Abstract
c-Maf is a bZip transcription factor expressed in developmental and cellular differentiation processes. Recently, a c-maf knockout mouse model, showing abnormal lens development, has been reported. In order to study the regulation mechanisms of c-maf gene expression during the differentiation process we have cloned and functionally characterized the rat c-maf (maf-2) gene. The rat c-maf gene is an intronless gene, covering a length of 3.5 kb. Transient transfection analysis of the 5′-flanking region of the c-maf gene using luciferase as the reporter gene shows that Pax6, a master transcription factor for lens development, strongly activates the c-maf promoter construct. Endogenous c-maf is also activated by the Pax6 expression vector. Electrophoresis mobility shift assay and DNase I footprinting analysis show that at least three Pax6-binding sites are located in the 5′-flanking and 5′-non-coding regions of the rat c-maf gene. The c-maf gene was also markedly activated by its own product, c-Maf, through the MARE (Maf recognition element), suggesting that a positive autoregulatory mechanism controls this gene. In situ hybridization histochemical detection of Pax6 and c-Maf in the E14 lens showed that both mRNAs are expressed in the lens equator where lens epithelial cells are differentiating to lens fiber cells. These results suggest that a Pax6/c-Maf transcription factor cascade is working in lens development.
INTRODUCTION
The maf oncogene (v-maf) was initially identified from an avian oncogenic retrovirus, AS42, which induces musculoaponeurotic fibrosarcoma in vivo and transforms chicken embryo fibroblast cells in vitro (1). Frequent translocations of the cellular c-maf gene to the Ig locus have been found in multiple myelomas (2). Several maf-related genes have been identified so far (i.e. mafB, mafK, mafG, mafF and Nrl) (3–5). The maf gene family includes the large maf genes (c-maf, mafB and Nrl) and the small maf genes (mafK, mafG and mafF), which lack the N-terminal half of the large maf genes which contains the transactivation domain. Members of both subfamilies possess a well-conserved bZip DNA-binding domain in the C-terminus. The large Maf proteins form homodimers as well as heterodimers with other members of the bZip transcription factor family, such as Jun and Fos (6,7). The small Maf proteins heterodimerize with the CNC family of proteins, including NF-E2, Nrf1 and Nrf2, and work as positive transcription factors (8–10). The consensus binding sequences of Maf family proteins [the Maf recognition element, MARE, -TGCTGAC(G)TCAGCT-] overlaps with the 12-O-tetradecanoylphorbol 13-acetate responsive element (TRE) and cAMP-responsive element (CRE) (7,11). Recent studies indicate that the Maf-related factors play important roles in development and cellular differentiation (12). MafB interacts with Ets-1, a transcription factor containing a helix–turn–helix DNA-binding domain, and inhibits Ets-1 activity and, hence, interrupts erythroid differentiation (13). The mouse mafB gene was identified as a gene responsible for segmentation of the hindbrain during early development, as shown from an analysis of kreislar (Kr) mutant mice (14). The c-maf gene product activates transcription from two sites in the L7 gene, which is expressed in all adult cerebellar Purkinje cells (15). Furthermore, c-Maf regulates tissue-specific expression of IL-4 in T helper 2 (Th2) cells and controls Th2 cell differentiation (16). c-Maf physically interacts with c-Myb to regulate transcription of the genes of early myeloid cells and elevated c-Maf–c-Myb complexes induce monocytic differentiation (17,18). It has also been demonstrated that c-Maf activates the p53 gene and its overexpression leads to p53-dependent apoptosis (19). We have previously isolated the rat maf-related cDNA clones c-maf and mafB (originally named maf-1 and maf-2 for rat mafB and c-maf, respectively) and shown that both genes are specifically expressed in cartilage, lens, spinal cord and kidney in a differentiation-specific manner (20–22). Furthermore, when precise expression patterns were compared, expression of c-maf and mafB were spatially and temporally different from each other. These findings indicate that the large maf gene family products play an important role in cellular differentiation of several tissues in a related but distinct manner.
More recently, targeted disruptions of the c-maf gene have been reported as displaying abnormal lens development (23–25). The differentiation of lens epithelial cells to fiber cells and crystallins expression were impaired. Since many of the crystallin family genes and other lens-specific genes have MARE-like sequence in their regulatory regions, these genes might be a direct target of the c-Maf transcription factor (25–28).
The upstream factor that regulates c-maf expression is a critical point for the elucidation of lens development. It has been shown that the transcription factors Pax6, Sox1 and L-Maf play a pivotal role in lens development (26,29,30). Induction of lens differentiation by ectopic expression of Pax6 or L-maf (26,31) suggests that these factors may be placed upstream of c-Maf in the transcription factor cascade of lens development.
To determine the upstream factor that controls c-maf expression in lens development and other processes of cellular differentiation, we have analyzed the cis-acting elements and the factors that regulate this gene.
Here we present evidence that Pax6 activates the c-maf gene and that this gene is positively autoregulated by its own product, c-Maf.
MATERIALS AND METHODS
Cloning of the rat c-maf gene
A rat genomic library (kindly provided by Dr K. Watanabe, Metropolitan Institute for Gerontology, Tokyo) constructed in the EMBL-3 λ phage vector (Stratagene, La Jolla, CA), was screened using a rat maf-2 (rat c-maf) cDNA. Approximately 5 × 106 phage were screened and one of the positive clones containing the longest 5′-flanking sequence was used for analysis. The various restriction fragments of the cDNA were subcloned into vector plasmid pBluescript II (Stratagene, La Jolla, CA) and subjected to sequencing. The nucleotide sequences of the cloned fragments were determined with an ABI Prism dye terminator cycle sequencing kit and ABI 373S DNA sequencer (Applied Biosystems, CA).
