Abstract
FLOWERING LOCUS T (FT) is a key flowering integrator in Arabidopsis (Arabidopsis thaliana), with homologs that encode florigens in many plant species regardless of the type of photoperiodic response. We identified 10 FT homologs, which were arranged as five pairs of linked genes in different homoeologous chromosomal regions, in soybean (Glycine max), a paleopolyploid species. Two of the FT homologs, GmFT2a and GmFT5a, were highly up-regulated under short-day (SD) conditions (inductive for flowering in soybean) and had diurnal expression patterns with the highest expression 4 h after dawn. Under long-day (LD) conditions, expression of GmFT2a and GmFT5a was down-regulated and did not follow a diurnal pattern. Flowering took much longer to initiate under LD than under SD, and only the GmFT5a transcript accumulated late in development under LD. Ectopic expression analysis in Arabidopsis confirmed that both GmFT2a and GmFT5a had the same function as Arabidopsis FT, but the effect of GmFT5a was more prominent. A double-mutant soybean line for two PHYTOCHROME A (PHYA) genes expressed high levels of GmFT2a and GmFT5a under LD, and it flowered slightly earlier under LD than the wild type grown under SD. The expression levels of GmFT2a and GmFT5a were regulated by the PHYA-mediated photoperiodic regulation system, and the GmFT5a expression was also regulated by a photoperiod-independent system in LD. Taken together, our results suggest that GmFT2a and GmFT5a coordinately control flowering and enable the adaptation of soybean to a wide range of photoperiodic environments.
A florigen is a hypothetical leaf-produced signal that induces floral initiation at the shoot apex. Recent progress toward understanding the regulatory network for flowering in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) has led to the conclusion that the product of FLOWERING LOCUS T (FT), FT protein, is a florigen that moves through the phloem to the shoot apex (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007; Notaguchi et al., 2008). Overexpression of FT orthologs causes extremely early flowering in dicots such as Populus trees (Populus deltoids and Populus trichocarpa; Böhlenius et al., 2006; Hsu et al., 2006), tomato (Solanum lycopersicum; Lifschitz et al., 2006), and morning glory (Ipomoea nil; formally Pharbitis nil; Hayama et al., 2007) as well as in monocots such as rice (Izawa et al., 2002; Kojima et al., 2002) and wheat (Triticum aestivum; Yan et al., 2006). Ectopic expression analysis indicated that some of these FT orthologs also induce premature flowering in Arabidopsis (Hsu et al., 2006; Lifschitz et al., 2006; Hayama et al., 2007). These results indicate that FT and its orthologs are essential for flowering and that their functions are highly conserved among unrelated species.
Photoperiod response is one of the important pathways in the regulation of FT mRNA abundance. In Arabidopsis, CONSTANS (CO) is a key protein in photoperiod sensing; it directly induces the expression of FT and the closely related gene TWIN SISTER OF FT (TSF) under long-day (LD) conditions (Samach et al., 2000; Suárez-López et al., 2001; Yamaguchi et al., 2005; for review, see Lagercrantz, 2009). The expression of CO is controlled by a circadian clock, with a diurnal peak of expression during the night under short-day (SD) conditions and at the end of the day under LD conditions (Suárez-López et al., 2001). Light regulates CO protein stability: red light acting through PHYTOCHROME B (PHYB) promotes the degradation of CO by the proteasome, whereas far-red and blue light acting through PHYA and CRYPTOCHROME 2 (CRY2), respectively, increase the stability of CO (Valverde et al., 2004). Under LD conditions, PHYB-promoted degradation of the CO protein is antagonized toward the end of the day by the action of PHYA and CRY2, resulting in the stabilization of CO, which in turn induces the expression of FT (Valverde et al., 2004).
The CO-dependent pathway also regulates the expression of several rice FT orthologs, Heading date3a (Hd3a), Rice flowering locus T-like1 (RFT1), and FT-like (Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003). However, the role of the rice CO ortholog Hd1 is more complex than the role of CO in Arabidopsis (for review, see Turck et al., 2008). Hd1 promotes the expression of Hd3a under SD (inductive) conditions and represses it under LD (noninductive) conditions. The repression of Hd3a expression under LD has been explained by an interaction of Hd1 protein with the active Pfr (far-red) form of phytochrome (Izawa et al., 2002). Night-break experiments further demonstrated that light signal transduction by phytochromes is the primary determinant of Hd3a transcription, because circadian-regulated Hd1 expression is not affected (Ishikawa et al., 2005, 2009).
Another model SD plant, morning glory, possesses two FT orthologs, PnFT1 and PnFT2 (Hayama et al., 2007). These genes exhibit circadian rhythms that are set by the onset of darkness and are up-regulated at the end of the night under SD only if the night is sufficiently long. Night-break treatment inhibits floral induction of morning glory but does not influence the circadian rhythm of expression of its CO ortholog, PnCO (Liu et al., 2001). PnCO thus appears not to directly regulate PnFT1 and PnFT2, although the role of phytochromes, as suggested by night-break experiments in rice (Ishikawa et al., 2005, 2009), has not been fully examined. Despite the conserved functions of FT homologs, their expression may be controlled by different systems in different species.
Soybean (Glycine max) is basically an SD plant: flowering is induced when the daylength becomes shorter than a critical length. Soybean is grown at a wide range of latitudes from equatorial to up to 50°, although the cultivation area of each cultivar is restricted to a very narrow range of latitudes. This wide adaptability has most likely been created by genetic diversity at a large number of the major genes and quantitative trait loci controlling flowering behavior. For example, soybean cultivars adapted to high latitudes have weak or no photoperiod sensitivity. Four major loci in soybean (E1, E3, E4, and E7) are known to be involved in the response to LD artificially induced by fluorescent and incandescent lamps (Buzzell, 1971; Buzzell and Voldeng, 1980; Saindon et al., 1989; Cober et al., 1996a; Cober and Voldeng, 2001a). Of these, E3 and E4 encode PHYA proteins, and their loss-of-function alleles promote photoperiod insensitivity (Liu et al., 2008; Watanabe et al., 2009), which enables soybean plants to flower under LD during early summer and complete seed production in the limited frost-free season at high latitudes. On the other hand, soybean cultivars adapted to equatorial regions possess a trait that suppresses the photoperiod response in seedling stages, enabling a longer juvenile period (Sinclair and Hinson, 1992; Tomkins and Shipe, 1996). One or two recessive genes are known to control the “long juvenile period” trait (Ray et al., 1995; Carpentieri-Pípolo et al., 2002), which enables the plant to retain sufficient vegetative growth until flowering under SD conditions. The genetic diversity in flowering-related loci, therefore, may contribute to the wide adaptability of soybean to diverse environmental conditions. Despite the economic importance of soybean, our knowledge about its molecular mechanisms of flowering is still limited. Here, we identified FT orthologs in soybean and analyzed their diversity in terms of gene structure and expression. We report that products of two FT orthologs have florigen-like functions and coordinately control flowering through both PHYA-mediated photoperiodic regulation and photoperiod-independent regulation.
