, and 0.054 cells cell−1 h−1 for scr, shr, and wild-type leaves, corresponding to mean cell cycle durations of 21.8, 20.7, and 18.6 h, respectively, indicating that cell cycle duration increased in both mutants. Curiously, our results suggest that in the scr mutant cell division continues at a very slow rate for a few more days before stopping completely, which partly compensates the reduced cell division rate during proliferation. The appearance of guard cells marks the exit from proliferation. Consistent with an earlier exit from mitosis, the first guard cells are observed at day 6 in the mutants compared with day 7 in the wild type. In mature shr and scr leaves, however, the stomatal index, the fraction of guard cells in the total number of epidermal cells, is comparable to that of the wild type (Fig. 4E). Curiously, we found that the mature guard cell area is reduced, with 38% and 23% in shr and scr leaves, respectively, indicating that mutation of SHR and SCR somehow specifically affects the expansion of guard cells (Supplemental Fig. S3). Due to their relatively small size, this reduction in guard cell area has a negligible effect on the average cell size and whole leaf area. In conclusion, the results of the kinematic analysis indicate that both shr and scr reduce leaf growth primarily by inhibition of cell division rates and an earlier exit of cell proliferation for shr.
Figure 4.
Kinematic analysis of leaf growth in shr and scr mutants. Characteristics of the abaxial epidermis of leaves 1 and 2 of shr-6, scr-3, and wild-type (WT) plants are shown. A, Leaf blade area. B, Average cell size. C, Number of cells per leaf. D, Cell division rate. E, Stomatal index. Error bars represent se (n = 5).
Progression of the Cell Division Gradient in shr and scr Mutants
To further investigate the effect of the shr mutation on cell division, we analyzed its effect on the spatiotemporal pattern of cell cycle activity during leaf development. In Arabidopsis leaves, cell division stops in a tip-to-base gradient, as can be shown by expression of the B-type cyclin CYCB1;1 (Ferreira et al., 1994; Donnelly et al., 1999; Kang and Dengler, 2002). Therefore, we crossed the cell division marker CYCB1;1:GUS (Ferreira et al., 1994; Donnelly et al., 1999) into the shr and scr background. The expression of this cell cycle marker illustrated that retraction of the cell division gradient in shr occurs significantly earlier than in scr and wild-type leaves (Fig. 5). Furthermore, in wild-type and scr leaves, the GUS signal was elevated at the position of the hydathodes, which is much less pronounced in the shr mutant. After retraction of the cell division gradient, CYCB1;1 expression in wild-type plants remained in the dispersed meristematic cells, where divisions occur in the stomatal lineage, and in cells near the vascular tissue (White, 2006). In shr plants, this expression was absent. The analyses of CYCB1;1 expression patterns confirm the early exit of the cell proliferation phase in shr shown by kinematic analysis of leaf growth.
Figure 5.
Spatiotemporal pattern of cell cycle activity during leaf development in shr, scr, and wild-type (WT) plants. GUS expression driven by the CYCB1;1 promoter in shr-6, scr-3, and wild-type leaves from 10 to 12 DAS is shown. Note the early retraction of CYCB1;1 expression in the shr-6 background.
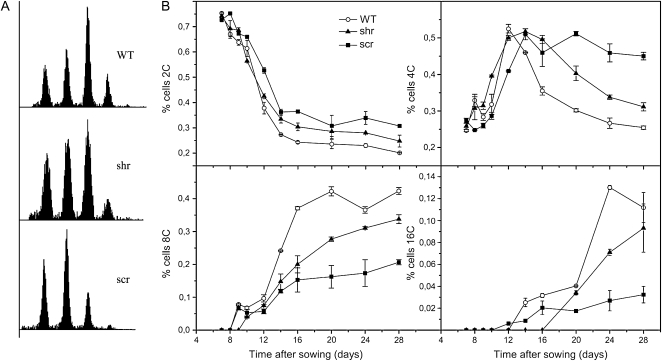
SHR and SCR Affect Endoreduplication
In leaves, exit from the cell proliferation phase coincides with the onset of endoreduplication (Beemster et al., 2005). Because mutants affecting cell division often show altered endoreduplication levels, we performed flow cytometry. Leaves 1 and 2 of shr, scr, and wild-type plants were harvested throughout leaf development. In mature leaves, at day 28, endoreduplication levels of shr and scr leaves were lower than those of wild-type leaves, as can be seen from the reduced fraction of 8C (shr and scr) and 16C (scr) in ploidy distributions (Fig. 6A). The corresponding endoreduplication index, which is the average number of endoreduplication cycles per nucleus, is 1.47, 1.31, and 0.97 for the wild type, shr, and scr, respectively. The transition from proliferation to endoreduplication around day 10 is evident from the strong increase in cells with 4C DNA content at that time (Fig. 6B). Consistent with the early exit of the proliferation phase, this transition occurred earlier in the shr mutant than that in scr and the wild type. After this stage, the decrease of the fraction of nuclei with a 4C DNA content was slower in shr than in the wild type, together with a later and slower accumulation of 8C and 16C cells. At late time points, when the leaf was mature, the amount of 8C and 16C nuclei in wild-type leaves reached a plateau, whereas in the shr mutant, the amount of 8C and 16C nuclei continued to increase. This result suggests that in the shr mutant endoreduplication starts off earlier than in the wild type but proceeds at a lower rate, so that the wild type reaches its final level before shr does. Thus, in addition to an effect on the mitotic cell cycle, shr and scr also perturb the endoreduplication cycle without affecting cell size.