RNase protection mapping and 5′-rapid amplification of cDNA end (RACE) analysis
We analyzed the transcription initiation site by the RNase protection mapping and 5′-RACE methods. Total cellular RNA from cartilage cells, induced by subcutaneous implantation of bone morphogenetic protein (BMP) on a fibrous glass membrane (20), was prepared by the acid guanidine thiocyanate/phenol method using the ISOGEN RNA extraction kit (Nippon Gene, Toyama, Japan). RNAs from rat fibroblast cell line 3Y1 and Wister rat heart were also extracted. RNase protection analysis was performed as described (20). To construct the template plasmid for synthesis of the antisense RNA probe (riboprobe) a DNA fragment containing the transcription initiation site (–125 to +110) was subcloned into pBluescript. The riboprobe was synthesized with T7 RNA polymerase (Takara, Kyoto, Japan) with [α-32P]UTP in vitro. The riboprobe for the coding region of the c-maf gene (the XhoI–StyI fragment of c-maf cDNA; 20) was also synthesized and used as a positive control in the RNase protection analysis. The cap site of the c-maf gene was also analyzed by 5′-RACE using a 5′/3′RACE kit (Boehringer Mannheim, Germany) according to the supplier’s protocol. The cDNA was synthesized using total RNA of cartilage cells with a primer complementary to –90/–110 (-CAAGGCAAGGCCGAGAGCAA-). The cDNA was tailed with poly(A) and amplified using a primer complementary to –63/–84 (-TGGGGGGAGAGGCCGAGCGGA-) and a poly(T) anchor primer (-GACCACGCGTATCGATGTCGACT16V-, V = A, C or T). The PCR products were further amplified with nested primer –40/60 (-AGCAGCTCGAGCATCAGCTC-) and the anchor primer. The PCR products were cloned into plasmid vector T-Easy (Promega, Madison, WI) and sequenced.
For RNase protection analysis to detect Pax6 mRNA, the Pax6 cDNA (–110/+136 with respect to the ATG initiation codon) was amplified by reverse transcription and PCR (RT–PCR) and subcloned into pBluescript II. The mouse c-maf cDNA fragment (corresponding to the 974–1284 region of rat c-maf cDNA) was amplified by RT–PCR and subcloned into pBluescript II. These plasmids were used as the templates for riboprobe synthesis. The mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA (initiation codon ATG–HindIII site, 190 nt) was amplified by RT–PCR, inserted into pBluescript II and used to synthesize an antisense riboprobe for control experiments.
Construction of plasmids
Various DNA fragments from the c-maf genomic clone, prepared by restriction cleavage or PCR, were inserted into the promoterless luciferase plasmid vector pGVB2 (Nippon Gene). To analyze the enhancer activity of the various regions of the c-maf gene, the fragments were inserted into vector pGVB2 having the promoter sequence of the rat glutathione S-transferase P (GST-P) gene (–50 to +36, a minimal promoter containing a TATA box and a GC box; 32). One or three copies of the consensus binding sequence of the Pax6 paired domain (-GATCCAATTTCACGCTTGAGTTCACAGCT-; 33) was inserted into a plasmid containing the luciferase gene with the GST-P promoter to construct the Pax6 control reporter plasmid, PaxCon1/Luc or PaxCon3/Luc, respectively.
The expression plasmid for rat Pax6 (pAct/Pax6) was prepared as follows. The coding region of the Pax6 cDNA was isolated by RT–PCR with primers 5′-CCGGATCCAGCATGCAGAACAGTCAC- 3′ and 5′-CCAAGCTTACTGTAATCGAGGCCAGT-3′, using total RNA from newborn rat lens as template. This cDNA was joined to the expression vector pAct2, containing the human β-actin enhancer and promoter (34). The expression plasmids for rat c-maf (maf-2), mafB (maf-1) and c-jun have been described previously (20).
Cell culture and transient transfection analyses
The mouse fibroblast cell line C3H10T1/2 was obtained from the Japanese Collection of Research Bioresources, Tokyo, and maintained in Dulbecco’s modified minimal essential medium (DMEM; Nissui, Tokyo) supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, the cells were plated at a density of 2 × 105 cells/plate. Transient transfection experiments were performed by the calcium phosphate co-precipitation method according to Chen and Okayama (35). A total of 5 µg DNA, including 1 µg reporter plasmid, 0.5 µg β-galactosidase expression plasmid (pSV-β-gal; Promega, WI) as an internal control, with or without various amounts of expression plasmid of the effector gene, and pUC18 DNA, was co-transfected into C3H10T1/2 cells. After 45 h incubation, cells were harvested and extracted in 100 µl of lysis buffer (Nippon Gene) by freezing and thawing. The cell extracts were assayed for luciferase activity using a luciferase assay kit (Nippon Gene) and for β-galactosidase activity (36). Luciferase activity was normalized to β-galactosidase activity and all experiments were repeated at least twice.
Electrophoretic mobility shift assay (EMSA) and DNase I footprinting analysis
The paired domain of Pax6 was synthesized in Escherichia coli as a fusion protein with E.coli maltose-binding protein (MBP). The cDNA fragment corresponding to the paired domain (N-terminal 131 amino acids) was synthesized by PCR, using primers 5′-CCGGATCCAGCATGCAGAACAGTCAC-3′ and 5′-GAGAAGCTTCTAGCCAGGTTGCGAAGAACT-3′, and inserted into pMalc2, an expression vector for the fusion protein with MBP (New England Biolabs, Beverly, MA). The plasmid for c-Maf–MBP fusion protein was synthesized as follows. At first we tried to construct a c-Maf–MBP fusion protein, but were unsuccessful. We attributed this failure to possible toxicity of the fusion protein. We found that the N-terminal region of c-Maf was harmful to E.coli. Then we replaced this region of the c-maf cDNA with the corresponding region of the highly homologous gene mafB. The C-terminal half of c-maf cDNA, including the DNA-binding domain (amino acids 159–369) was joined to the N-terminal region of mafB cDNA (amino acids 83–157) by a common ApaI site. The chimeric cDNA was inserted into vector pMalc2. The fusion protein was expressed in E.coli and purified by amylose affinity column chromatography according to the supplier’s protocol (New England Biolabs). The probes for EMSA and DNase I footprinting analysis were prepared by filling in the 5′-overhang end of appropriate restriction enzyme fragments of the c-maf gene using Klenow DNA polymerase I (Takara, Kyoto) and [α-32P]dCTP.
EMSA was performed essentially as described previously (7). About 100 ng Pax6 paired domain or c-Maf protein, fused with MBP, was preincubated with 0.5 µg poly(dI·dC) (Pharmacia, Stockholm) at room temperature for 10 min in 10 µl of binding buffer (20 mM HEPES pH 7.9, 20 mM KCl, 1 mM EDTA, 5 mM DTT, 4 mM MgCl2, 15% glycerol, 100 µg/ml bovine serum albumin). 32P-labeled DNA probe (∼2 × 104 c.p.m.) was added to the binding mixture and incubated for another 10 min. Protein–DNA complexes were analyzed by 4% PAGE in 0.5× TBE (25 mM Tris–borate, 0.5 mM EDTA, pH 8.2). For the footprinting analysis, DNA–protein binding reactions were carried out as for the EMSA but at five times the scale (50 µl volume) and with about five times the protein concentration. Aliquots of 50 µl 5 mM MgCl2, 5 mM CaCl2 and 20 ng DNase I (Takara) were added and incubated for 2 min at room temperature. The reaction was terminated by addition of 100 µl of 20 mM EDTA, 1% SDS, 0.2 M NaCl. After proteinase K treatment (100 µg/ml for 15 min) the DNA was extracted and analyzed on 6% polyacrylamide gels containing 8 M urea. Adenine and guanine bases of the same probes were modified and digested by the Maxam–Gilbert method and loaded as a marker.
