Abstract
In rice (Oryza sativa), trans-acting small interfering RNA (ta-siRNA) is essential for shoot development, including shoot apical meristem (SAM) formation and leaf morphogenesis. The rice wavy leaf1 (waf1) mutant has been identified as an embryonic mutant resembling shoot organization1 (sho1) and sho2, homologs of a loss-of-function mutant of DICER-LIKE4 and a hypomorphic mutant of ARGONAUTE7, respectively, which both act in the ta-siRNA production pathway. About half of the waf1 mutants showed seedling lethality due to defects in SAM maintenance, but the rest survived to the reproductive phase and exhibited pleiotropic phenotypes in leaf morphology and floral development. Map-based cloning of WAF1 revealed that it encodes an RNA methyltransferase, a homolog of Arabidopsis (Arabidopsis thaliana) HUA ENHANCER1. The reduced accumulation of small RNAs in waf1 indicated that the stability of the small RNA was decreased. Despite the greatly reduced level of microRNAs and ta-siRNA, microarray and reverse transcription-polymerase chain reaction experiments revealed that the expression levels of their target genes were not always enhanced. A double mutant between sho and waf1 showed an enhanced SAM defect, suggesting that the amount and/or quality of ta-siRNA is crucial for SAM maintenance. Our results indicate that stabilization of small RNAs by WAF1 is indispensable for rice development, especially for SAM maintenance and leaf morphogenesis governed by the ta-siRNA pathway. In addition, the inconsistent relationship between the amount of small RNAs and the level of the target mRNA in waf1 suggest that there is a complex regulatory mechanism that modifies the effects of microRNA/ta-siRNA on the expression of the target gene.
Small RNAs are 20 to 30 nucleotides (nt) long and function in diverse eukaryotes as regulatory molecules for gene expression in a sequence-dependent manner. They are classified into three types: microRNA (miRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA). miRNAs are 21- to 24-nt RNAs that are processed from long single-stranded precursors (Chapman and Carrington, 2007; Carthew and Sontheimer, 2009). siRNAs are 21- to 24-nt RNAs derived from long perfect double-stranded RNAs (dsRNAs), and based on differences in their primary precursors, types of their targets, and protein components in their biogenesis, they are further categorized into trans-acting siRNA (ta-siRNA), natural-antisense transcript-derived siRNA, and heterochromatic siRNA (Vaucheret, 2006). piRNAs are a distinct class of small RNAs that are 25 to 30 nt long and found only in the germlines of animals (Malone and Hannon, 2009).
During miRNA biogenesis in Arabidopsis (Arabidopsis thaliana), the RNase III enzyme DICER-LIKE1 (DCL1) produces a 21-nt miRNA/miRNA* duplex from a hairpin-like RNA precursor that is transcribed from noncoding RNA genes (Carthew and Sontheimer, 2009; Voinnet, 2009). The miRNA/miRNA* duplex is then methylated at the 2′ OH of the 3′ end of the terminal nucleotide by a small RNA methyltransferase, HUA ENHANCER1 (HEN1; Yu et al., 2005; Yang et al., 2006). This methylation prevents uridylation and subsequent degradation (Li et al., 2005). After the stabilization of the miRNA/miRNA* duplex, the sense strand, miRNA, is selectively assembled into an RNA-induced silencing complex (RISC; Carthew and Sontheimer, 2009). In Arabidopsis, miRNA-RISC is associated with ARGONAUTE1 (AGO1), which cleaves the target mRNA at the center of the base-pairing site of the miRNA/mRNA (Baumberger and Baulcombe, 2005).
The ta-siRNAs are endogenous siRNAs that are found only in plants. They are derived from noncoding TAS genes that are themselves targets of miRNAs. TAS transcripts undergo specific miRNA-mediated cleavage, and then an RNA-dependent RNA polymerase is recruited to generate long dsRNAs using the cleaved transcripts as templates (Allen et al., 2005). In Arabidopsis, the long dsRNAs are processed by DCL4 into multiple 21-nt ta-siRNA duplexes (Xie et al., 2005). In this step, HEN1 contributes to the stabilization of ta-siRNA by methylating its 3′ terminal nucleotide (Li et al., 2005). Mature ta-siRNA is incorporated into RISC and then plays the role of a guide RNA for target RNA cleavage.