Figure 6.
Endoreduplication in leaves of shr, scr, and wild-type (WT) plants. A, Ploidy distribution of shr-6, scr-3, and wild-type plants at 28 DAS. B, 2C, 4C, 8C, and 16C contents of shr-6, scr-3, and wild-type plants from 7 to 28 DAS. Error bars represent se of two biological repeats.
Microarray Analysis
After finding that SHR and SCR affect cell division in the leaf, we set out to investigate the molecular basis of this inhibition. Therefore, a genome-wide expression analysis was performed using Affymetrix ATH1 microarrays. To this end, shr, scr and wild-type plants were grown in vitro on agar-solidified medium, and complete shoots were harvested at rosette developmental stage 1.04 (Boyes et al., 2001), which consists of roughly equal fractions of proliferating, expanding, and mature tissue. From each genotype, RNA was extracted, labeled, and hybridized to the microarrays. Using a false discovery rate of 0.05 and a 2-fold expression level cutoff, 100 and 349 genes were differentially expressed in scr and shr shoots, respectively. There was a significant overlap between the genes that were affected in scr and shr: 88% of the genes differentially expressed in scr were also differentially regulated in shr, and all of them changed expression in the same direction, with a fold change that was on average 1.75 times higher in the shr mutant. Seven genes from the 10 most differentially expressed genes in shr were also present among the 10 most affected genes in scr (Supplemental Table S1). Furthermore, microarray data showed that SCR was differentially down-regulated in shr leaves, which could be confirmed by Q-reverse transcription (RT)-PCR (data not shown). These data, together with the comparable expression pattern in the leaf, are consistent with SCR acting downstream of SHR in the shoot, which is equivalent to the situation in the root (Helariutta et al., 2000; Levesque et al., 2006).
Genes differentially regulated in both shr and scr shoots with false discovery rate of 0.05 or less and a fold change greater than 1.5 were considered to be part of the regulatory SHR/SCR pathway in the shoot. This set of genes was used as a starting point for further analysis. To get a global overview of these differentially expressed genes, we first investigated which Gene Ontology categories were statistically overrepresented by means of a BiNGO analysis (Maere et al., 2005). Gene Ontology categories overrepresented among the genes down-regulated in both shr and scr shoots were “response to auxin,” “response to brassinosteroid,” and “chromatin assembly.” The latter showed that this was due to differential expression of histone H2A and H2B genes. Besides a down-regulation of auxin and brassinosteroid signaling, these data also suggest an effect on nucleosome assembly.
Among the genes up-regulated in both the shr and scr shoot transcriptomes was an overrepresentation of the flavonoid biosynthetic process, involved in protection against various biotic and abiotic stresses, the response to several stresses, and abscisic acid, suggesting a general stress response upon mutation of the SHR and SCR genes. Furthermore, the CCAAT-binding factor family was the only transcription factor family differentially regulated in the shr and scr shoot transcriptomes. This family, better known as the nuclear transcription factor Y (NF-Y) family, is, as in mammals, subdivided into A-, B-, and C-type subunits (Siefers et al., 2009). Strikingly, in shr and scr, the same seven out of 10 A-type subunits were up-regulated, compared with none of the B- and C-type subunits (Supplemental Table S2). This indicates a specific up-regulation of the A-type subunits of the CCAAT-binding factor complex, which function as transcriptional activators and cell cycle regulators by chromatin remodeling (van Ginkel et al., 1997; Elkon et al., 2003; Gurtner et al., 2008).
Differential expression of histone H2A and H2B genes and A-type NF-Y genes indicates a role for SHR and SCR in chromatin-related cell cycle regulation. Consistently, the cell cycle regulators CYCD6;1, CDKB2;1, CDKB2;2, KRP5, and CYCD3;3 have been identified as targets of SHR (Levesque et al., 2006; Sozzani et al., 2010). Nevertheless, of the 61 core cell cycle genes (Vandepoele et al., 2002), only CYCB1;1 in shr was differentially expressed in our transcriptome data. Using synchronized Arabidopsis cell cultures, Menges et al. (2003) identified a much larger set of 1,372 genes that are differentially expressed in one of the cell cycle phases. Comparing this set of cell cycle-regulated genes, we found a 2.5- and 2.6-fold overrepresentation of cell cycle-regulated genes in the scr and shr shoot transcriptomes, respectively. These data indicate that cell cycle regulation in whole shoot samples is affected upon mutation of the SHR and SCR genes.