RESULTS
Soybean Orthologs of Arabidopsis FT
We began by screening soybean ESTs deposited in the GenBank/EMBL/DDBJ database with the cDNA sequence of Arabidopsis FT (NM105222) and identified two EST sequences, BU548465 (designated GmFT1a) and TC300311 (GmFT2c). These EST sequences were then used for PCR screening of a soybean bacterial artificial chromosome (BAC) library of cv Williams 82. Two BAC clones were detected, each of which contained one of the EST sequences (clone WBb135L8 contained the GmFT1a sequence, and WBb127D9 contained the GmFT2c sequence). Detailed sequence analysis revealed that GmFT2c in WBb127D9 was a chimeric sequence that possessed only the fourth exon of the FT homologs, so further analysis on that clone was stopped.
A cDNA covering the entire coding region of GmFT1a was amplified from RNAs of trifoliate leaves of cv Harosoy plants grown under SD by reverse-transcription (RT)-PCR. We searched the databases with the cDNA sequence and found a BAC clone (AC121763) containing two tandemly linked FT homologs (designated GmFT3a and GmFT5a), which were originally identified as TERMINAL FLOWER1 (TFL1) homologs (Cannon et al., 2003). We then found seven additional FT homologs (designated GmFT1b, GmFT2a, GmFT2b, GmFT3b, GmFT4, GmFT5b, and GmFT6) by screening the Williams 82 genomic sequence database (Phytozome; http://www.phytozome.net/soybean) using the predicted amino acid sequence of FT.
In total, we identified 10 FT homologs in the soybean EST and genome sequence databases and recovered five BAC clones that included all 10 of the homologs (Fig. 1). Shotgun sequencing analyses revealed that each of the five clones contained two FT homologs: GmFT1a and GmFT1b in BAC clone WBb135L8, GmFT2a and GmFT2b in WBb214D14, GmFT3a and GmFT5a in WBb111F4, GmFT3b and GmFT5b in WBb174H15, and GmFT4 and GmFT6 in WBb132N10. All of the FT homologs identified were thus arranged in linked gene pairs. The cDNA sequences synthesized from RNAs extracted from trifoliate leaves of SD-grown Harosoy plants were determined for six of the 10 homologs. No cDNAs were obtained for GmFT2b, GmFT4, GmFT5b, and GmFT6, because these genes were not expressed or were expressed at a very low level; the cDNA sequences for these homologs were taken from the Phytozome database. The gene structures were determined on the basis of the alignment between the genomic sequences and the cDNA sequences. This structural analysis indicated that all 10 FT homologs contained four exons, as in Arabidopsis and other plant species (Supplemental Fig. S1). These FT homologs correspond to the translated gene model sequences in the database: Glyma18g53680 (GmFT1a), Glyma18g53690 (GmFT1b), Glyma16g26660 (GmFT2a), Glyma16g26690 (GmFT2b), Glyma16g04840 (GmFT3a), Glyma19g28390 (GmFT3b), Glyma08g47810 (GmFT4), Glyma16g04830 (GmFT5a), Glyma19g28400 (GmFT5b), and Glyma08g47820 (GmFT6).
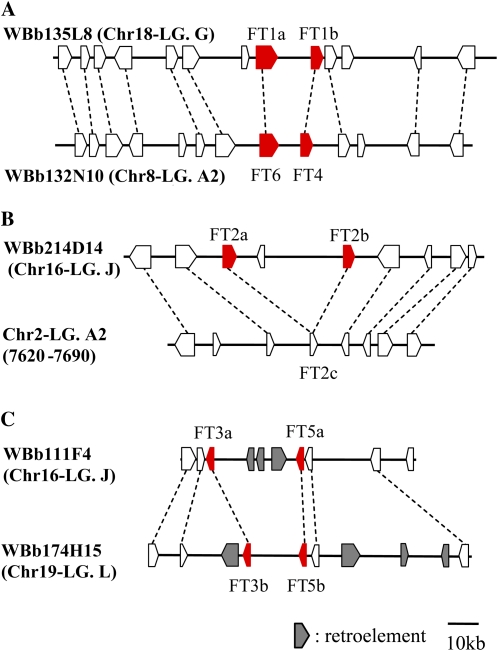
Figure 1.
Soybean FT homologs arranged as tandemly linked genes in different pairs of homoeologous BAC clones. A, Linked pairs GmFT1a/GmFT1b in WBb135L8 (Chr18-LG G) and GmFT4/GmFT6 in WBb132N10 (Chr8-LG A2). B, A linked pair, GmFT2a/GmFT2b in WBb214D14 (Chr16-LG J), and a pseudogene of FT (GmFT2c) in chromosome 2 (Chr2-LG D1b). C, Linked pairs GmFT3a/GmFT5a in WBb174H15 (Chr16-LG J) and GmFT3/GmFT5b in WBb111F4 (Chr19-LG L).
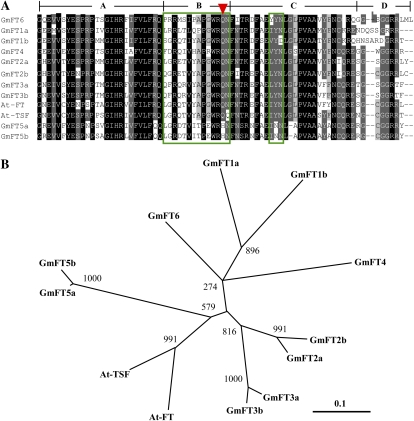
The percentage identity in predicted amino acid sequences of the 10 FT homologs compared with Arabidopsis FT ranged from 72.5% for GmFT3a and GmFT3b to 61.0% for GmFT1a. All of the FT homologs identified shared a crucial amino acid (Tyr) in position 85 of FT at the entrance of the ligand-binding pocket, responsible for the differences in function between FT and TFL1 (Hanzawa et al., 2005; data not shown). GmFT2a further possessed the most similar sequence to Arabidopsis FT in the 14-amino acid stretch referred to as “segment B” and in the LYN triad in segment C (Ahn et al., 2006; Fig. 2A). Both of the two regions are almost invariant in all known FT orthologs and form the external loop crucial for binding to a bZIP transcription factor, FLOWERING LOCUS D (Abe et al., 2005; Wigge et al., 2005). In contrast, GmFT5a and GmFT5b contained six amino acid differences in segment B and in the LYN triad. They also contained His in place of Gln in position 140 of FT, another crucial amino acid that is conserved in FT orthologs (Ahn et al., 2006).
Figure 2.