Isolation of Pax6 overexpressed in cell lines
The Pax6 expression plasmid (pAct/Pax6) was co-transfected with a plasmid containing the neomycin resistance gene (pSV2-neo) into C3H10T1/2 cells. The neomycin-resistant clones were isolated after 2 weeks culture in medium containing 400 µg/ml G418 (Gibco BRL, NY).
In situ hybridization histochemistry
In situ hybridization was carried out as previously described (20,22). Briefly, sections of the eyes from E14 mouse embryos were dried and immersed in 1% Triton X-100 in 50 mM Tris–HCl, 25 mM EDTA, pH 8.0, and acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine. The sections were dehydrated and hybridized with the probes in 50% formamide, 10% dextran, 1× Denhart’s solution, 12 mM EDTA pH 8.0, 10 mM Tris–HCl pH 8.0, 30 mM NaCl, 0.5 mg/ml yeast tRNA and 10 mM DTT at 55–60°C overnight. The slides were rinsed in 4× SSC, digested with RNase A (20 µg/ml) for 30 min at 37°C and washed sequentially in 2× SSC, 1× SSC and 0.5× SSC, then for 30 min in 0.1× SSC at 60°C. The sections were exposed to X-ray film (Kodak X-Omat) for 4 days, then dipped in NTB2 nuclear emulsion (1:1 with water; Kodak) and exposed for 3 weeks before being developed. Counter-staining of cells was with 0.001% bisbenzimide. Radioactive RNA probes for c-maf (nt 195–410) and Pax6 (–110/+136 with respect to the ATG initiation codon) were synthesized with [α-35S]UTP (NEN Life science) by T7 RNA polymerase (Takara).
In control experiments adjacent sections were treated with RNase before hybridization or sense strand cRNA probes were used for hybridization.
RESULTS
Cloning and sequencing of the rat c-maf gene
A rat genomic library was screened with a fragment of the rat maf-2 (c-maf) cDNA (EcoRI–PstI fragment, 5′-untranslated region). We obtained three positive clones from ∼5 × 106 recombinant phage. The clone with the longest 5′-flanking region (λc-maf) was further analyzed. The restriction map and the nucleotide sequence of the 5′-flanking region of this clone are shown in Figure 1. Nucleotide sequence analysis indicates that this clone contains ∼9.5 kb of the 5′-flanking sequence, the entire coding sequence and 0.3 kb of the 3′-non-coding sequences. Comparison of the genomic clone with the cDNA clone indicates that this gene has no intron, at least in the 5′-non-coding and coding sequences.
Figure 1.
Structure of the rat c-maf gene. (A) Restriction map of the rat c-maf gene. Restriction maps of the λc-maf and c-maf cDNAs are presented. (B) Nucleotide sequences of the 5′-flanking and 5′-non-coding regions of the rat c-maf gene. Arrows indicate transcription initiation sites. The putative transcription factor-binding sites and restriction enzyme recognition sites are represented above and under the sequence, respectively. The major transcription initiation site is designated +1.
To identify the transcription initiation site of the rat c-maf gene, RNase protection mapping was carried out as described in Materials and Methods. As shown in Figure 2, an ∼115 nt protected band was detected in all RNAs from different tissues with the 5′-end probe (probe A). A minor band at ∼87 nt was also detected. Some non-specific bands were seen on the gel, which may be due to the high GC content of probe A. Thus an internal probe (probe B) was used with the same RNA samples as a control. The expected 171 nt bands were detected by probe B. The intensities of the 5′-end probe bands were almost proportional to that of the internal probe, indicating that both probes detect the same c-maf mRNA. To confirm this result we performed 5′-RACE analyses as described in Materials and Methods. The final PCR products were cloned and sequenced. As predicted from the RNase protection analysis, two types of cDNA clones were obtained, one corresponding to the +1 site in Figure 1 and another 28 nt shorter than the longer clone. The sizes of the protected bands were several nucleotides longer than expected from the RACE analysis. However, taking into account that RNase A, used in this experiment, cleaves only 3′ of a pyrimidine residue (C or U), the results of the RNase protection analysis agree with those of the 5′-RACE analysis. Of the 22 clones determined, 14 corresponded to the longer clone. This suggests that the major mRNA is at twice the concentration of the minor mRNA. From the results of the RNase protection mapping and 5′-RACE analyses we assigned the major and minor transcription initiation sites to +1 and +29, respectively, as shown in Figure 1. A TATA box (-TATAAA-) is located 28 nt upstream of the major cap site.
Figure 2.
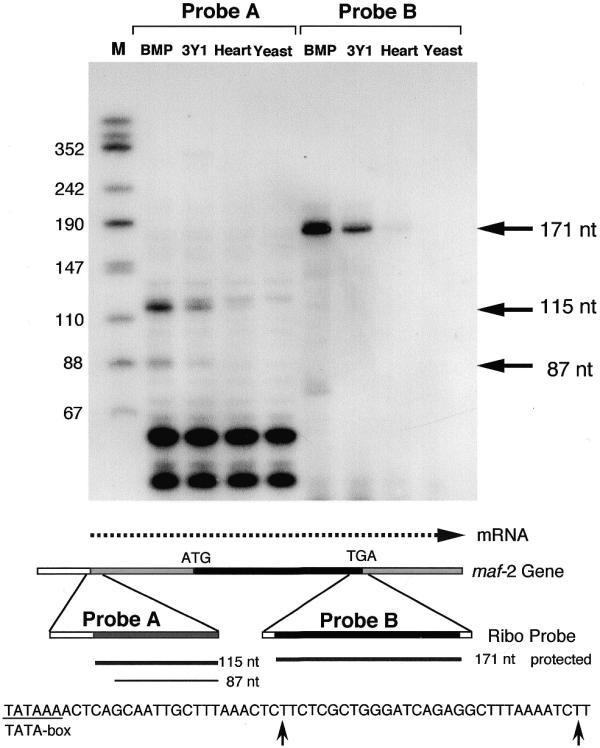
Analysis of the transcription initiation site of the rat c-maf gene. RNase protection mapping was carried out as described in Materials and Methods. RNA was extracted from BMP-induced chondrocytes (BMP), a rat fibroblast cell line (3Y1) and rat heart (Heart). The same RNA sample was analyzed with two probes (5′-end region, probe A; internal region, probe B) as illustrated in the lower panel. Yeast RNA was used as a negative control. End-labeled HinfI-digested pUC18 DNA was used as a size maker (M). A schematic representation of the experiment and the nucleotide sequence around the cap sites are shown in the lower panel.