Cell Cycle Regulation
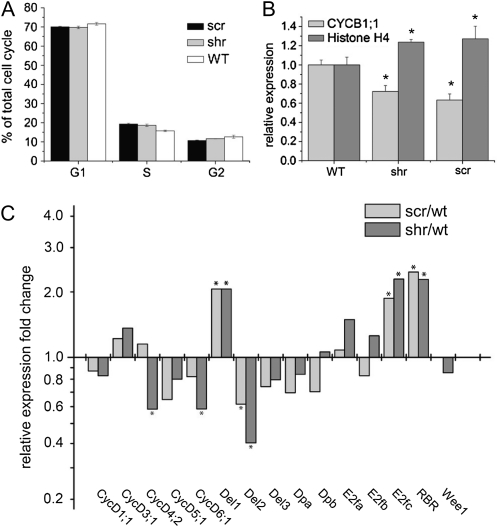
Kinematic leaf growth and transcriptome analysis suggest that cell cycle regulation is downstream of the SHR/SCR pathway. Therefore, cell cycle progression in scr and shr mutants was analyzed in more detail. The relative duration of the different cell cycle phases was determined in both mutants using flow cytometry. To this end, leaves 1 and 2 were harvested at 7 DAS when they were fully proliferating and contained only 2C and 4C nuclei (Fig. 6B), allowing estimation of the relative amount of cells in G1-, S-, and G2-phase of the cell cycle (Fig. 7A). Data show that shr and scr have an enrichment of cells in S-phase, suggesting a relative increase of the S-phase duration in shr (P = 0.036) and scr (P = 0.004). Since kinematic analysis showed a prolongation of the total cell cycle duration with 2 to 3 h in the mutants, this enrichment reflects a drastic impact on S-phase duration, which was estimated at 3.9 h in shr and 4.2 h in scr compared with only 2.9 h in the wild-type leaves. This suggests that S-phase takes significantly longer in the mutants than in the wild type, which is also consistent with the reduced endoreduplication rate.
Figure 7.
Cell cycle regulation in shr and scr mutants. A, Relative abundance of G1-, S-, and G2-phase cells in proliferating leaves 1 and 2 determined by flow cytometry at 7 DAS (n ≥ 3). Note that cells in mitosis cannot be detected due to the absence of a nuclear membrane. B, Relative expression of the S-phase-specific gene, Histone H4, and the M-phase-specific gene, CYCB1;1, in shr, scr, and wild-type (WT) leaves (P < 0.05; n = 2). C, Relative expression of core cell cycle genes involved in the G1/S transition during the plant cell cycle in shr and scr compared with their expression in wild-type leaves. Asterisks indicate differentially expressed genes (fold change > 2; P < 0.05; n = 2).
To support the increased S-phase duration, suggested by flow cytometry, we tested the relative expression of the Histone H4 and CYCB1;1 genes, which are specifically expressed in S-phase and M-phase of the plant cell cycle, respectively (Menges et al., 2003). Q-RT-PCR analysis on mRNA from proliferating leaves of 7-d-old shr, scr, and wild-type plants (Fig. 7B) indicated that expression of Histone H4 was 27% higher in scr and 24% higher in shr than in the wild type. Inversely, expression of CYCB1;1 was reduced by 37% and 28% for scr and shr, respectively. The lower expression of CYCB1;1 suggests that there is no arrest in M-phase in the shr and scr mutants because this would increase the fraction of cells in this phase and therefore increase the relative expression levels of M-phase marker genes. Inversely, the higher expression of the S-phase-specific Histone H4 gene confirms that S-phase duration in shr and scr would be prolonged.
To investigate which cell cycle genes could be involved in the prolonged S-phase duration, the expression of a number of core cell cycle genes involved in the G1/S transition and S-phase progression, CYCD1;1, CYCD3;1, CYCD4;2, CYCD5;1, CYCD6;1, DEL1, DEL2, DEL3, DPa, DPb, E2Fa, E2Fb, E2Fc, RBR, and WEE1, was (in contrast to the above whole shoot-based microarray analysis) determined in proliferating leaves of 7-d-old shr, scr, and wild-type plants (Fig. 7C). Q-RT-PCR analysis revealed that in scr leaves the expression of DEL1, E2Fc, and RBR, all negative regulators of S-phase entry (del Pozo et al., 2002; Vlieghe et al., 2005; Wildwater et al., 2005), was significantly up-regulated. The same expression differences were observed for the shr leaves. Additionally, CYCD4;2 and CYCD6;1 (both negative regulators of RBR and, therefore, positive regulators of S-phase entry) were down-regulated. Expression of the transcription factor E2Fa, which controls the transcription of early S-phase genes involved in cell cycle regulation and DNA replication and is controlled by RBR (Ramirez-Parra et al., 2003), did not change significantly. These data suggest that in the leaf, SHR and SCR control the cell cycle in a similar way as in the root (Wildwater et al., 2005), by inhibiting RBR and thereby stimulating E2F-mediated S-phase gene expression.