Similarity of the predicted amino acid sequences of Arabidopsis FT and TSF and 10 soybean FT homologs. A, Multiple alignment of the predicted amino acid sequences in exon 4 of Arabidopsis FT (NM105222) and TSF (AF152907) and soybean FT homologs. The cDNAs for six FT homologs were isolated from cv Harosoy in this study: GmFT1a (AB550120; Glyma18g53680), GmFT1b (AB550121; Glyma18g53690), GmFT2a (AB550122; Glyma16g26660), GmFT3a (AB550124; Glyma16g04840), GmFT3b (AB550125; Glyma19g28390), and GmFT5a (AB550126; Glyma16g04830). The cDNA sequences for GmFT2b (Glyma16g26690), GmFT4 (Glyma08g47810), GmFT5b (Glyma19g28400), and GmFT6 (Glyma08g47820) were taken from the Williams 82 genome database. Highly conserved amino acids are in black, dark gray, or light gray depending on the level of identity (darker = higher level). Green boxes and the red arrowhead indicate the 14-amino acid stretch (segment B) and the LYN triad in segment C and a crucial amino acid identified as diagnostic of FT genes (Ahn et al., 2006). B, Phylogenetic relationships of Arabidopsis and soybean FT proteins constructed using the neighbor-joining method with the program ClustalW. Bootstrap percentage supports are indicated at the branches of the tree.
The phylogenetic tree constructed by the neighbor-joining method classified the 10 FT homologs into four clades and two singletons (GmFT4 and GmFT6; Fig. 2B). In three of the four clades, the two FT homologs within the clade exhibited a high degree of amino acid identity: 92.0% between GmFT2a and GmFT2b, 94.3% between GmFT3a and GmFT3b, and 96.5% between GmFT5a and GmFT5b. The homologs in the fourth clade, GmFT1a and GmFT1b, had a relatively low degree of amino acid identity (78.5%).
Pairs of Linked FT Homologs Are Located in Homoeologous Regions of Different Linkage Groups
Soybean is a paleopolyploid species with a complex genome (Lackey, 1980; Hymowitz, 2004; Shoemaker et al., 2006), and homoeologous duplicated genes are scattered across different linkage groups (LGs; Schmutz et al., 2010). Shotgun sequencing and gene prediction in all five BACs identified 48 genes including the 10 FT homologs: 13 in WBb135L8, nine in WBb214D14, seven in WBb111F4, six in WBb174H15, and 13 in WBb132N10 (Fig. 1). The gene order and orientation were well conserved between WBb135L8 (GmFT1a/GmFT1b) and WBb132N10 (GmFT6/GmFT4) and between WBb111F4 (GmFT3a/GmFT5a) and WBb174H15 (GmFT3b/GmFT5b), suggesting that these FT homologs were located in different pairs of homoeologous BACs. The homology between BACs WBb135L8 (GmFT1a and GmFT1b) and WBb132N10 (GmFT4 and GmFT6) was very high; most of the predicted genes were conserved in both order and orientation. Both gene order and orientation were also similar between BACs WBb111F4 (GmFT3a and GmFT5a) and WBb174H15 (GmFT3b and GmFT5b), although the repetitive element sequences in the intergenic regions differed between the two BAC clones. The GmFT2a-GmFT2b region in BAC WBb214D14 exhibited a similar gene order and orientation to a region (Glyma02g07620 to Glyma02g07690) in chromosome 2 (Chr2) in the Williams 82 genome database, in which a chimeric sequence (GmFT2c) was located (Fig. 1).
We mapped the FT homologs to soybean LGs using recombinant inbred lines (RILs) derived from a cross between breeding line TK780 and wild accession Hidaka 4 (Supplemental Fig. S2). Two pairs of linked genes, GmFT3a/GmFT5a and GmFT3b/GmFT5b, embedded in the homoeologous BAC clones, were mapped to LGs J (Chr16) and L (Chr19), respectively. Two other pairs of linked genes in the homoeologous BAC clones, GmFT1a/GmFT1b and GmFT4/GmFT6, were mapped to LGs G (Chr18) and A2 (Chr8), respectively. A survey on the Williams 82 genome database revealed that, in both cases, the linked genes were located in “recently duplicated segments.” GmFT2a and GmFT2b were mapped 35.4 centimorgan from the GmFT3a/GmFT5a linked pair in LG J.
Two FT Homologs Are Highly Expressed and Exhibit a Circadian Rhythm under SD Conditions
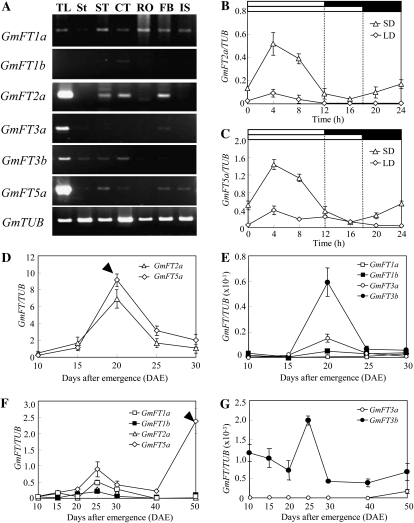
We analyzed transcription profiles of six of the FT homologs in various tissues of Harosoy plants grown under inductive SD conditions (Fig. 3A) by RT-PCR. The GmFT1a transcript accumulated to low but nearly identical levels in all of the tissues except the stem. The GmFT1b transcript was detected faintly and only in cotyledons. In contrast, GmFT2a and GmFT5a were strongly expressed in trifoliate leaves. GmFT2a was also detected at moderate levels in stem tips, cotyledons, and flower buds, where GmFT5a expression was very low. GmFT3a and GmFT3b were also expressed at relatively high levels in trifoliate leaves.
Figure 3.
Transcription profiles of soybean FT homologs. A, Tissue-specific expression in SD. Tissues tested are trifoliate leaf (TL), stem (St), stem tip (ST), cotyledon (CT), root (RO), flower bud (FB), and immature seed (IS). B and C, Diurnal expression of GmFT2a and GmFT5a under SD and LD. Trifoliate leaves were sampled every 4 h at 15 DAE. White and black bars at the top represent light and dark phases, respectively. D to G, Time course-dependent expression in SD (D and E) and LD (F and G). Relative transcript levels were analyzed by quantitative RT-PCR and normalized to β-tubulin (TUB). Average and se values for three replications are given for each data point. Arrowheads indicate the time of flower bud formation.
The diurnal circadian rhythm of FT gene expression was then analyzed by quantitative real-time PCR for GmFT2a and GmFT5a in trifoliate leaves sampled at 15 d after emergence (DAE). The two genes exhibited a diurnal circadian rhythm under SD condition (Fig. 3, B and C), suggesting that their expression was partly regulated by circadian clock genes. In both genes, the expression level increased beginning at dawn, reached a maximum 4 h later, and decreased toward dusk. The diurnal rhythm observed in SD conditions was thus different from those in rice (Kojima et al., 2002) and morning glory (Hayama et al., 2007), in which FT transcripts are most abundant at the end of the night. Rather, it resembled the diurnal rhythm observed in Chenopodium rubrum (SD plant), which produced the highest expression at 8 h after dawn in the light phase and low expression during the night (Cháb et al., 2008). Therefore, a different mechanism may be involved in the circadian rhythm-dependent expression of FT genes of soybean from that in some other SD plants (e.g. rice and morning glory). A similar diurnal rhythm of GmFT2a and GmFT5a expression was also observed in LD (18-h light) conditions, but the expression level was very low (Fig. 3, B and C).