In Arabidopsis, many genes are associated with the biogenesis of miRNAs and ta-siRNAs and their metabolic pathways. Among them, DCL1 affects the accumulation of mature miRNAs (Park et al., 2002). Although complete loss-of-function mutants of DCL1 are embryo lethal, hypomorphic dcl1 alleles show pleiotropic phenotypes and reduced accumulation of all miRNAs (Park et al., 2002; Schauer et al., 2002). Similarly, mutations in another class of Dicer-like gene, DCL4, abolish 21-nt ta-siRNA production but not miRNA production (Gasciolli et al., 2005; Xie et al., 2005). In contrast to the embryonic lethality in null alleles of dcl1, a loss-of-function mutant of DCL4 is viable but exhibits abnormal phase changes and leaf morphogenesis (Xie et al., 2005). HEN1 is involved in the accumulation of both miRNAs and ta-siRNA (Li et al., 2005). hen1 mutants show reduced miRNA and ta-siRNA accumulation and share developmental defects with hypomorphic dcl1 mutants (Park et al., 2002).
Studies of these Arabidopsis mutants have provided significant insights into the regulatory mechanisms of miRNA and ta-siRNA biogenesis and metabolic pathways and have revealed how miRNA and ta-siRNA are profoundly associated with development. In this context, it is important to analyze loss-of-function mutants of small RNA-related genes in other plant species for a comprehensive understanding of the significant role of small RNAs in plant development. Recently, ta-siRNA biogenesis mutants were reported in the monocots rice (Oryza sativa) and maize (Zea mays). In rice, loss-of-function mutations of orthologs of DCL4, RNA-DEPENDENT RNA POLYMERASE6 (RDR6), and AGO7 are severely affected in key processes of plant development (Nagasaki et al., 2007). shootless2 (shl2) and shl4, which are loss-of-function mutants of RDR6 and AGO7 orthologs, respectively, completely lack embryonic shoots but show no effects in other embryonic organs. shoot organization1 (sho1) and sho2, which are a loss-of-function mutant of a DCL4 ortholog and a hypomorphic mutant of an AGO7 ortholog, respectively, are defective in shoot meristem maintenance and leaf morphogenesis (Itoh et al., 2000, 2008). Similar phenotypes have also been reported in loss-of-function mutants of a maize homolog of SUPPRESSOR OF GENE SILENCING3 and AGO7, components of the ta-siRNA production pathway (Nogueira et al., 2007; Douglas et al., 2010). These studies have shown that mutations in the ta-siRNA pathway exert more profound effects on plant development in grasses (monocots) than in Arabidopsis. However, no loss-of-function mutants associated with miRNA biogenesis and metabolism pathways have been reported in plants other than Arabidopsis. Thus, our knowledge of the role of miRNAs in plant development is still limited.
We identified two allelic mutants, wavy leaf1-1 (waf1-1) and waf1-2, showing phenotypes that are partially similar to those of sho mutants, which are involved in the ta-siRNA production pathway. The waf1 mutants show milder and more pleiotropic phenotypes than the sho mutants. WAF1 encodes an ortholog of Arabidopsis HEN1, an RNA methyltransferase targeting small RNAs. Our results reveal that WAF1 is required for a wide range of developmental events by stabilizing miRNAs and ta-siRNAs.