DISCUSSION
Effects of Cell Cycle Inhibition on Leaf Growth
Leaf growth is the result of cell division and cell expansion. Kinematic analysis generates detailed data on the dynamics of these parameters during development (De Veylder et al., 2001a). shr and scr have a strong cell division defect, resulting in a severe reduction in final leaf area. Two mechanisms underlie the reduced cell number in shr and scr leaves: reduction in cell division rate, and an early exit of the proliferation phase. Reduction in cell division rate (i.e. increased cell cycle duration) is also observed in overexpression lines of the core cell cycle genes KRP2 and CKS1, in which this is accompanied by an increased proliferative cell size (De Veylder et al., 2001a, 2001b). A faster cell cycle progression, on the other hand, is reported for E2Fb overexpression, which was accompanied by a decreased cell size (Magyar et al., 2005; Sozzani et al., 2006). In shr, scr, and hub1 (Fleury et al., 2007) mutants, however, which also affect cell cycle regulation, a decreased cell division rate did not result in a change in cell size. There is evidence that cell cycle progression depends on reaching a critical cell size, or cytoplasm-to-DNA ratio (Killander and Zetterberg, 1965; Carter and Jagadish, 1978; Donnan and John, 1983; Mitchison, 2003; Dolznig et al., 2004). These data suggest the existence of a cell growth coordination acting upstream of the cell cycle machinery. This mechanism would not be affected by mutations upstream of the cell cycle regulation, such as shr, scr, and hub1, where a change in cell cycle duration did not affect cell growth, but would be disrupted by misexpression of the core cell cycle genes, such as KRP2, CKS1, and E2Fb.
In addition to the reduction in cell division rate, shr mutants also show an early exit of cell proliferation. Evidence that exit from proliferation is a mechanism underlying leaf size variation comes from a number of genotypes with increased leaf size, which is associated with an extension of proliferation phase (Mizukami and Fischer, 2000; Bögre et al., 2008; Li et al., 2008; Horiguchi et al., 2009). Furthermore, study of leaf series in wild-type plants shows that leaf size reaches a pronounced maximum for leaves 4 to 6 of the rosette (Fig. 1D; Supplemental Fig. S1). In shr and scr mutants, consecutive leaves are very similar in size. Assuming that shr and scr also affect cell division and not cell expansion in higher order leaves suggests that the larger size difference in subsequent rosette leaves is due to even bigger changes in cell proliferation.
Despite the fact that the overall phenotype is less severe in the scr mutant than in shr, the effect on cell division rate, S-phase duration, and endoreduplication is at least as strong in the scr mutant as in shr. Inversely, the reduced expansion of guard cells and the early exit of the cell proliferation phase are more pronounced in shr. These observations suggest that the reduced rate of cell cycle progression in shr is mainly regulated through its effect on SCR, whereas the developmental transitions seem to be at least partly regulated through other SHR targets, making it possible to functionally discriminate SCR-dependent and -independent downstream pathways in leaf development.
Regulation of S-Phase Initiation and Progression in shr and scr
Our phenotypic analysis of shr and scr leaves confirms the drastic effect of these mutations on cell division reported previously for the QC and stem cell niche in roots. Local reduction of the expression of RBR, an inhibitor of the E2F/DP transcriptional complex, in the root meristem restores stem cell maintenance in scr and transiently in shr (Wildwater et al., 2005). Q-RT-PCR analysis on a number of core cell cycle genes (Vandepoele et al., 2002) involved in the G1/S transition revealed that DEL1, E2Fc, and RBR were up-regulated in scr and shr leaves. Compared with E2F, DEL proteins contain two DNA-binding domains and lack the transcription activation domain, which allow them to block the E2F/DP-binding sites as monomers and to repress the transcription of E2F target genes needed for S-phase progression (Kosugi and Ohashi, 2002; Mariconti et al., 2002; De Veylder et al., 2003). Notably, DEL1 has been demonstrated to regulate the expression of only a subset of E2F-dependent genes (Vlieghe et al., 2005). Also, E2Fc lacks the activation domain, allowing it to act as a repressor (Vandepoele et al., 2002), and binding of RBR to E2F represses the transcription of E2F target genes (Sekine et al., 1999; Durfee et al., 2000). Thus, all up-regulated cell cycle genes in both shr and scr inhibit S-phase entry. Additionally, CYCD4;2 and CYCD6;1 were down-regulated in shr leaves. The latter has recently been identified as a direct target of SHR in the root (Sozzani et al., 2010). D-type cyclins in complex with cyclin-dependent kinases drive the inactivation of RBR proteins through hyperphosphorylation, releasing E2F target genes, and mark E2Fc for degradation (Sozzani et al., 2010). Curiously, CYCD4;2 and CYCD6;1 are the only Arabidopsis D-type cyclins that lack the RBR-binding motif (Vandepoele et al., 2002; Menges et al., 2007). Despite this, overexpression of CYCD4;2 has effects similar to those of other D-type cyclins (Kono et al., 2006), suggesting that somehow they are still positive regulators of E2F function. This suggests that in the shr and scr mutants, E2F action, needed for S-phase progression, is repressed at the protein level by a dual mechanism: up-regulation of inhibitors and simultaneous down-regulation of positive regulators, without affecting E2Fa expression. Furthermore, a 2.4- and 2.3-fold overrepresentation of E2F target genes (Vandepoele et al., 2005), for SHR and SCR, respectively, was observed in a recent microarray time-course experiment inducing SHR and SCR in the ground tissue of their respective mutant backgrounds (Sozzani et al., 2010). This mechanism is also consistent with the RBR data obtained in the root meristem (Wildwater et al., 2005). These results indicate that SHR and SCR could have comparable effects on the cell cycle regulation both in roots and leaves.