The time course-dependent expression patterns of six FT homologs were analyzed in Harosoy plants grown under SD and LD conditions, using RNAs isolated from trifoliate leaves that were sampled at 4 h after dawn. The levels of GmFT2a and GmFT5a transcripts under SD conditions were low at 10 DAE but increased sharply to their maximum levels at 20 DAE (the time of flower bud formation) and thereafter decreased (Fig. 3D). In contrast, the other FT genes showed extremely low levels (Fig. 3E), although GmFT3b exhibited a similar pattern of expression to GmFT2a and GmFT5a. Under LD conditions, the expression of all of the FT homologs except for GmFT5a was very low (Fig. 3, F and G). The transcript levels of GmFT5a and GmFT1a increased slightly at 25 DAE and decreased thereafter. The GmFT5a transcript reached a relatively high level at 50 DAE (the time of flower bud formation), but GmFT2a did not similarly increase. Since FT expression is up-regulated by conditions that induce flowering in both SD and LD plants, GmFT2a and GmFT5a may be possible candidates for soybean orthologs of Arabidopsis FT.
Ectopic Expression Induces Premature Flowering in Arabidopsis
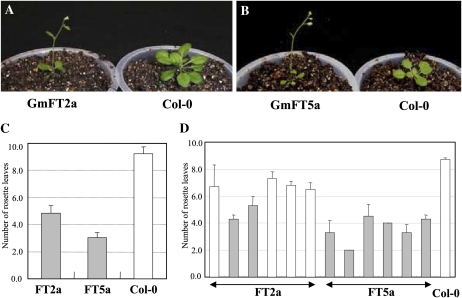
To confirm the function of GmFT2a and GmFT5a in flowering, we conducted an ectopic expression experiment in Arabidopsis ecotype Columbia (Col-0). The ectopic expression of GmFT2a and GmFT5a was confirmed by RT-PCR with homolog-specific primers (data not shown). The T1 plants carrying the GmFT2a or GmFT5a construct flowered earlier (Fig. 4, A and B) and had fewer rosette leaves at flowering than the wild-type Col-0 plants (Fig. 4C) in the 16-h light condition. The flowering date was significantly earlier in the T1 plants carrying GmFT5a (21.8; t = 13.26, P < 0.001) or GmFT2a (28.2; t = 3.75, P < 0.005) than in wild-type plants (32.0). The average number of rosette leaves produced was significantly fewer in the T1 plants carrying GmFT5a (3.0; t = 9.63, P < 0.001) or GmFT2a (4.8; t = 5.74, P < 0.001) than in wild-type plants (9.2; Fig. 4C). T2 plants also showed earlier flowering than wild-type plants (Fig. 4D). T2 plants in all of the six independent T1 families for GmFT5a produced significantly fewer rosette leaves at flowering than wild-type plants. T2 plants in the T1 families for GmFT2a, as a whole, flowered earlier than wild-type plants; two families produced significantly fewer rosette leaves before flowering. These results indicate that the GmFT2a and GmFT5a products function as florigens. In addition, the data obtained in the T1 and T2 generations suggest that the strength of floral induction differs between GmFT2a and GmFT5a: GmFT5a had a more prominent effect than GmFT2a on Arabidopsis flowering (Fig. 4, C and D).
Figure 4.
Effect of ectopic expression analyses of GmFT2a and GmFT5a on Arabidopsis. A and B, Premature flowering induced by ectopic expression of GmFT2a and GmFT5a. C, Number of rosette leaves when the first flower bud appears in T1 and Col-0 plants. Average and se values are calculated from six independent T1 plants for each construct and 10 Col-0 plants. D, Number of rosette leaves in T2 and Col-0 plants. PCR with primers specific to the 35S promoter was carried out to confirm the presence of the transgene in each T2 plant derived from independent T1 plants. Gray bars indicate mean values significantly different from Col-0 by the Tukey-Kramer method.
Expression of GmFT2a and GmFT5a Is Regulated under PHYA-Mediated Photoperiodic Control
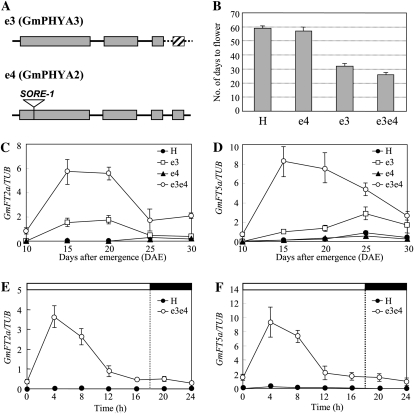
The soybean maturity genes E3 and E4 encode the PHYA proteins GmPHYA3 and GmPHYA2, respectively (Liu et al., 2008; Watanabe et al., 2009). The two genes control flowering under LD artificially induced with different light sources of different red-to-far-red quantum (R:FR) ratios (Buzzell, 1971; Buzzell and Voldeng, 1980; Saindon et al., 1989; Cober et al., 1996b). The recessive alleles e3 and e4 are loss-of-function alleles; the mutation in e3 is caused by the deletion of a 13-kb region including the fourth exon (Watanabe et al., 2009), whereas e4 contains an insertion of a Ty1/copia-like retrotransposon (SORE-1) in the first exon (Liu et al., 2008; Kanazawa et al., 2009; Fig. 5A). The e3e3 recessive homozygote can initiate flowering under R-enriched LD, but the e4 mutant allele is necessary for e3e3 plants to flower under FR-enriched LD. The e4 allele alone cannot induce flowering under either R- or FR-enriched LD conditions (Buzzell and Voldeng, 1980; Saindon et al., 1989; Cober et al., 1996b).
Figure 5.
Expression of GmFT2a and GmFT5a in Harosoy and its PHYA mutant isolines under LD conditions. A, Dysfunctional alleles at the E3 and E4 loci encoding the PHYA genes, GmPHYA3 and GmPHYA2, respectively. The mutation in e3 is caused by the deletion of a 13-kb region including the fourth exon (Watanabe et al., 2009), whereas e4 contains an insertion of a Ty1/copia-like retrotransposon (SORE-1) in the first exon (Liu et al., 2008; Kanazawa et al., 2009). B, Flowering dates under LD of Harosoy (E3E3E4E4; H) and its NILs for e3 (e3), e4 (e4), and e3 and e4 (e3e4). C and D, Time course-dependent expression of GmFT2a and GmFT5a. E and F, Diurnal expression of GmFT2a and GmFT5a. White and black bars at the top represent light and dark phases, respectively. Relative transcript levels were analyzed by quantitative RT-PCR and normalized to β-tubulin (TUB). Average and se values for three replications (six plants for the evaluation of flowering time) are given for each data point.