RESULTS
The waf1 Embryo Resembles That of sho Mutants
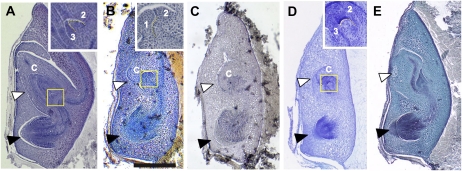
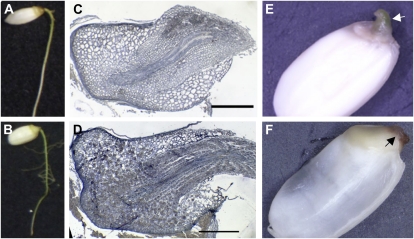
In the wild-type embryo, a shoot and a radicle form on the ventral side of the embryo, and a scutellum forms on the dorsal side adjacent to the endosperm. The wild-type embryonic shoot contains a coleoptile, three leaves, and a shoot apical meristem (SAM; Fig. 1A). In most waf1-1 and waf1-2 mature embryos, the coleoptile was rudimentary and the leaves were morphologically abnormal (Fig. 1, B and C). In addition, the number of leaves was reduced to two or one, in contrast to the three seen in the wild type (Fig. 1B). The shapes of the SAMs in waf1-1 and waf1-2 were flattened compared with that of the wild type (Fig. 1, A and B, insets). These embryonic phenotypes are very similar to those of sho embryos, in which coleoptile deficiency, leaf abnormalities, and a flattened SAM are commonly observed (Fig. 1, D and E; Itoh et al., 2000). Very rarely, the waf1 embryo lacked the embryonic shoot, as does the shl embryo. Thus, the embryonic phenotypes of waf1 overlap considerably with those of the ta-siRNA mutants sho and shl. However, waf1 embryos exhibited a malformed radicle as well (Fig. 1, B and C), which is not observed in sho mutants.
Figure 1.
Phenotypes of waf1 and sho embryos. A, The wild type. B, waf1-1. C, waf1-2. D, sho1-1. E, sho2. The insets in A, B, and D show higher magnification views of the shoot apex of the wild type, waf1-1, and sho1-1, respectively. The yellow dotted line indicates the outline of the SAM. White and black arrowheads indicate embryonic shoot and radicle, respectively. 1, First leaf; 2, second leaf; 3, third leaf; c, coleoptile. Bar = 500 μm.
Vegetative Development in waf1
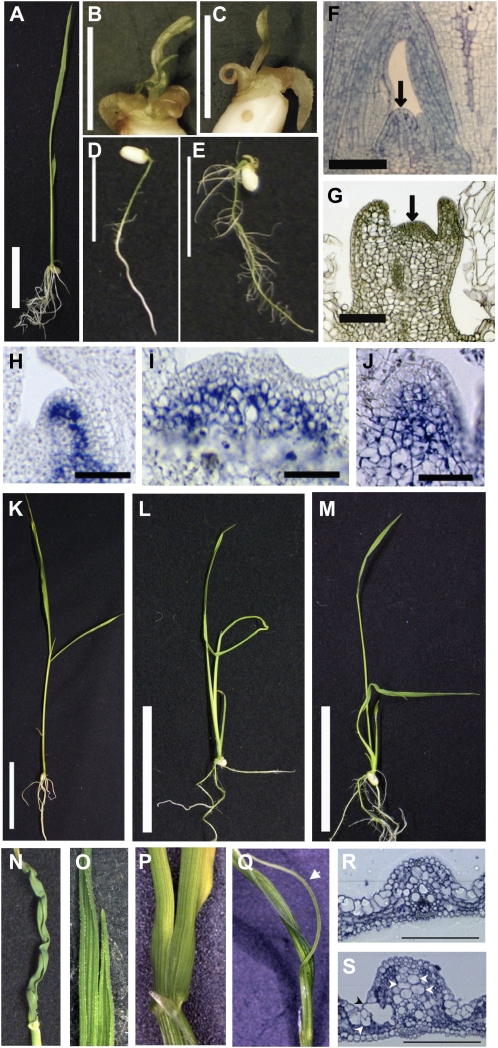
About half of waf1-1 (50.0%) and waf1-2 (55.8%) seedlings stopped growing before 7 d after germination (DAG) and died after the emergence of a few morphologically abnormal leaves by 10 DAG (Fig. 2, A–C), although their roots were sometimes still growing (Fig. 2, D and E). Longitudinal sections of severe waf1 seedlings showed flattened and severely malformed SAMs, indicating that one of the main causes of seedling lethality is a defect in SAM maintenance (Fig. 2, F and G). To confirm this, OSH1 expression in the SAM was observed in severe waf1-1 seedlings at 5 DAG. Although OSH1 expression was detected uniformly below the L1 layer of the wild-type SAM (Fig. 2H), OSH1 expression in the waf1-1 SAM was restricted to the L3 or inner cells, which were frequently enlarged and vacuolated (Fig. 2, I and J). Thus, the SAM structure and indeterminate state of the cells are not properly maintained in severe waf1-1 seedlings.