The functional promoter of the c-maf gene
To determine the functional promoter region of the c-maf gene we made a series of 5′-deletion constructs joined to the luciferase gene and introduced these into C3H10T1/2 cells.
The mouse fibroblast cell line C3H10T1/2 expresses the c-maf gene at low level and is able to differentiate to muscle cells, adipocytes and chondrocytes on treatment with 5-aza-cytidine, an inhibitor of DNA methylation (37). As previously reported, c-maf is expressed in the final stage of chondrocyte differentiation (20). c-maf expression is down-regulated during adipocyte differentiation and muscle differentiation also correlates with c-maf expression (M.Sakai, J.Hirokawa and S.Nishi manuscript in preparation). These findings suggest that the differentiation process from mesenchymal fibroblast cell to other cell types is accompanied by an alteration in c-maf expression. The C3H10T1/2 cell line is an advantageous one in which to study expression and function of the c-maf gene during differentiation.
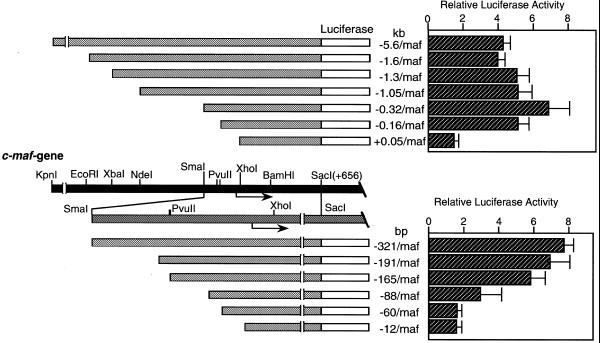
Figure 3 shows luciferase analysis of the 5′-deletion constructs. –5.6 k/Luc has strong promoter activity, inducing ∼100-fold higher luciferase activity than the negative control plasmid (pGVB2). Deletion of the distal region from –5.6 to –1.0 kb produces essentially no change in promoter activity. Progressive deletion of the 5′-sequence –1050 to –321 results in a slight increase in promoter activity. However, a drastic decrease in luciferase activity was observed when the region from –165 to –60 bp was deleted. These results indicate that the region between –165 and –60 bp of the c-maf gene contains elements that are essential in directing basic transcription of the reporter gene in C3H10T1/2 cells.
Figure 3.
Functional promoter analysis of the rat c-maf gene. A series of 5′-deletion constructs joined to the luciferase gene (1 µg) were transfected into mouse fibroblast cell line C3HT101/2. The lower panel indicates fine deletion constructs of the proximal region. Constructs are designated by the nucleotide length of the 5′-flanking regions. The gene structure is schematically represented in the middle panel.
Pax6 and c-Maf activate the c-maf promoter
c-maf is expressed in the differentiation process of lens fiber cells. Many lens-specific genes, such as the crystallin family genes, have a MARE in their regulatory region and are activated by c-Maf (27). In order to determine the upstream factor of c-Maf we have examined Pax6, the master transcription factor in lens development, as well as c-Maf itself.
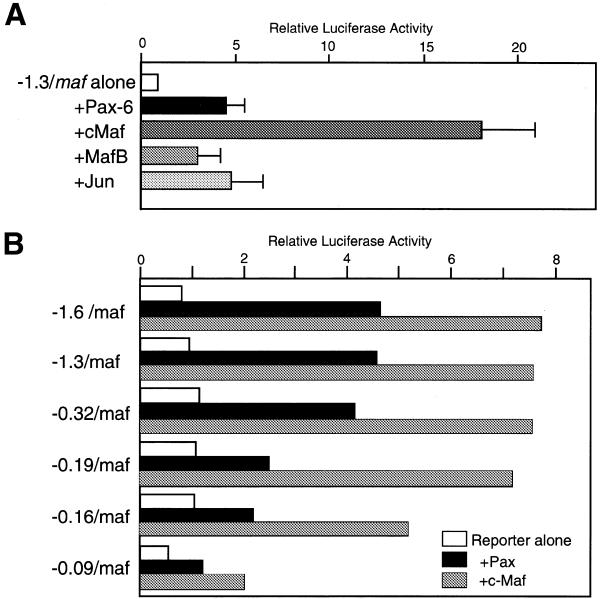
We co-transfected expression vectors for Pax6, c-maf, mafB and c-jun with a c-maf–luciferase reporter gene into C3H10T1/2 cells. Figure 4A shows that both Pax6 and c-Maf markedly activate the c-maf promoter. MafB and c-Jun also activate the c-maf promoter, but this activation is weaker than that of c-Maf. Since both MafB and c-Jun recognize similar sequence elements to c-Maf, both factors may act through the same elements as c-Maf. Pax6(5a), an alternative splice variant of Pax6 (38), has no significant effect on c-maf expression (data not shown).
Figure 4.
(A) Activation of the c-maf gene by transcription factors. The luciferase gene having 1.3 kb of the 5′-flanking region of the c-maf gene (1 µg) was co-transfected with various expression plasmids (1 µg), as indicated, into C3H10T1/2 cells. (B) Activation of 5′-deletion constructs of the c-maf gene by Pax6 and c-Maf. An aliquot of 1 µg of each deletion construct was co-transfected with Pax6 expression plasmid (1 µg) or c-maf expression plasmid (0.5 µg).
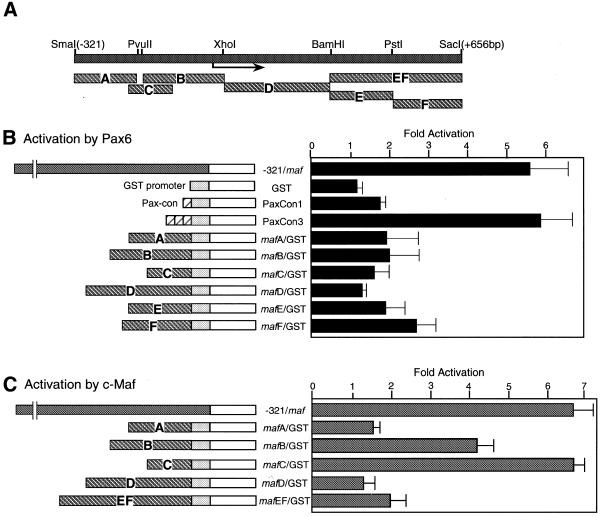
To identify the required element for Pax6 activation of the c-maf gene, 5′-deletion constructs were analyzed (Fig. 4B). The construct deleted to –0.32 kb shows strong activation, while activation gradually decreases with further deletions. The deletion to –90 was still activated by Pax6. This suggests that there are multiple binding sites in the c-maf promoter. Next, various 5′-fragments were joined to a luciferase gene having a GST-P gene promoter (32), a minimal promoter containing a TATA box and a GC box, and were co-transfected with a Pax6 expression vector (Fig. 5B). The –321 maf construct was activated by Pax6 ∼6-fold, but each of the fragments derived from the c-maf promoter region had rather weak activity (up to 2.5-fold). Activation of the luciferase gene containing a single Pax consensus element was not strong (Fig. 5B, PaxCon1). However, the construct having three tandemly repeated elements was strongly activated (Fig. 5B, PaxCon3). This suggests that multiple binding sites are required for strong activation by Pax6.