Harosoy (E3E3E4E4) grown under LD conditions with a R:FR ratio of 1.2 (18 h of light) took an average of 58 d until the first flower appeared, 30 d longer than under SD conditions (12 h of light; Fig. 5B). The allelic difference between e4 and E4 was relatively slight in the presence of E3, with the difference between Harosoy and its near isogenic line (NIL) for e4 (E3E3e4e4) being only 2 d. In contrast, the allelic difference between e3 and E3 was relatively large: the NIL for e3 (e3e3E4E4) flowered 24 d earlier than Harosoy. The double-recessive homozygote (e3e3e4e4) initiated flowering at 24 DAE, which was slightly earlier than the flowering time of SD-grown Harosoy. The timing of flowering of Harosoy and its NILs for the E3 and E4 mutant alleles (H-e3, H-e4, and H-e3e4) was in good accordance with previous reports (Cober et al., 1996b, Cober and Voldeng, 2001b).
The time course-dependent expression analysis revealed that the expression pattern of GmFT2a and GmFT5a correlated well with the earliness of flowering (Fig. 5, C and D). The expression levels of GmFT2a and GmFT5a in the double-mutant NIL increased sharply, reached the maximum at 15 DAE, and thereafter decreased, similar to the expression pattern observed in SD-grown Harosoy (Fig. 3D). The expression levels in the NIL for e3 also increased gradually and reached the maximum at 20 DAE for GmFT2a and at 25 DAE for GmFT5a. In contrast, very little expression of either GmFT2a or GmFT5a was detected in Harosoy or its NIL for e4.
GmFT2a and GmFT5a expression in the double-mutant NIL grown under LD conditions exhibited a diurnal rhythm (Fig. 5, E and F), similar to that observed in SD-grown wild-type Harosoy (Fig. 3, B and C). Expression levels reached a maximum at 4 h after dawn and decreased to their original levels by dusk. The time of maximum expression was thus the same between the SD-grown wild-type Harosoy and the LD-grown double-mutant NIL.
GmFT2a and GmFT5a Respond Differently to Photoperiodic Changes
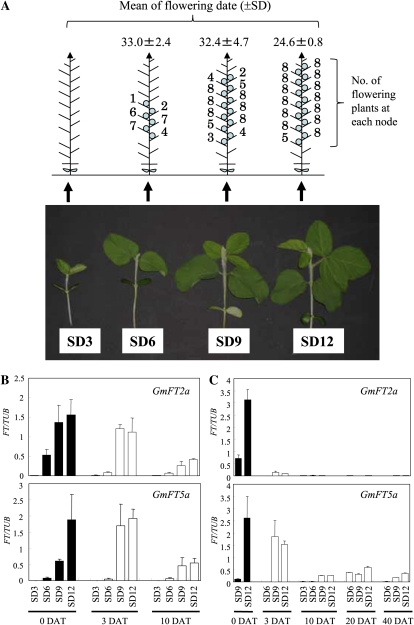
Some soybean cultivars can respond to photoperiod at the time the primary leaves are fully expanded (Borthwick and Parker, 1938; Thomas and Raper, 1984; Thakare et al., 2010). We analyzed the dynamics of GmFT2a and GmFT5a expression in Harosoy plants exposed to different durations of SD (12 h of light) after emergence followed by transfer to LD conditions (20 h of light). A 12-d exposure to SD was sufficient to induce flowering despite subsequent LD treatment; all of the plants tested began flowering at the fifth or sixth node (24.6 DAE) and continued until 40 d after transfer (DAT; flowers at the 18th node), when the experiment was stopped (Fig. 6A). Flowers were formed at the sixth through 11th nodes of the plants exposed to SD for 6 d and at the fifth through 16th nodes of the plants exposed to SD for 9 d, but no flowers were produced at higher nodes. In those plants, the growth phase at lateral meristems later reverted from reproductive growth to vegetative growth. The plants grown under SD for only 3 d did not flower at all.
Figure 6.
Expression of GmFT2a and GmFT5a in soybean plants upon transferring from SD to LD conditions. A, Flowering responses of Harosoy exposed to different durations of SD after emergence in LD. Seedlings of Harosoy grown in SD conditions for 3, 6, 9, and 12 d, from left to right, after emergence were transferred to LD. B, Expression of GmFT2a and GmFT5a in primary leaves. C, Expression of GmFT2a and GmFT5a in trifoliate leaves. DAT, Days after transferring to LD conditions. SD3, SD6, SD9, and SD12 indicate that plants were exposed to SD conditions for 3, 6, 9, and 12 d, respectively. Relative transcript levels were analyzed by quantitative RT-PCR and normalized to β-tubulin (TUB). Average and se values for three replications are given for each data point.
The expression levels of GmFT2a and GmFT5a increased with increasing exposure to SD in both the primary leaves (Fig. 6B, 0 DAT) and the first trifoliate leaves (Fig. 6C, 0 DAT). In primary leaves, a pronounced increase in the transcript level of GmFT2a was detected during SD treatment for 3 to 6 d and 6 to 9 d, whereas that of GmFT5a was detected during SD treatment for 6 to 9 d and 9 to 12 d (Fig. 6B). Similar earlier response of GmFT2a expression to SD condition than GmFT5a expression was also detected in trifoliate leaves (Fig. 6C). After transfer to LD, the transcription levels of both GmFT2a and GmFT5a decreased gradually in the primary leaves as time after transfer increased (Fig. 6B), but the changes in the trifoliate leaves were quite different between the two genes (Fig. 6C). The GmFT2a transcript was lost rapidly and could barely be detected by 3 DAT to LD, even in plants grown for 12 d under SD before transfer. In contrast, the GmFT5a transcript level increased during the first few DAT in plants that had been treated with SD for 9 d (Fig. 6C, 0 DAT and 3 DAT); in plants treated with SD for 12 d, it decreased gradually during exposure to LD. GmFT5a expression was detected at a low level until 40 DAT to LD. These results suggest that the expression of GmFT2a and GmFT5a is regulated differently between primary and trifoliate leaves and that GmFT2a expression in trifoliate leaves responds more sensitively to photoperiodic changes than GmFT5a expression.
DISCUSSION
Diversity of FT Homologs in the Soybean Genome
FT plays a central role as a florigen in floral induction, and its function is conserved across different plant species. The number of FT homologs present in each plant species, however, varies from two in Arabidopsis (FT and TSF) to 13 in rice (Izawa et al., 2002) and 15 in maize (Zea mays; Danilevskaya et al., 2008), although most FT homologs remain undetermined for their functions. In this study, we identified 10 FT homologs in soybean, a paleopolyploid species that has evolved through two rounds of polyploidization (approximately 59 and 13 million years ago; Schmutz et al., 2010). These 10 genes were arranged as five pairs of linked genes. Several of these pairs mapped to homoeologous regions located in different LGs. Both tandem duplications and subsequent chromosomal duplications due to paleopolyploidy, therefore, may have created a large number of FT homologs in the soybean genome. Different levels of gene conservation in order and orientation, different insertions of retroelement sequences between homoeologous BAC clones, and different amino acid identities between linked homologs or between homoeologs in different LGs all suggest that the genomic regions surrounding the five tandemly linked FT homologs have been exposed to independent and complex evolutionary forces.