Figure 2.
Phenotypes of waf1 in the early vegetative phase. A to J, Wild-type and severe waf1 plants. A, Wild-type seedling at 5 DAG. B and C, waf1-1 and waf1-2 seedlings at 5 DAG, respectively. D and E, Elongated waf1-1 and waf1-2 seminal roots at 10 DAG, respectively. F and G, Wild-type and waf1-1 SAMs at 5 DAG, respectively. Arrows indicate SAMs. H to J, In situ expression pattern of OSH1 in wild-type (H) and waf1-1 (I and J) SAMs at 5 DAG. K to S, Wild-type and mild waf1 plants at 10 DAG. K, The wild type. L and M, waf1-1 and waf1-2, respectively. N to S, Leaf phenotypes in waf1. N, Wavy leaf. O, Bifurcation of the waf1-1 leaf blade at the tip. P, Bifurcation of the waf1-1 leaf blade from the base. Q, waf1-1 leaf showing a separation of the filamentous structure (arrow) from the leaf blade. R and S, Cross-sections of the leaf blade in the wild type and waf1-1, respectively. Arrowheads indicate abnormally enlarged cells. Bars = 1 cm (A–E), 50 μm (H–J), 5 cm (K–M), and 100 μm (R and S).
The remaining portion of the waf1 seedlings continued growing, but their leaves were morphologically abnormal (Fig. 2, K–M). Although the leaf phenotype varied greatly among individuals, the most frequent leaf phenotype was severely wavy leaves (Fig. 2N). Other characteristic phenotypes were a bifurcation of the leaf blade (Fig. 2, O and P) and a separation of a filamentous structure from the abaxial side of the blade (Fig. 2Q). These abnormalities were also commonly observed in sho mutants (Itoh et al., 2000, 2008). Bulliform cells were frequently enlarged and ill arranged, seemingly due to unusual (periclinal) cell divisions. Similar defects were also observed in bundle sheath extension cells (Fig. 2, R and S).
Root Development
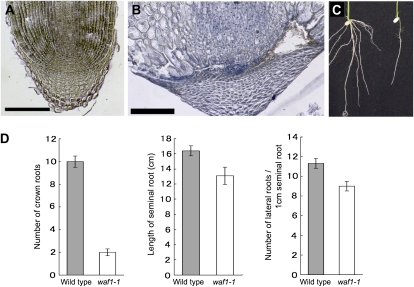
Abnormalities were also detected in roots. Seminal roots of severe waf1 seedlings often arrested soon after germination; the cell size and arrangement at the tip were irregular (Fig. 3, A and B). In mild waf1 seedlings, the seminal roots were relatively short and the numbers of both the crown and lateral roots were reduced (Fig. 3, C and D). These phenotypes indicate that waf1 is defective not only in root apical meristem maintenance of the seminal root but also in adventitious and lateral root apical meristem formation.
Figure 3.
Root phenotypes of waf1-1. A, Longitudinal section of the wild-type seminal root tip. B, Longitudinal section of the waf11-1 arrested seminal root tip. C, Root systems of the wild-type (left) and waf1-1 (right) plants. Crown roots do not elongate in waf1-1. D, Root characters in waf1. From left to right: number of crown roots counted at 10 DAG, length of seminal roots at 10 DAG, and number of lateral roots per 1 cm of seminal root. Error bars indicate sd. [See online article for color version of this figure.]