The E2F pathway controls the expression of genes required for both the G1/S transition and S-phase progression. E2F target genes are involved in cell cycle regulation, transcription, chromatin dynamics, defense responses, signaling, and DNA replication (Ramirez-Parra et al., 2003; Vandepoele et al., 2005). Also, many key regulators of DNA licensing and initiation of DNA replication, such as origin recognition complex subunits (Diaz-Trivino et al., 2005), members of the prereplication complexes, cell division control proteins (Castellano et al., 2001; Vandepoele et al., 2005) and minichromosome maintenance proteins (Stevens et al., 2002; Vandepoele et al., 2005; Shultz et al., 2009), genes involved in DNA replication and DNA synthesis, such as the DNA polymerase processivity factor PCNA (Chabouté et al., 2000; Egelkrout et al., 2002), ribonucleotide reductase subunits RNRs (Chabouté et al., 2000, 2002), replication factors, and DNA polymerases (Vandepoele et al., 2005), contain E2F-binding sites in their promoters. Inhibition of E2F action, therefore, will down-regulate the expression of these E2F target genes. Our results show that down-regulation of E2Fa activity in shr and scr did not lead to a block in G1-phase but rather to a slower S-phase progression, indicated by the relative duration of the G1-phase and the S-phase during proliferation, the up-regulation of S-phase-specific genes, and an effect on endoreduplication (which involves repeated cycles of S-phase in the absence of mitosis). This result suggests that the expression of E2F targets involved in DNA replication and DNA synthesis is rate limiting for S-phase progression in scr and shr mutants. The G1/S transition itself, driven by DNA licensing and initiation of DNA replication, either has a lower threshold for E2F activity or is controlled by additional regulators.
Stem Cell Maintenance and Cell Cycle Regulation Disentangled
Stem cell maintenance in the root meristem is needed to ensure indeterminate growth of plant organs. Mutation of SHR, SCR, and PLETHORA (PLT) genes results in a loss of the stem niche, followed by differentiation of the root meristem (Benfey et al., 1993; Scheres et al., 1995; Aida et al., 2004). Both SHR and SCR are required for QC identity, as SCR expression in the QC alone cannot rescue root growth in the shr background (Sabatini et al., 2003). Also, the PLT genes act in parallel with the SHR/SCR pathway in root stem cell specification (Aida et al., 2004). In addition, a number of core cell cycle genes have been shown to affect stem cell fate in roots. Overexpression of CYCD3;1, KRP2, RBR, and E2Fa/DPa results in differentiation or accumulation of stem cells, in agreement with their roles in the plant E2F pathway (Wildwater et al., 2005). RBR regulation occurs downstream of SCR, which provides a mechanistic link between the SHR/SCR pathway and cell cycle regulation in stem cell maintenance (Wildwater et al., 2005). Our data strongly suggest that SCR expression in the ground tissue contributes to overall meristem activity but cannot support indeterminate growth and maintenance of the stem cell niche. Interestingly, in the scr mutant background, restitution of SCR expression in the ground tissue, excluding the QC, contributes to overall meristem activity but not to stem cell maintenance (Sabatini et al., 2003). These data are in line with our observations indicating a role for SHR and SCR in proliferative cell division. However, to date, this has not been studied in detail in the root, presumably because the consumption of the meristem due to the loss of the stem cell niche has a more pronounced effect in this organ. In contrast to the leaf, the expression of SCR and SHR in the root tip does not appear in all cells but is restricted to the stem cell niche, endodermis, and, in the case of SHR, the stele (Di Laurenzio et al., 1996; Helariutta et al., 2000; Wysocka-Diller et al., 2000). The question raised whether this more general cell cycle regulatory function is specific for the leaf (and possibly other aerial structures) and whether other genes perform this function in proliferating root cells or cell cycle regulation in the root is independent of this regulatory mechanism.