Of the 10 FT homologs identified, GmFT2a and GmFT5a exhibited the expression patterns that were strongly controlled by photoperiod. Under inductive SD conditions, the two genes were highly up-regulated, with the peak of transcript abundance at the flower bud formation stage; under LD conditions, the expression of both genes was suppressed. Using ectopic expression analyses in Arabidopsis, we confirmed that these two genes have the expected florigen function of FT. The other homologs showed no or very low levels of expression in the tissues and environments tested. In particular, two homoeologs, GmFT2b and GmFT5b, of GmFT2a and GmFT5a, respectively, produced no transcripts in the SD-grown Harosoy. It is tempting to speculate that an optimal level of FT expression has been gained through pseudogenization and/or subfunctionalization of these homoeologs.
GmFT2a and GmFT5a Expression Is Controlled by PHYA-Mediated Photoperiod Response
Transcriptional profiles in Harosoy, a photoperiod-sensitive cultivar, and its NILs for PHYA double mutants, e3 and e4, suggest that the inhibition of GmFT2a and GmFT5a expression under LD is mainly controlled by the PHYA genes, GmPHYA2 and GmPHYA3. This is in sharp contrast to Arabidopsis, in which PHYA, together with CRY2, promote flowering through the stabilization of the CO protein (Valverde et al., 2004). This promotive function of PHYA in flowering is also observed in pea (Pisum sativum; LD plant) and rice (SD plant); the phyA mutants delayed flowering under inductive light conditions (Weller et al., 2001; Takano et al., 2005), in contrast to the e3 and e4 mutant alleles that result in no flowering delay under inductive SD conditions (Cober et al., 1996b; Cober and Voldeng, 2001b). Furthermore, night-break experiments in rice demonstrate that Hd3a (rice FT ortholog) transcription is determined mainly by light signal transduction dependent on PHYB, not PHYA (Ishikawa et al., 2005, 2009).
The possible involvement of PHYB and CRYs in the photoperiodic pathway of flowering has still not been fully addressed in soybean. Zhang et al. (2008) revealed that a soybean CRY1 ortholog, GmCRY1a, rescued the Arabidopsis late-flowering cry2 mutant in ectopic expression analysis with a 35S::GFP-GmCRY1a construct, suggesting that the GmCRY1a protein promotes floral initiation. Furthermore, GmCRY1a exhibited photoperiod-dependent rhythmic production, which correlated with the photoperiodic flowering and the latitudinal distribution cline of soybean cultivars. However, the genetic variations affecting the circadian expression pattern remain unsolved and are suggested to reside outside of the GmCRY1a gene itself (Zhang et al., 2008). Actually, there are no known major genes or quantitative trait loci for flowering around the mapped positions of the CRY family (Matsumura et al., 2009) and PHYB genes (Tasma and Shoemaker, 2003), suggesting that neither CRY nor PHYB genes contributed significantly to the diversification of flowering behavior in soybean. Our data rather support the idea that two PHYA genes, GmPHYA2 (E4) and GmPHYA3 (E3), mainly contribute to diverse responses of flowering to LDs.
GmFT2a and GmFT5a Respond Differently to Photoperiod, and GmFT5a Induces Flowering under LD Conditions
The SD-to-LD transfer experiment indicates that Harosoy possesses no distinct juvenile stage in which photoperiodic response is inhibited. The 12-d exposure to SD after emergence was sufficient to induce and maintain flowering under the LD condition; flowering started around 12 DAT to LD (Fig. 6A; 24.6 DAE) and continued for 27 d until the experiment terminated at 40 DAT. Both GmFT2a and GmFT5a were expressed at a high level in both primary and trifoliate leaves of 12-d-old plants in SD, but 10 DAT to LD no noticeable expression of GmFT2a was detected, and only GmFT5a was expressed at a low level in trifoliate leaves (Fig. 6, B and C). GmFT2a and GmFT5a, therefore, may serve as signals to induce and maintain flowering in soybean; once induced by the two FT proteins, the flowering may continue independently of transcript levels of both GmFT2a and GmFT5a. Alternatively, the lower level of GmFT5a expression observed at 20 to 40 DAT may allow the persistence of flowering.
The SD-to-LD transfer experiment further demonstrated that both GmFT2a and GmFT5a have different responses to photoperiod. The expression of GmFT2a is strictly regulated by photoperiodic changes. In contrast, the response of GmFT5a to photoperiodic changes was gradual, and its expression was conserved at low levels even after the plants were transferred to LD. Furthermore, GmFT5a expression was up-regulated later in development in LD-grown Harosoy (carrying functional PHYA genes; Fig. 3F). These findings suggest that in addition to PHYA-mediated photoperiod response, a second regulatory mechanism may be involved in the control of GmFT5a expression. Under the PHYA-mediated photoperiodic regulation system, GmFT2a and GmFT5a may redundantly and strongly induce flowering in shorter daylengths, but in longer daylengths GmFT5a may solely condition flowering in a photoperiod-independent manner. Therefore, these two FT homologs may coordinately control flowering in soybean.
The expression of rice FT orthologs Hd3a and RFT1 is controlled through two pathways regulated by Hd1 (rice CO ortholog) and Early heading date1 (Ehd1), a rice-specific floral inducer that functions independently of Hd1 (Komiya et al., 2008). Under SD, Hd3a is up-regulated mainly by the Hd1-dependent pathway, and RFT1 expression is very low. Under LD, however, RFT1 is regulated by Ehd1, which in turn is activated by OsMAD50, the rice ortholog of SUPPRESSOR OF OVEREXPRESSION OF CO1 in Arabidopsis, and is inhibited by PHYB (Komiya et al., 2009). RFT1 is further known to be partly up-regulated by chromatin modification; there was a marked increase in RFT1 expression by the increased level of H3K9 histone acetylation around the transcription initiation site of RFT1, particularly when Hd3a expression was suppressed by RNA interference (Komiya et al., 2008). These two pathways regulate the accumulation of both Hd3a and RFT1 transcripts under a range of photoperiods, resulting in the adaptation of rice to a wide range of latitudes by genotypic variation at each of the two loci (Hagiwara et al., 2009).