Reproductive Development
Surviving waf1 plants eventually entered into the reproductive phase. The panicle of waf1 was shortened, and the bract embracing the basal-most primary rachis branch was abnormally elongated (Fig. 4B) compared with the rudimentary development in the wild type (Fig. 4A). In waf1 spikelets, a variety of phenotypes were observed. One of the conspicuous phenotypes of waf1 spikelets was an awn-like protrusion of the central domain of the lemma (Fig. 4, B and D), a phenotype also observed in sho mutants (Itoh et al., 2000). Most frequently observed were the lack of lemma and/or palea and a reduced number of floral organs (Fig. 4, C–E). In the most severe case, only a rod-like terminal structure whose epidermis resembled that of the lemma formed and no other floral organs formed (Fig. 4F), indicating that the floral meristem is converted into a determinate organ due to the loss of meristem activity in early floral development.
Figure 4.
Phenotypes of waf1 in the reproductive phase. A, Wild-type panicle. B, waf1-1 panicle. Arrowhead indicates an elongated bract, and arrows indicate awn-like structures protruded from the lemmas. C, Wild-type spikelet. Lateral halves of the palea and lemma are artificially removed to see inner floral organs. D, waf1-1 spikelet lacking palea. Lemma is reduced in size but protrudes an awn-like structure (arrow), and the stamens lack anthers (arrowheads). E, waf1-1 spikelet lacking palea and floral organs. F, Severe waf1-2 spikelet. A rod-like terminal structure (arrow) is formed. [See online article for color version of this figure.]
Identification of the WAF1 Gene
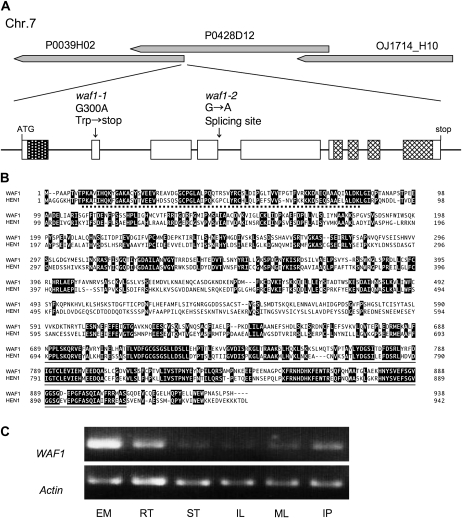
We isolated the WAF1 gene by a map-based cloning strategy. Using the F2 population of a cross between WAF1-1/waf-1 and cv Kasalath, the WAF1 locus was mapped in three bacterial artificial chromosome and P1-derived artificial chromosome clones (OJ1714_H10, P0428D12, and P0039H02) on the short arm of chromosome 7 (Fig. 5A). In this region, the Rice Genome Automated Annotation System (Rice Genome Research Program) predicts about 120 genes. Among these candidate genes, we found a gene annotated as a putative HEN1 of Arabidopsis, which acts in small RNA metabolism. Because the waf1 phenotypes overlapped with those of sho mutants, which are affected in the ta-siRNA production pathway, the putative HEN1 was a likely candidate for WAF1. We compared the genomic sequence of the gene between the wild type and waf1-1 and detected a single base substitution from G to A at the second exon of the gene in waf1-1. This substitution generates a premature stop codon. In waf1-2, a single base substitution was detected at the splicing site of the fourth intron (Fig. 5A). Sequence analysis of the waf1-2 mRNAs revealed that they were spliced incorrectly (data not shown). The introduction of a genomic DNA fragment containing the coding region plus 4 kb upstream and 1.5 kb downstream of the predicted gene rescued the phenotypes of waf1-1 plants (Supplemental Fig. S1). Therefore, we conclude that the predicted HEN1 homolog is the causal gene of waf1.
Figure 5.
Molecular characterization of the WAF1 gene. A, Map position and structure of WAF1. Exons in pitted and checked boxes indicate the positions of the dsRNA-binding domain and the methyltransferase domain, respectively. Locations of the two mutations are indicated. B, Deduced amino acid alignments of WAF1 and HEN1. Dotted and solid lines indicate the dsRNA-binding motif and the methyltransferase motif, respectively. C, RT-PCR analysis of WAF1. EM, Embryos at 5 DAP; RT, roots; ST, stems; IL, immature leaves; ML, mature leaves; IP, young panicle at the primary rachis branch differentiation stage.