RBR complexes also interact with chromatin-remodeling factors, which have recently been identified as key players in the maintenance of stem cell identity in mammals (Hennig et al., 2003; Narita et al., 2003; Lee et al., 2006; Jullien et al., 2008). Our transcriptome analysis of scr and shr shoots revealed the differential expression of histones and NF-Ya factors, which function as transcriptional activators and cell cycle regulators by chromatin remodeling (van Ginkel et al., 1997; Elkon et al., 2003; Gurtner et al., 2008). In human, it was shown that NF-Ya factors regulate G1/S gene expression, related to the loss of dividing potential during replicative senescence, and self-renewal of stem cells (Matuoka and Chen, 2000; Zhu et al., 2005). These data are consistent with the molecular phenotypes observed in the leaves of shr and scr mutants and suggest that the SHR/SCR pathway affects chromatin remodeling in plants and that this function is not restricted to stem cells.
Our data suggest the inhibition of E2F action as the primary target of core cell cycle regulation in shr and scr mutants. E2F action is crucial for both cell cycle progression and stem cell maintenance. Overexpression of E2Fa together with its dimerization partner, DPa, not only induces enhanced division and endoreduplication in leaves but also the accumulation of stem cells in roots (De Veylder et al., 2002; Kosugi and Ohashi, 2003). Moreover, transient overexpression of E2Fa and DPa can even induce nondividing mesophyll cells to reenter S-phase (Rossignol et al., 2002). Comparably, increasing E2F activity in leaves by inducible inactivation of RBR leads to epidermal cell hyperplasia during the late proliferation phase and allows differentiated pavement cells to reenter cell division later during development (Desvoyes et al., 2006). These data emphasize the role of E2F activity on both cell proliferation rate and the transition between stem cell, proliferative, and nonproliferative states. Consistent with a role upstream of E2F, our data demonstrate that SHR and SCR not only function in differentiation and stem cell maintenance in the root meristem but also play a role in the control of proliferative cell division in developing leaves.
CONCLUSION
The shr and scr mutants have been instrumental in unraveling the mechanism of stem cell maintenance in the root meristem (Benfey et al., 1993; Scheres et al., 1995; Di Laurenzio et al., 1996; Helariutta et al., 2000; Sabatini et al., 2003). In both shr and scr, cell fate changes result in the differentiation of the stem cell niche, causing an arrest of formative divisions and ultimately root growth. This indicates that in the root the function of SHR and SCR in differentiation is instrumental for growth. In leaves, shr and scr mutants affect differentiation of the bundle sheath (Wysocka-Diller et al., 2000) and the expansion of guard cells. These differentiation effects, however, contribute little to the overall leaf growth phenotype. Indeed, our kinematic data show that the reduced leaf growth in shr and scr mutants is primarily determined by an effect on cell proliferation. Furthermore, we show that this is a direct consequence of the lack of SHR and SCR function in the shoot rather than an indirect consequence of compromised root growth. These phenotypes cannot readily be understood, given the absence of a stem cell niche in the developing leaf (Tsukaya, 2002), indicating that SHR and SCR play a direct role in the control of proliferative cell division in developing leaves. Furthermore, these data indicate a much more pronounced function for SHR and SCR in the regulation of cell division than has been identified in root studies. Our transcriptome, endoreduplication, flow cytometric, and expression analyses reveal that SHR and SCR primarily drive S-phase progression, presumably by a stimulation of E2F action. However, future research is needed to unravel the molecular details of cell cycle regulation by the SHR/SCR pathway, taking into account the possible difference between proliferating cells in the root and leaf.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
In this study, we used Arabidopsis (Arabidopsis thaliana) to study the mutants shr-6 (SALK_002744), shr-3 (Helariutta et al., 2000), and scr-3, (Fukaki et al., 1996) and the promoter fusions CYCB1;1:GUS (Ferreira et al., 1994) and SHR:GUS (Helariutta et al., 2000). These lines were all in the Columbia (Col-0) background. Seeds for in vitro analysis were sterilized in 3% bleach for 15 min and sown on medium containing 0.5× Murashige and Skoog medium (Duchefa) solidified with 0.9 g L−1 plant tissue culture agar (Lab M) on round plates (1013; Becton-Dickinson). For the root experiments, we used 1.0 g L−1 agar and square plates (Greiner Bio-One). After a stratification period of 2 d, the plates were placed horizontally (leaf growth) or inclined vertically (root growth) in a growth chamber under long-day conditions (16 h of light, 8 h of darkness) at 22°C with a light intensity of 80 to 100 mE m−2 s−1 supplied by cool-white fluorescent tubes (Spectralux Plus 36W/840; Radium).