Further studies are needed to answer how GmFT5a transcription is controlled by factors other than the photoperiodic (PHYA-mediated) pathway. As suggested in rice, the functions of soybean CO orthologs and the involvement of other regulatory mechanisms should be addressed. The loss-of-function alleles of the two PHYA genes (e3 and e4) have contributed to the adaptation of soybean, in particular, to high latitudes. However, the insertion of the SORE-1 retrotransposon into the GmPHYA2 gene (E4) is most likely of evolutionarily recent origin, because distribution of the cultivars carrying the e4 allele is restricted to northern Japan (Kanazawa et al., 2009). The two differently regulated FT genes, GmFT2a and GmFT5a, therefore, may be another key component that makes it possible for soybean to flower in a wide range of photoperiods under the PHYA-mediated photoperiod regulation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Soybean (Glycine max ‘Harosoy’ [L58-266; E3E3E4E4]) and its NILs for two maturity genes, e3 (L62-667; H-e3), e4 (OT94-41; H-e4), and both (OT89-5; H-e3e4), were used in this study. Harosoy is an early-maturing cultivar and belongs to the maturity group II. All plants were grown in 1/5000-a Wagner pots in a growth chamber at a consistent air temperature of 25°C and an average photon flux of 300 μmol m–2 s–1 supplied by fluorescent and incandescent lights. The irradiance ratio of the light at 660 and 730 nm (R:FR) was 1.2 as measured using a LI-COR quantum sensor (model LI-4800C). Daylength regimes were 12 h of light/12 h of dark for SD and 18 h of light/6 h of dark or 20 h of light/4 h of dark for LD. RILs, which were derived from a cross between the breeding line TK780 and the wild soybean line Hidaka 4 (Liu et al., 2007), were used for mapping the FT homologs into LGs.
Transfer from SD to LD
Four germinated seeds of Harosoy with tap roots of almost the same size were transplanted into Wagner pots in the growth chamber. Seeding was repeated four times every 3 d to prepare seedlings of different plant ages, 3, 6, 9, and 12 d old, when the daylength was changed from SD to LD (20 h of light). Primary leaves and the trifoliate leaves were sampled at 4 h after dawn as a bulk from four SD-grown plants of each growing stage and from those transferred into LD at 3, 10, 20, and 40 DAT. The dates of the first flower appearance and flower bud formation at each node were recorded individually.
Sequence Analyses of cDNA and BAC Clones, Assembly, and Annotation
The transcripts covering the entire coding region of the FT homologs were amplified by RT-PCR from cDNAs synthesized from RNA of trifoliate leaves of Harosoy grown under SD. Total RNA was isolated and cDNA was synthesized as described by Koseki et al. (2005). Amplification reactions used the cDNAs as templates and sets of homolog-specific primers (Supplemental Table S1). Amplified products were cloned and sequenced.
The whole nucleotide sequence of each BAC insert was determined by the bridging shotgun method (Sato et al., 2001) with some modification. Plasmids of shotgun clones were sequenced and assembled by Phred-Phrap software (Phil Green, University Washington, Seattle). The lower threshold of acceptability for the generation of consensus sequences was set at a Phred score of 20 for each base. Assignment of the protein-coding regions was performed by similarity searches and computer prediction as described by Sato et al. (2001). The transcribed regions were assigned by comparison of the nucleotide sequences with the Dana-Farber Cancer Institute Soybean Gene Index database by the BLASTN algorithm. All results were compiled with the aid of the Kazusa Annotation Pipeline for Lotus japonicus (Sato et al., 2008). Potential protein-coding genes were assigned by taking into consideration both similarity to known genes and computer prediction.
Genetic Mapping
The FT homologs were mapped in a linkage map constructed using the RILs derived from the cross between TK780 and Hidaka 4, which covered 2,383 centimorgan in length with 282 markers (Liu et al., 2007). Markers developed and primers used in PCR are listed in Supplemental Table S2. The linkage data were incorporated into the map using the Map Manager program QTXb17. Marker order and distance were determined using the Kosambi function and a criterion of 0.001 probability.
Expression Analyses
Tissue-specific expression was analyzed for Harosoy grown under SD. Total RNA was isolated from root, cotyledon, trifoliate leaf, stem, stem tip, flower bud, and immature seed. In diurnal expression analyses, pieces of young fully developed trifoliate leaves were sampled every 4 h starting at dawn for 24 h as a bulk of three plants grown under SD or LD (18 h of light) at 15 DAE. In time course-dependent expression analyses, pieces of young fully developed trifoliate leaves were sampled at 4 h after dawn by bulk from four individual plants grown in SD (12 h of light) and LD (18 h of light) every 5 d starting at 10 DAE until 30 DAE and thereafter every 10 d in the plants grown in LD until 50 DAE.
RT-PCR and Quantitative RT-PCR Analyses
RT-PCR of FT and β-tubulin (as a control) used cDNAs synthesized from total RNA. PCR conditions were one cycle of 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C to 58°C (depending on the gene), and 30 s at 72°C; and a final extension of 10 min at 72°C. RT-PCR was performed using homolog-specific forward primers in the 5′ untranslated region or exon 1 and homolog-specific reverse primer in the 3′ untranslated region to easily separate the RT-PCR products (approximately 500 bp) from the fragments amplified from genomic DNAs (greater than 1 kb). The RT-PCR products were separated by electrophoresis on a 1% agarose gel and visualized with ethidium bromide under UV light. Quantitative RT-PCR was performed as described by Liu et al. (2008). The quantitative RT-PCR mixture was prepared by mixing a 1-μL aliquot of the reaction mixture of cDNA synthesis, 5 μL of 1.2 μm primer premix, 10 μL of SYBR Premix ExTaq Perfect Real Time (TaKaRa Bio), and water to a final volume of 20 μL. The analysis was done using the DNA Engine Opticon 2 System (MJ Research). The PCR cycling conditions were as follows: 95°C for 10 s, 52°C to 56°C (depending on the gene) for 20 s, 72°C for 20 s, and 78°C for 2 s. This cycle was repeated 40 times. Fluorescence quantification was carried out before and after the incubation at 78°C to monitor the formation of primer-dimers. The mRNA level of the β-tubulin gene was used as a control for the analysis. A reaction mixture without reverse transcriptase was also used as a control to confirm that no amplification occurred from genomic DNA contaminants in the RNA sample. In all PCR experiments, amplification of a single DNA species was confirmed by both melting curve analysis of quantitative PCR and gel electrophoresis of PCR products. The primers used for RT-PCR and quantitative RT-PCR are listed in Supplemental Table S1.
Ectopic Expression of GmFT2a and GmFT5a in Arabidopsis
The cDNA sequences of Harosoy GmFT2a and GmFT5a were first cloned into the pGEM-T Easy vector (Promega). SacΙ/XbaI-digested fragments were then inserted in place of the intron-GUS in the pMDC100IG vector, so that the transgene was driven by the cauliflower mosaic virus 35S promoter. Arabidopsis (Arabidopsis thaliana) Col-0 plants were transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog medium with 50 μg mL−1 kanamycin. Six plants selected for each of the two constructs, together with 10 Col-0 plants, were grown in a growth room at a constant air temperature of 23°C and an average photon flux of 270 μmol m–2 s–1 with a daylength of 16 h. T2 plants from each T1 plant, together with 12 wild-type Col-0 plants, were germinated in the Murashige and Skoog medium without kanamycin and grown in the growth chamber with the same temperature and photoperiod but lower photon flux (120 μmol m–2 s–1). PCR with primers specific to the 35S promoter was carried out to confirm the presence of the transgene.