The coding sequence of WAF1 consists of 2,814 bp encoding 938 amino acid residues, and the gene is composed of nine exons (Fig. 5A). It was reported that the Arabidopsis HEN1 protein contains an N-terminal dsRNA-binding motif and a C-terminal methyltransferase motif (Fig. 5B; Tkaczuk et al., 2006). Between WAF1 and HEN1, the amino acid identities were 41.7% along the entire amino acid sequence, 57.9% in the dsRNA-binding motif, and 66.5% in the methyltransferase motif (Fig. 5B). Because both waf1-1 and waf1-2 completely lack the methyltransferase domain, they are likely null alleles (Fig. 5A).
To investigate the expression profile of the WAF1 gene, we performed a reverse transcription (RT)-PCR experiment using several organs (Fig. 5C). WAF1 mRNA was detected in all organs, including embryo, root, leaf, stem, and panicle, and a relatively high level of expression was observed in the developing embryo. We also examined the spatial expression patterns of WAF1 by in situ hybridization. However, no tissue- or organ-specific expression was detected (data not shown), indicating that the WAF1 gene is constitutively expressed in the whole plant.
WAF1 Functions in miRNA and ta-siRNA Accumulation
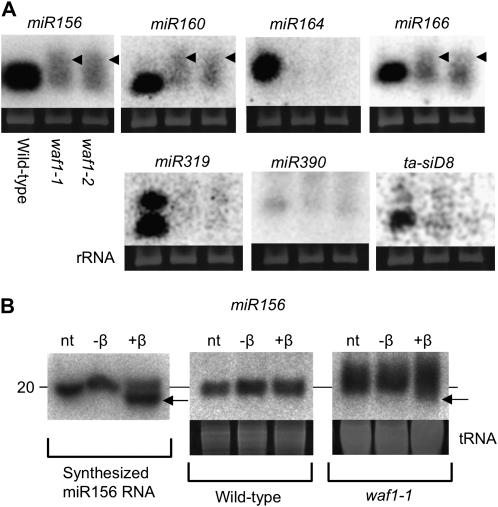
Arabidopsis HEN1 stabilizes miRNA and siRNA by methylating small RNA duplexes at their 3′ termini (Yu et al., 2005). In hen1 mutants, accumulation of these small RNAs is drastically decreased (Park et al., 2002). To clarify whether waf1 mutations also affect small RNA accumulation, we performed northern hybridization for six miRNAs (miR156, miR160, miR164, miR166, miR318, and miR390) and one ta-siRNA (tasiR-ARF/ta-siD8), using total RNA extracted from wild type, waf1-1, and waf1-2 seedlings at 2 weeks after germination. The accumulation of all of the miRNAs and the ta-siRNA was greatly reduced or nondetectable in both waf1-1 and waf1-2 (Fig. 6A). In addition to the reduced accumulation level, smeared bands larger than the band in the wild type were observed in waf1-1 and waf1-2 (Fig. 6A, arrowheads). These smeared bands were also observed in hen1 of Arabidopsis, and it has been proven that the heterogeneity in size of the small RNA is caused by additional uridylation at their 3′ ends (Li et al., 2005). Since HEN1 prevents uridylation by methylating the Rib of the last nucleotide of miRNAs and ta-siRNAs, it is expected that miRNAs and ta-siRNAs are not methylated in waf1 as in hen1 of Arabidopsis. To test whether the 3′ terminal nucleotide of miRNAs in waf1 is modified, we treated small RNAs of the wild type and waf1-2 with NaIO4, and we also treated a synthesized miR156 RNA oligonucleotide with NaIO4 as a control. If the last nucleotide of a small RNA is unmodified, it is sensitive to the chemical reaction β-elimination, resulting in increased mobility of the RNAs. The result was that synthesized RNA oligonucleotide and miR156 in waf1-1 treated with NaIO4 migrated faster in gel electrophoresis than nontreated synthesized RNA (Fig. 6B, left) and miR156 (Fig. 6B, right), whereas the migration of treated and nontreated miR156 in the wild type did not change (Fig. 6B, center). This result indicates that miRNAs in the wild type, but not in waf1-1, were modified at the last nucleotide.