Figure 5.
Analysis of the required region of the c-maf gene for Pax6 and c-Maf activation. (A) The fragments of the c-maf gene used for luciferase constructs are illustrated. (B) The indicated fragments joined to a GST–luciferase gene construct (1 µg) were co-transfected with Pax6 expression plasmid (1 µg) into C3H10T1/2 cells. (C) The indicated constructs (1 µg) were co-transfected with c-maf expression plasmid (0.5 µg). Activities are expressed as fold activation by co-transfection with the expression plasmids.
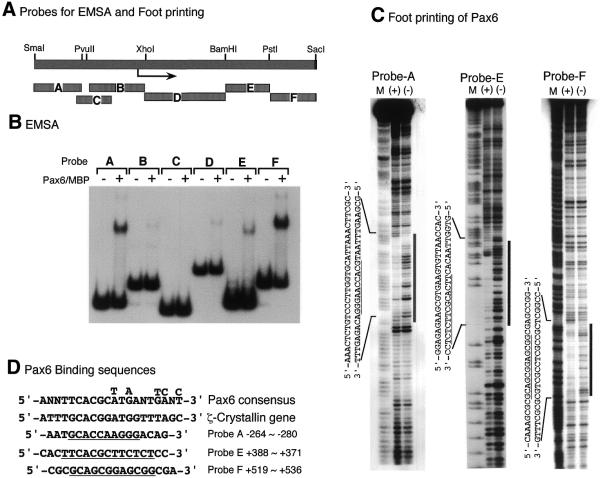
To identify the binding site for Pax6 in the promoter of the c-maf gene, we carried out EMSA and footprinting analyses using various fragments derived from the c-maf promoter (Fig. 6A). The paired domain of Pax6 fused with MBP was used for the EMSA and footprinting analyses as described in Materials and Methods. Surprisingly, five of the six fragments of the 5′-flanking and 5′-non-coding regions of the c-maf gene examined bound to the Pax6 paired domain (Fig. 6B). Fragments A, E and F had strong and B and D had weak binding activities. To further identify the binding sequence, footprinting analyses were carried out. Fragments B and D weakly bind to Pax6 and have no clear footprints. As shown in Figure 6C, the other three fragments (A, E and F) show clear footprints. The nucleotide sequences of the binding sites are aligned in Figure 6D. All three binding sites have similar sequences to that of the Pax6 consensus (5′-T/G-T/G-C-A-C-G-C/G-3′). These results indicate that the c-maf promoter has multiple Pax6-binding elements in the 5′-flanking and 5′-non-coding regions and that these elements are required for full activation by Pax6.
Figure 6.
Identification of Pax6-binding sites of the c-maf gene. (A) Schematic representation of the probes used for EMSA and footprinting analyses. (B) EMSA of c-maf promoter fragments with Pax6–MBP fusion protein. The terminally labeled fragments indicated were bound with Pax6–MBP protein (100 ng) and analyzed as described in Materials and Methods. (C) Footprinting analysis of c-maf promoter fragments. Probe A, E or F was mixed with Pax6–MBP protein (1 µg), treated with DNase I and analyzed by denatured polyacrylamide gel electrophoresis. The same probes, modified and digested by the Maxam–Gilbert method, were used as makers (M). (+) and (–) indicate with or without Pax6–MBP protein, respectively. Footprinting regions are indicated by vertical bars and lined under the sequences. (D) Pax6-binding sequences. Pax6 paired domain binding consensus sequence obtained by PCR-based oligonucleotide selection (33) and that of the promoter region of the ζ-crystallin gene (28) are aligned together with binding sequences of the c-maf promoter.
Localization of c-Maf-responsive elements in the c-maf promoter
As described above, c-Maf dramatically activates the c-maf promoter. To identify the responsive elements for c-Maf activation, we have dissected the promoter region into small fragments and analyzed each fragment by transient transfection assay. As shown in Figure 5B, fragments B (–165/+51) and C (–190/–56) were activated by c-maf co-transfection. The other fragments displayed slight activation. As expected, fragments B and C bind to a c-Maf–MBP fusion protein in an EMSA (Fig. 7B). Using the same c-Maf–MBP protein, footprinting analysis showed that c-Maf protein binds to the sequence –177 to –163, which has two adjacent PvuII sites. This sequence contains a 17 bp complete palindromic sequence with a 1 bp insert (-CAGCTGACAGTCAGCTG-) and is almost identical to the CRE-type MARE (-TGCTGACGTCAGCA-) (11). Another binding site was detected on fragment B (Fig. 7C) at –26 to –15, just downstream of the TATA box. This sequence, 5′-TGCTGAGTT-3′ is located in the opposite orientation and is also similar to the MARE. A weak binding site was found at –106 to –90 (Fig. 7C, thin lines), the region of overlap for the two probes, although we could not find a typical MARE-like sequence in this region.
Figure 7.