The sequences reported in this paper have been deposited in the GenBank/EMBL/DDBJ database with accession numbers AP011804 (WBb111F4), AP011805 (WBb132N10), AP011806 (WBb135L8), AP011807 (WBb174H15), and AP011808 (WBb214D14) for BAC clones of cv Williams and accession numbers AB550120 (GmFT1a), AB550121 (GmFT1b), AB550122 (GmFT2a), AB550124 (GmFT3a), AB550125 (GmFT3b), and AB550126 (GmFT5a) for cDNA sequences of cv Harosoy.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene structures of FT homologs in soybean.
Supplemental Figure S2. Linkage map positions of soybean FT homologs in the RILs of a cross between a soybean breeding line, TK780, and a wild soybean accession, Hidaka 4.
Supplemental Table S1. List of primers for gene isolation/RT-PCR and real-time PCR analysis used in this study.
Supplemental Table S2. List of primers for genetic mapping used in this study.
Supplementary Material
Acknowledgments
We thank E.R. Cober and R. Nelson for supplying the seeds of the Harosoy isolines for maturity genes.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Borthwick HA, Parker MW. (1938) Effectiveness of photoperiodic treatments of plants of different age. Bot Gaz 100: 245–249 [Google Scholar]
- Buzzell RI. (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13: 703–707 [Google Scholar]
- Buzzell RI, Voldeng HD. (1980) Inheritance of insensitivity to long day length. Soybean Genet Newsl 7: 26–29 [Google Scholar]
- Cannon SB, McCombie WR, Sato S, Tabata S, Denny R, Palmer L, Katari M, Young ND, Stacey G. (2003) Evolution and microsynteny of the apyrase gene family in three legume genomes. Mol Genet Genomics 270: 347–361 [DOI] [PubMed] [Google Scholar]
- Carpentieri-Pípolo V, Almeida LA, Kiihl RAS. (2002) Inheritance of a long juvenile period under short-day conditions in soybean. Genet Mol Biol 25: 463–469 [Google Scholar]
- Cháb D, Kolár J, Olson MS, Storchová H. (2008) Two flowering locus T (FT) homologs in Chenopodium rubrum differ in expression patterns. Planta 228: 929–940 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cober ER, Tanner JW, Voldeng HD. (1996a) Genetic control of photoperiod response in early-maturing near-isogenic soybean lines. Crop Sci 36: 601–605 [Google Scholar]
- Cober ER, Tanner JW, Voldeng HD. (1996b) Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci 36: 606–610 [Google Scholar]
- Cober ER, Voldeng HD. (2001a) A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci 41: 698–701 [Google Scholar]
- Cober ER, Voldeng HD. (2001b) Low R:FR light quality delays flowering of E7E7 soybean lines. Crop Sci 41: 1823–1826 [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR. (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara WE, Uwatoko N, Sasaki A, Matsubara K, Nagano H, Onishi K, Sano Y. (2009) Diversification in flowering time due to tandem FT-like gene duplication, generating novel Mendelian factors in wild and cultivated rice. Mol Ecol 18: 1537–1549 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Agashe B, Luley E, King R, Coupland G. (2007) A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19: 2988–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hsu CY, Liu Y, Luthe DS, Yuceer C. (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18: 1846–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz T. (2004) Speciation and cytogenetics. Boerma HR, Specht JE, , Soybeans: Improvement, Production, and Uses, Ed 3. Agronomy Monograph No. 16. American Society of Agronomy/Crop Science Society of America/Soil Science Society of America, Madison, WI, pp 97–136 [Google Scholar]
- Ishikawa R, Shinomura T, Takano M, Shimamoto K. (2009) Phytochrome dependent quantitative control of Hd3a transcription is the basis of the night break effect in rice flowering. Genes Genet Syst 84: 179–184 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K. (2005) Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17: 3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Kanazawa A, Liu B, Kong F, Arase S, Abe J. (2009) Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J Mol Evol 69: 164–175 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A. (2005) The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol 46: 1879–1883 [DOI] [PubMed] [Google Scholar]
- Lackey JA. (1980) Chromosome numbers in the Phaseoleae (Fabaceae: Faboideae) and their relation to taxonomy. Am J Bot 67: 595–602 [Google Scholar]
- Lagercrantz ULF. (2009) At the end of the day: a common molecular mechanism for photoperiod responses in plants? J Exp Bot 60: 2501–2515 [DOI] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fujita T, Yan ZH, Sakamoto S, Xu D, Abe J. (2007) QTL mapping of domestication-related traits in soybean (Glycine max). Ann Bot (Lond) 100: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J. (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yu J, McIntosh L, Kende H, Zeevaart JAD. (2001) Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol 125: 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Kitajima H, Akada S, Abe J, Minaka N, Takahashi R. (2009) Molecular cloning and linkage mapping of cryptochrome multigene family in soybean. Plant Genome 2: 1–11 [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T. (2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Ray JD, Hinson K, Mankono EB, Malo FM. (1995) Genetic control of a long-juvenile trait in soybean. Crop Sci 35: 1001–1006 [Google Scholar]
- Saindon G, Beversdorf WD, Voldeng HD. (1989) Adjusting of the soybean phenology using the E4 loci. Crop Sci 29: 1361–1365 [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sato S, Kaneko T, Nakamura Y, Asamizu E, Kato T, Tabata S. (2001) Structural analysis of a Lotus japonicus genome. I. Sequence features and mapping of fifty-six TAC clones which cover the 5.4 Mb regions of the genome. DNA Res 8: 311–318 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter JA, Doyle JJ. (2006) Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Hinson K. (1992) Soybean flowering in response to the long-juvenile trait. Crop Sci 32: 1242–1248 [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H. (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tasma IM, Shoemaker RC. (2003) Mapping flowering time gene homologs in soybean and their association with maturity (E) loci. Crop Sci 43: 319–328 [Google Scholar]
- Thakare D, Kumudini S, Dinkins RD. (2010) Expression of flowering-time genes in soybean E1 near-isogenic lines under short and long day conditions. Planta 231: 951–963 [DOI] [PubMed] [Google Scholar]
- Thomas JF, Raper CD., Jr (1984) Photoperiod regulation of floral initiation for soybean plants at different ages. Crop Sci 24: 611–614 [DOI] [PubMed] [Google Scholar]
- Tomkins JP, Shipe ER. (1996) Soybean growth and agronomic performance in response to the long-juvenile trait. Crop Sci 36: 1144–1149 [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, et al. (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Beauchamp N, Kerckhoffs LH, Platten JD, Reid JB. (2001) Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J 26: 283–294 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li H, Li R, Hu R, Fan C, Chen F, Wang Z, Liu X, Fu Y, Lin C. (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc Natl Acad Sci USA 105: 21028–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.