Figure 6.
Analysis of small RNAs. A, Northern hybridization for seven small RNAs. Small RNA accumulations are greatly reduced or nondetectable in both waf1-1 and waf1-2. Arrowheads indicate larger smearing signals detected only in waf1. Ethidium bromide-stained gels corresponding to 5S rRNA are shown at the bottom. B, Modification of the 3′ end of miR156. Synthesized RNA oligonucleotide (left) does not show increased mobility after incubation without (−β) NaIO4/β-elimination but gains mobility (arrow) after incubation with (+β) NaIO4/β-elimination. nt, RNA without incubation. Wild-type miR156 (center) does not show increased mobility after incubation without (−β) and with (+β) NaIO4/β-elimination. miR156 in waf1-1 (right) gains mobility (arrow) after incubation with NaIO4/β-elimination, indicating that the 3′ end of miR156 in waf1-1 is unmodified. Ethidium bromide-stained gels corresponding to tRNA are shown at the bottom.
Accordingly, WAF1 is required for the stabilization of small RNAs through methylating the last nucleotide, as is Arabidopsis HEN1.
Expression Patterns of miRNA and ta-siRNA Target Genes in waf1
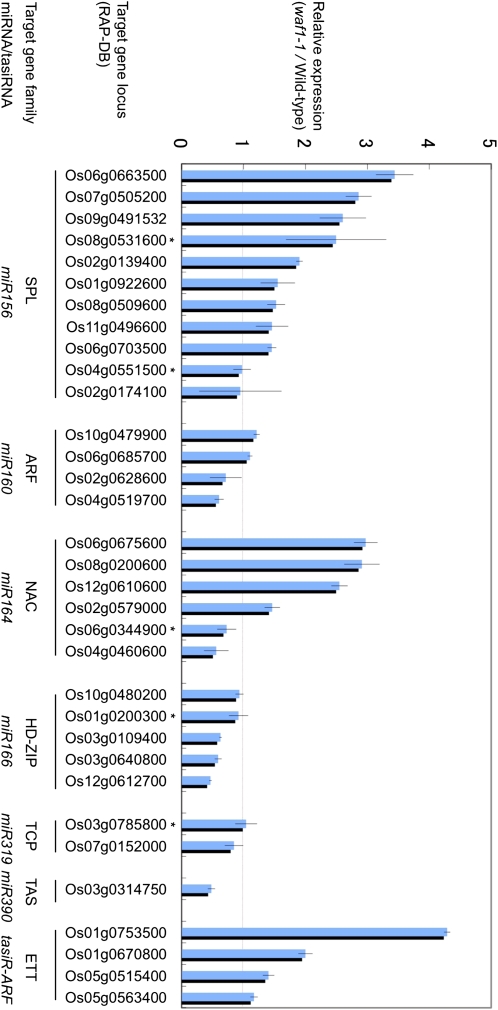
To understand the effects of lower miRNA levels on global gene expression, we performed Affymetrix microarray experiments using seedlings of the wild type and waf1-1 at 2 weeks after germination. Since most miRNAs down-regulate target gene expression by RISC-dependent cleavage, it is expected that lower amounts of miRNAs cause overexpression of their target genes. From the expression data of the microarray experiment, we extracted expression data for genes belonging to six gene families and the TAS3 gene targeted by six miRNAs and one ta-siRNA, whose accumulation was greatly reduced (Figs. 6A and 7). Unexpectedly, the expression level of the target genes varied greatly even within a gene family, and both up-regulation and down-regulation were observed. Among 11 SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) genes regulated by miR156, nine were up-regulated but the expression of two genes was unchanged. Among the up-regulated genes, the expression level of the most up-regulated SPL gene (Os06g0663500) remained less than 3.5-fold that of the wild type, in spite of the drastic reduction in the level of miR156 (about 1/25th that of the wild type; Supplemental Fig. S2A). Two of four ARF transcription factor family genes that are regulated by miR160 were slightly up-regulated, but the rest were down-regulated. Among the NAC transcription factor family genes targeted by miR164, four of six genes were up-regulated and the rest were down-regulated. The expression of five class III homeodomain Leu zipper (HD-ZIP III) genes, two TCP transcription factor family genes, and TAS3, targeted by miR166, miR319, and miR390, respectively, was either unchanged or down-regulated. In contrast, all ETTIN (ETT) genes regulated by tasiR-ARF (ta-siD7 and ta-siD8) were up-regulated. OsETT3 (Os01g0753500) showed the highest level of up-regulation among the putative targets examined.