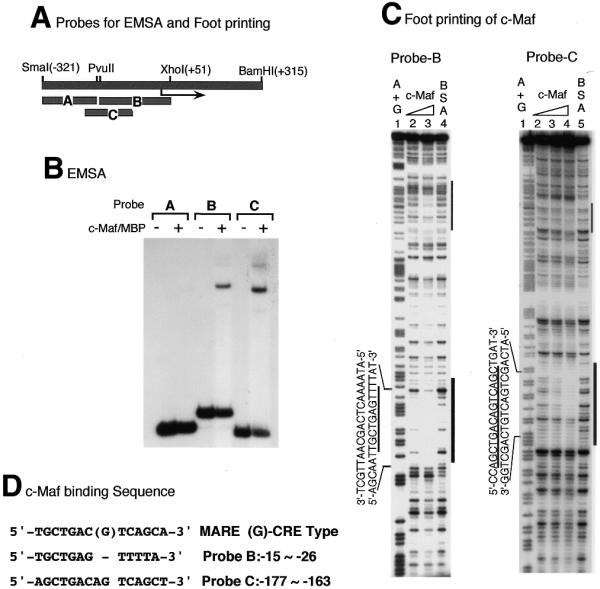
Identification of c-Maf-binding sequences on the c-maf gene. (A) Schematic representation of probes used for EMSA and footprinting analyses. (B) EMSA of c-maf promoter probes with c-Maf–MBP protein. EMSA was carried out as in the legend to Figure 6 and Materials and Methods. (C) Footprinting analysis of the c-maf gene promoter with c-Maf–MBP protein. Probe B or C was mixed with increasing amounts of c-Maf–MBP and digested with DNase I as in the legend to Figure 6. c-Maf–MBP protein was used at 0.3 and 1 µg for lanes 2 and 3 with probe B, at 0.1, 0.3 and 1 µg for lanes 2–4 with probe C. A and G residues were modified and cleaved and used as markers (lanes 1 for both probes). (C) c-Maf-binding sequences are aligned with the MARE consensus sequence. TRE-type consensus and CRE-type consensus (G in the middle position) sequences are presented.
These findings indicate that the c-maf gene is positively autoregulated by its own product, c-Maf, through the MARE sequence.
Pax6 activates the endogenous c-maf gene
In order to confirm activation of the c-maf gene by Pax6, we established a Pax6 overexpressing cell line by permanent introduction of a Pax6 expression vector into C3H10T1/2 cells. The plasmid containing the Pax6 gene under control of the human β-actin promoter and enhancer was transfected together with a plasmid, pSV2neo, expressing the neomycin resistance gene. G418-resistant clones were isolated and expression of Pax6 and c-maf mRNA was quantified by RNase protection analysis. The Pax6 riboprobe used in this experiment covered 110 nt of the 5′-non-coding region and 136 nt of the coding region. Since the Pax6 expression vector contains only the coding region, as described in Materials and Methods, the probe must be protected at nt 246 and 136 by transcripts from the endogenous and transfected genes, respectively. The results are shown in Figure 8. C3H10T1/2 cells do not express detectable Pax6 mRNA (Fig. 8, neo). c-maf mRNA was increased in Pax6 overexpressing cell clones. This result indicates that introduction and overexpression of the Pax6 gene activates the endogenous c-maf gene in C3H10T1/2 cells.
Figure 8.
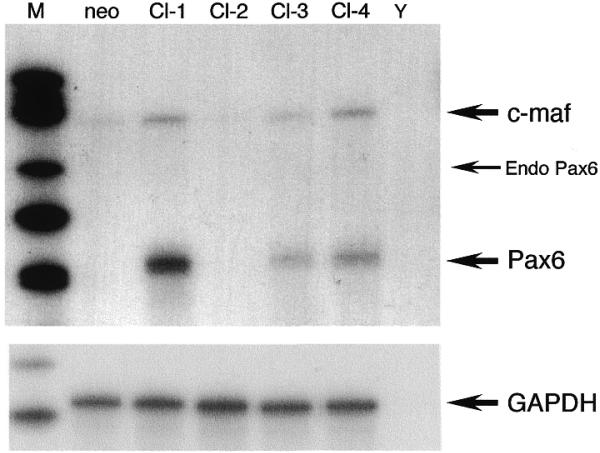
Activation of endogenous c-maf gene by overexpression of Pax6. The Pax6 expression plasmid was permanently introduced into C3H10T1/2 cells. Expression of Pax6 and c-maf mRNAs in four independent cell clones (Cl-1–Cl-4, each 20 µg) were determined by RNase protection analysis. RNA from the cell clone transfected only with the neomycin resistance gene and yeast RNA were used as controls (neo and Y, respectively). The same RNA samples (each 5 µg) were probed with a GAPDH probe as controls. Arrows indicate protected bands of c-maf, endogenous Pax6, transfected Pax6 mRNA and the GAPDH probe, respectively. Size markers were used as in Figure 2 (M).
In situ hybridization histochemistry of pax6 and c-maf in the E14 embryonic lens
To determine the precise expression patterns of Pax6 and c-Maf in the lens, we have carried out in situ hybridization histochemistry with E14 mouse embryonic lens. Pax6 was expressed in lens epithelial cells on the anterior surface of the lens and in the lens equator (Fig. 9A). However, c-maf was not expressed in lens epithelial cells but was expressed in the equator and lens fiber cells (Fig. 9B, arrowhead). These results agree with previous reports (20,21,23–25,39,40). The expression pattern of the two genes did not completely overlap. Lens fiber cells differentiate from actively proliferating lens epithelial cells in the lens equator region. Pax6 was expressed in lens epithelial cells and cells differentiating from epithelial to fiber cells, while c-maf was expressed in differentiating cells and finally differentiated fiber cells. These results support the hypothesis that Pax6 activates the c-maf gene in lens equator cells and that c-maf expression continued in lens fiber cells by a positive autoregulation mechanism.
Figure 9.
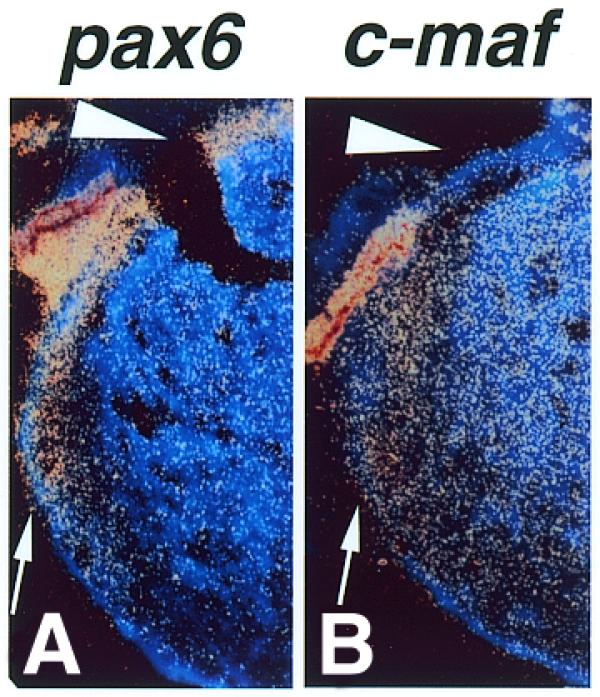
Expression of Pax6 (A) and c-maf (B) mRNAs in the lens of mouse E14 embryos. Pax6 and c-maf mRNAs were detected by in situ hybridization histochemistry. Pax6 mRNA was detected in the lens epithelium (A, arrowhead) and lens equator (A, arrow) whereas c-maf mRNA was not present in the lens epithelium (B, arrowhead) but was expressed in the lens equator and lens fiber cells (B, arrow).