Growth Analysis
Rosette growth was analyzed by taking photographs (Canon EOS 400D camera with an EFS 18-55 mm Canon ZOOM lens) of plants growing on petri dishes. Plants were followed for 18 DAS. Kinematic analysis of leaf growth was done as described previously (De Veylder et al., 2001a). Total leaf blade area of leaves 1 and 2 of five plants from 5 to 24 DAS was measured from dark-field binocular (days 8–24) or differential interference contrast light microscopy (days 5–7) images. Microscopic drawings containing about 100 cells, located 25% and 75% from the distance between the tip and the base of the leaf blade, of the abaxial epidermis of each leaf were made with a drawing tube attached to the microscope equipped with differential interference contrast optics.
Histochemical GUS Assays
Complete seedlings or tissue cuttings were stained on multiwell plates (3046; Becton-Dickinson). GUS assays were performed as described by Beeckman and Engler (1994). After staining, samples were mounted in lactic acid and observed and photographed with a stereomicroscope (MZ16; Leica) or with a differential interference contrast microscope (DMLB; Leica).
Flow Cytometry
For flow cytometry, leaves were lacerated with a razor blade and suspended in 1.5 mL of buffer (Galbraith et al., 1991). The supernatants was filtered over a 30-μm mesh, and 1 μL of 4′,6-diamidino-2-phenylindole from a stock of 1 mg mL−1 was added. The nuclei were analyzed with the CyFlow flow cytometer using FloMax software (Partec). On average, 10,000 nuclei were counted per sample. For cell cycle analysis on proliferative leaves, nuclei were detected on a linear scale and analyzed using MultiCYCle AV software (Phoenix Flow Systems; Kallioniemi et al., 1994). A debris fitting, a single cell cycle analysis, and a histogram reliability and confidence estimation were performed for each histogram. The model used for debris fitting and peak analysis was sliced background. For the relative cell cycle phase calculations, only samples with a good S-phase confidence were used.
Q-RT-PCR
RNA was extracted using the RNeasy kit (Qiagen) according to the supplier’s instructions. Poly(dT) cDNA was prepared from 1 mg of total RNA with SuperScript III reverse transcriptase (Invitrogen) and quantified on a LightCYCler 480 (Roche) with the qPCR LightCYCler 480 SYBR Green I Master (Roche). PCR was performed on 384-well reaction plates, which were heated for 10 min to 95°C, followed by 45 cycles of denaturation for 10 s at 95°C and annealing and extension for 15 s at 60°C and 72°C, respectively. Target quantifications were performed with specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International). All PCRs were done in three technical repeats, and at least two biological repeats were used for each sample. Expression levels were first normalized to CDKA, CBP20, and CKA2 expression levels, which did not show clear systematic changes in the cycle threshold value, and then to the respective expression levels in the wild type (Col-0). The primers used are listed in Supplemental Table S3.
Microarray Analysis and Data Processing
shr-2, scr-3, and Col-0 shoots were grown in vitro and harvested at stage 1.04 (Boyes et al., 2001). Each genotype was sampled in three biological replicates each containing approximately 20 rosettes from different petri dishes. RNA was extracted with the RNeasy kit (Qiagen). Arabidopsis ATH1 GeneChips (Affymetrix) were hybridized at the VIB Microarray Facility (www.microarrays.be) according to the manufacturer’s instructions. Expression levels were based on an improved custom-made Chip Description File (Casneuf et al., 2007). The multiarray analysis algorithm implemented in the BioConductor Affy package (www.bioconductor.org; Gautier et al., 2004) was used for background correction, normalization, and summary expression value computation. For statistical analysis and hierarchical clustering of the normalized expression files, we used the Multiexperiment Viewer of The Institute for Genomic Research (www.tm4.org/mev.html). Because one of the shr arrays (shr-3) clustered separately from the others, this array was removed from further analysis. The remaining eight arrays were subjected to an ANOVA analysis using the false discovery rate method (Benjamini and Hochberg, 1995). The corrected P value threshold was set to 0.05.
The microarray data are available in the public repository Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE21629. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SHR (AT4G37650) and SCR (AT3G54220). Sequence data for all other genes described in this article can be found in the relevant databases using the accession numbers given in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf series of shr and scr mutants.
Supplemental Figure S2. The effect of cutting the root system in wild-type plants on leaf growth is smaller compared with mutation of SHR and SCR genes.
Supplemental Figure S3. Guard cell areas in shr and scr mutants.
Supplemental Table S1. Top ten most differentially expressed genes in shr and scr shoot transcriptomes.
Supplemental Table S2. Expression of A-, B-, and C-type subunits of the NF-Y family.
Supplemental Table S3. Primer sequences used for Q-RT-PCR.