DISCUSSION
The rat c-maf gene
We have cloned the rat c-maf gene containing a long 5′-flanking, a 5′-non-coding, an entire coding and partial 3′-non-coding region. Like the mouse c-maf (24) and mouse mafB genes (41), the rat c-maf gene has no introns, at least in the 5′-non-coding and coding sequences. Two transcription initiation sites were found, at 958 and 930 nt upstream from the ATG translation initiation site. The sizes of the rat c-maf mRNAs are thus estimated to be 2849 and 2821 nt for the major and minor mRNA, respectively. The nucleotide sequence of the promoter region is almost completely conserved between rat and mouse (24), only 3 nt being different within 200 nt of the 5′-flanking region.
Pax6 and c-Maf activate the c-maf gene
As previously reported, c-Maf is expressed in various tissues in a differentiation-specific manner (20–22). Computer analysis of the rat c-maf gene suggests that huge amounts of transcription factors can potentially bind to the promoter region, but it is not known which factor might work on this gene. The mechanisms of c-maf gene regulation might not be simple in such many different tissues and developmental stages. Recent analyses of c-maf knockout mice show that lens fiber cell differentiation is impaired. Expression of Pax6, the master gene of eye development, in the lens is similar to that of the maf family genes, leading us to investigate the interrelationship between Maf and Pax6 transcription factors in lens development. We found that Pax6 activates the transiently transfected as well as the endogenous c-maf gene. We also detected multiple Pax6-binding elements in the c-maf promoter. c-Maf itself strongly activates the c-maf gene through MARE elements in the c-maf promoter. Pax6 and c-Maf expression partially overlap in epithelial cells near the lens equator during differentiation of lens fiber cells (Fig. 9). This result suggests that c-maf gene activation was initiated by Pax6 in the lens equator and that when Pax6 expression ceased c-maf gene expression continued in differentiated fiber cells due to a positive autoregulation mechanism.
MafB, a highly related Maf family large protein, is also expressed during lens differentiation (limited to the lens equator) and has a wider DNA-binding specificity than c-Maf (7). This suggests that both factors have related but distinct roles in lens differentiation. The promoter region of the mafB gene has some similarity to that of c-maf (41) and we surmise that MafB may also be a member of the Pax6 cascade in lens development. However, our preliminary results indicate that the mafB gene is not significantly activated by Pax6 (data not shown). In addition, abnormal lens development has not been reported in kreisler mice, a mafB gene mutant strain (14). Another Maf family large protein, Nrl, is also expressed in lens fiber cells (42). How these Maf-related transcription factors function in lens development remains to be elucidated.
Pax6 does not always activate the c-maf gene, because the expression patterns of Pax6 and c-maf are not identical. Pax6 mutant mice and rats (small eye, sey) (29,43) have been reported which have impaired eye development. Eyes do not develop in these mutant animals, therefore expression of c-maf cannot be detected in lens tissue. However, other organs in which both Pax and Maf are expressed, such as the hindbrain and spinal cord, would be valuable in order to determine whether c-maf is expressed or not in Pax6 mutant animals.
The c-maf gene is activated by its own product, c-Maf. The c-jun and mafB genes are also activated by their own products (41,44). Thus, positive autoregulation may be a common mechanism for gene regulation of bZip transcription factors for their continuous expression. The Maf family are bZip proteins that form homo- and heterodimers with one of the bZip factors to modulate transcription through various MARE-related binding elements, such as the TRE, ARE (antioxidant-responsive element of phase II drug metabolizing enzyme genes) and CRE. Therefore, it is likely that several bZip factors are capable of binding to c-maf MARE-related elements and regulating this gene in response to different cellular signals or environments.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr K.Watanabe (Tokyo Metropolitan Institute of Gerontology) for providing the rat genomic library and Dr J.L.Millan (La Jolla Cancer Research Foundation) for critical reading of the manuscript. We also thank the Japanese Collection of Research Bioresources, Tokyo, for providing the C3H10T1/2 cell line. This work was supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture, Japan and by YASUDA Medical Research Foundation.
DDBJ/EMBL/GenBank accession no. AY005471
References
- 1.Nishizawa M., Kataoka,K., Goto,N., Fujiwara,K.T. and Kawai,S. (1989) v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc. Natl Acad. Sci. USA, 86, 7711–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesi M., Bergsagel,P.L., Shonukan,O.O., Martelli,M.L., Brents,L.A., Chen,T., Schrock,E., Ried,T. and Kuehl,W.M. (1998) Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood, 91, 4457–4463. [PubMed] [Google Scholar]
- 3.Kataoka K., Fujiwara,K.T., Noda,M. and Nishizawa,M. (1994) MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol., 14, 7581–7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara K.T., Kataoka,K. and Nishizawa,M. (1993) Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene, 8, 2371–2380. [PubMed] [Google Scholar]
- 5.Swaroop A., Xu,J.Z., Pawar,H., Jackson,A., Skolnick,C. and Agarwal,N. (1992) A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl Acad. Sci. USA, 89, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerppola T.K. and Curran,T. (1994) Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene, 9, 675–684. [PubMed] [Google Scholar]
- 7.Matsushima-Hibiya Y., Nishi,S. and Sakai,M. (1998) Rat maf-related factors: the specificities of DNA binding and heterodimer formation. Biochem. Biophys. Res. Commun., 245, 412–418. [DOI] [PubMed] [Google Scholar]
- 8.Andrews N.C., Kotkow,K.J., Ney,P.A., Erdjument-Bromage,H., Tempst,P. and Orkin,S.H. (1993) The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl Acad. Sci. USA, 90, 11488–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi K., Kataoka,K., Itoh,K., Hayashi,N., Nishizawa,M. and Yamamoto,M. (1994) Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature, 367, 568–572. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka K., Igarashi,K., Itoh,K., Fujiwara,K.T., Noda,M., Yamamoto,M. and Nishizawa,M. (1995) Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol., 15, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kataoka K., Noda,M. and Nishizawa,M. (1994) Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol., 14, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blank V. and Andrews,N.C. (1997) The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci., 22, 437–441. [DOI] [PubMed] [Google Scholar]