Supplementary Material
Acknowledgments
We thank Yrjo Helariutta and Lieven De Veylder for fruitful discussions.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Engler G. (1994) An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep 12: 37–42 [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bögre L, Magyar Z, López-Juez E. (2008) New clues to organ size control in plants. Genome Biol 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BL, Jagadish MN. (1978) Control of cell division in the yeast Saccharomyces cerevisiae cultured at different growth rates. Exp Cell Res 112: 373–383 [DOI] [PubMed] [Google Scholar]
- Casneuf T, Van de Peer Y, Huber W. (2007) In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Philipps G. (2002) S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J Biol Chem 277: 17845–17851 [DOI] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Sekine M, Philipps G, Chaubet-Gigot N. (2000) Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12: 1987–2000 [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C. (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001a) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beemster GTS, Beeckman T, Inzé D. (2001b) CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J 25: 617–626 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Joubès J, Inzé D. (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6: 536–543 [DOI] [PubMed] [Google Scholar]
- Diaz-Trivino S, del Mar Castellano M, de la Paz Sanchez M, Ramirez-Parra E, Desvoyes B, Gutierrez C. (2005) The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Res 33: 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dolznig H, Grebien F, Sauer T, Beug H, Müllner EW. (2004) Evidence for a size-sensing mechanism in animal cells. Nat Cell Biol 6: 899–905 [DOI] [PubMed] [Google Scholar]
- Donnan L, John PC. (1983) Cell cycle control by timer and sizer in Chlamydomonas. Nature 304: 630–633 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Durfee T, Feiler HS, Gruissem W. (2000) Retinoblastoma-related proteins in plants: homologues or orthologues of their metazoan counterparts? Plant Mol Biol 43: 635–642 [DOI] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L. (2002) Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. (2003) Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res 13: 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. (1994) Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GTS, Neyt P, Anami S, Robles P, et al. (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Fujisawa H, Tasaka M. (1996) SGR1, SGR2, SGR3: novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol 110: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S. (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gurtner A, Fuschi P, Magi F, Colussi C, Gaetano C, Dobbelstein M, Sacchi A, Piaggio G. (2008) NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue. PLoS ONE 3: e2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W. (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Gonzalez N, Beemster GTS, Inzé D, Tsukaya H. (2009) Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J 60: 122–133 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol 6: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi OP, Visakorpi T, Holli K, Isola JJ, Rabinovitch PS. (1994) Automated peak detection and cell cycle analysis of flow cytometric DNA histograms. Cytometry 16: 250–255 [DOI] [PubMed] [Google Scholar]
- Kang J, Dengler N. (2002) Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216: 212–219 [DOI] [PubMed] [Google Scholar]
- Killander D, Zetterberg A. (1965) A quantitative cytochemical investigation of the relationship between cell mass and initiation of DNA synthesis in mouse fibroblasts in vitro. Exp Cell Res 40: 12–20 [DOI] [PubMed] [Google Scholar]
- Kono A, Ohno R, Umeda-Hara C, Uchimiya H, Umeda M. (2006) A distinct type of cyclin D, CYCD4;2, involved in the activation of cell division in Arabidopsis. Plant Cell Rep 25: 540–545 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2002) Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol 128: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132: 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim B, Song SK, Heo JO, Yu NI, Lee SA, Kim M, Kim DG, Sohn SO, Lim CE, et al. (2008) Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol 67: 659–670 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xie T. (2005) Stem cell niche: structure and function. Annu Rev Cell Dev Biol 21: 605–631 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW. (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L. (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D. (2002) The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Matuoka K, Chen KY. (2000) Possible role of subunit A of nuclear factor Y (NF-YA) in normal human diploid fibroblasts during senescence. Biogerontology 1: 261–271 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH. (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Menges M, Pavesi G, Morandini P, Bögre L, Murray JAH. (2007) Genomic organization and evolutionary conservation of plant D-type cyclins. Plant Physiol 145: 1558–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM. (2003) Growth during the cell cycle. Int Rev Cytol 226: 165–258 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Kai T, Decotto E, Spradling A. (2004) The stem cell niche: theme and variations. Curr Opin Cell Biol 16: 693–699 [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Fründt C, Gutierrez C. (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Rossignol P, Stevens R, Perennes C, Jasinski S, Cella R, Tremousaygue D, Bergounioux C. (2002) AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol Genet Genomics 266: 995–1003 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN. (1995) Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Sekine M, Ito M, Uemukai K, Maeda Y, Nakagami H, Shinmyo A. (1999) Isolation and characterization of the E2F-like gene in plants. FEBS Lett 460: 117–122 [DOI] [PubMed] [Google Scholar]
- Shultz RW, Lee TJ, Allen GC, Thompson WF, Hanley-Bowdoin L. (2009) Dynamic localization of the DNA replication proteins MCM5 and MCM7 in plants. Plant Physiol 150: 658–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, IV, Tayrose G, Holt BF., III (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R. (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C. (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2002) Leaf development. Somerville CR, Meyerowitz EM, eds, The Arabidopsis Book American Society of Plant Biologists; Rockville, MD, doi/ http://www.aspb.org/publications/arabidopsis/ [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ginkel PR, Hsiao KM, Schjerven H, Farnham PJ. (1997) E2F-mediated growth regulation requires transcription factor cooperation. J Biol Chem 272: 18367–18374 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L. (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- White DWR. (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. (2005) NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci USA 102: 11728–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.