- 13.Sieweke M.H., Tekotte,H., Frampton,J. and Graf,T. (1996) MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell, 85, 49–60. [DOI] [PubMed] [Google Scholar]
- 14.Cordes S.P. and Barsh,G.S. (1994) The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell, 79, 1025–1034. [DOI] [PubMed] [Google Scholar]
- 15.Kurschner C. and Morgan,J.I. (1995) The maf proto-oncogene stimulates transcription from multiple sites in a promoter that directs Purkinje neuron-specific gene expression. Mol. Cell. Biol., 15, 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho I.C., Hodge,M.R., Rooney,J.W. and Glimcher,L.H. (1996) The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell, 85, 973–983. [DOI] [PubMed] [Google Scholar]
- 17.Hedge S.P., Kumar,A., Kurschner,C. and Shapiro,L.H. (1998) c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol. Cell. Biol., 18, 2729–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegde S.P., Zhao,J., Ashmun,R.A. and Shapiro,L.H. (1999) c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood, 94, 1578–1589. [PubMed] [Google Scholar]
- 19.Hale T.K., Myers,C., Maitra,R., Kolzau,T., Nishizawa,M. and Braithwaite,A.W. (2000) Maf transcriptionally activates the mouse p53 promoter and causes a p53-dependent cell death. J. Biol. Chem., 275, 17991–17999. [DOI] [PubMed] [Google Scholar]
- 20.Sakai M., Imaki,J., Yoshida,K., Ogata,A., Matsushima-Hibaya,Y., Kuboki,Y., Nishizawa,M. and Nishi,S. (1997) Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene, 14, 745–750. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida K., Imaki,J., Koyama,Y., Harada,T., Shinmei,Y., Oishi,C., Matsushima-Hibiya,Y., Matsuda,A., Nishi,S., Matsuda,H. and Sakai,M. (1997) Differential expression of maf-1 and maf-2 genes in the developing rat lens. Invest. Ophthalmol. Vis. Sci., 38, 2679–2683. [PubMed] [Google Scholar]
- 22.Imaki J., Onodera,H., Tsuchiya,K., Imaki,T., Mochizuki,T., Mishima,T., Yamashita,K., Yoshida,K. and Sakai,M. (2000) Developmental expression of maf-1 messenger ribonucleic acids in rat kidney by in situ hybridization histochemistry. Biochem. Biophys. Res. Commun., 272, 777–782. [DOI] [PubMed] [Google Scholar]
- 23.Kim J.I., Li,T., Ho,I.C., Grusby,M.J. and Glimcher,L.H. (1999) Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl Acad. Sci. USA, 96, 3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawauchi S., Takahashi,S., Nakajima,O., Ogino,H., Morita,M., Nishizawa,M., Yasuda,K. and Yamamoto,M. (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem., 274, 19254–19260. [DOI] [PubMed] [Google Scholar]
- 25.Ring B.Z., Cordes,S.P., Overbeek,P.A. and Barsh,G.S. (2000) Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development, 127, 307–317. [DOI] [PubMed] [Google Scholar]
- 26.Ogino H. and Yasuda,K. (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science, 280, 115–118. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo I. and Yasuda,K. (1992) The cooperative interaction between two motifs of an enhancer element of the chicken alpha A-crystallin gene, alpha CE1 and alpha CE2, confers lens-specific expression. Nucleic Acids Res., 20, 3701–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharon-Friling R., Richardson,J., Sperbeck,S., Lee,D., Rauchman,M., Maas,R., Swaroop,A. and Wistow,G. (1998) Lens-specific gene recruitment of zeta-crystallin through Pax6, Nrl-Maf, and brain suppressor sites. Mol. Cell. Biol., 18, 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill R.E., Favor,J., Hogan,B.L., Ton,C.C., Saunders,G.F., Hanson,I.M., Prosser,J., Jordan,T., Hastie,N.D. and van Heyningen,V. (1991) Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature, 354, 522–525. [DOI] [PubMed] [Google Scholar]
- 30.Nishiguchi S., Wood,H., Kondoh,H., Lovell-Badge,R. and Episkopou,V. (1998) Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev., 12, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow R.L., Altmann,C.R., Lang,R.A. and Hemmati-Brivanlou,A. (1999) Pax6 induces ectopic eyes in a vertebrate. Development, 126, 4213–4222. [DOI] [PubMed] [Google Scholar]
- 32.Sakai M., Okuda,A. and Muramatsu,M. (1988) Multiple regulatory elements and phorbol 12-O-tetradecanoate 13-acetate responsiveness of the rat placental glutathione transferase gene. Proc. Natl Acad. Sci. USA, 85, 9456–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epstein J., Cai,J., Glaser,T., Jepeal,L. and Maas,R. (1994) Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem., 269, 8355–8361. [PubMed] [Google Scholar]
- 34.Gunning P., Leavitt,J., Muscat,G., Ng,S.Y. and Kedes,L. (1987) A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc. Natl Acad. Sci. USA, 84, 4831–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C. and Okayama,H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol., 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Assay for β-galactosidase in extracts of mammalian cells. In Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Taylor S.M. and Jones,P.A. (1979) Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell, 17, 771–779. [DOI] [PubMed] [Google Scholar]
- 38.Walther C. and Gruss,P. (1991) Pax-6, a murine paired box gene, is expressed in the developing CNS. Development, 113, 1435–1449. [DOI] [PubMed] [Google Scholar]
- 39.Richardson J., Cvekl,A. and Wistow,G. (1995) Pax-6 is essential for lens-specific expression of zeta-crystallin. Proc. Natl Acad. Sci. USA, 92, 4676–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan M.K., Haynes,J.I., Cvekl,A. and Piatigorsky,J. (1998) Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol. Cell. Biol., 18, 5579–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang K., Serria,M.S., Nakabayashi,H., Nishi,S. and Sakai,M. (2000) Molecular cloning and functional characterization of the mouse mafB gene. Gene, 242, 419–426. [PubMed] [Google Scholar]
- 42.Liu Q., Ji,X., Breitman,M.L., Hitchcock,P.F. and Swaroop,A. (1996) Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene, 12, 207–211. [PubMed] [Google Scholar]
- 43.Matsuo T., Osumi-Yamashita,N., Noji,S., Ohuchi,H., Koyama,E., Myokai,F., Matsuo,N., Taniguchi,S., Doi,H., Iseki,S. et al. (1993) A mutation in the Pax-6 gene in rat small eye is associated with impaired migration of midbrain crest cells. Nature Genet., 3, 299–304. [DOI] [PubMed] [Google Scholar]
- 44.Angel P., Hattori,K., Smeal,T. and Karin,M. (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell, 55, 875–885. [DOI] [PubMed] [Google Scholar]