Figure 7.
Expression changes of miRNA/ta-siRNA putative targets in the seven gene families in waf1-1 revealed by the Affymetrix GeneChip rice genome array. Vertical bars indicate expression levels of waf1-1 relative to that of the wild type. Their Rice Annotation Project Database (RAP-DB) locus, target gene family, and miRNA/ta-siRNA are indicated. Error bars indicate sd. A RAP locus with an asterisk is not verified by the degradome data as a miRNA target (Li et al., 2010). [See online article for color version of this figure.]
Since HD-ZIP III and ETT are regulated competitively by miR166 and tasiR-ARF and are responsible for the key process of shoot development (Nogueira et al., 2007), we further confirmed their expression patterns by real-time RT-PCR experiments (Supplemental Fig. S2). The accumulation of both miR166 and ta-siD7 was reduced (Supplemental Fig. S2A). Four of the five HD-ZIP III genes were slightly down-regulated, and the last one was unchanged (Supplemental Fig. S2B, left). A similar result was observed in three HD-ZIP III genes using primers that can amplify a fragment upstream miR166-directed cleavage site (Supplemental Fig. S2B, right). Two of the ETT genes were up-regulated, but the others were unchanged (Supplemental Fig. S2C). These results indicate that the expression levels of target genes are not always negatively correlated with the miRNA accumulation levels in waf1. This is typically shown in the expression levels of HD-ZIP III genes in waf1, none of which showed enhanced expression in spite of the drastic reduction in miR166 accumulation.
Genetic Interaction between WAF1 and ta-siRNA Pathway Genes
It has been reported that SHO1, SHL2, and SHO2/SHL4 are involved in ta-siRNA production in rice (Nagasaki et al., 2007). Strong alleles of shl2 and shl4 cause a complete loss of the embryonic shoot, but sho1 (Fig. 1D) and sho2, a weak allele of SHO2/SHL4 (Fig. 1E), form abnormal shoots in the embryo. OsETTs are up-regulated in sho1 and sho2 due to the deficiency of TAS3-derived ta-siRNAs (Nagasaki et al., 2007). Since the accumulation level of ta-siRNA was reduced in waf1 (Fig. 6A), it is expected that WAF1 plays a role in a developmental process regulated by ta-siRNAs. To understand the genetic interaction between WAF1 and these ta-siRNA biogenesis genes, we constructed the double mutants waf1-2 sho1-1 and waf1-1 sho2. SHO1 and SHO2/SHL4 are rice orthologs of DCL4 and AGO7 in Arabidopsis, respectively. Both double mutants showed identical phenotypes, a complete lack of a shoot, and an elongated radicle (Fig. 8, A–D), although some seedlings exhibited an arrested seminal root (Fig. 8, E and F). This phenotype is comparable to that of the strong alleles shl2 and shl4. Since a shootless phenotype was rarely observed in waf1-1 and waf1-2 embryos and was never observed in the single mutants sho1 and sho2 (Fig. 1, D and E), it is thought that waf1 mutations enhance the SAM phenotype of sho mutants. This is consistent with the reduced expression of HD-ZIP III and the elevated level of ETT genes in waf1 (Fig. 7; Supplemental Fig. S2, B and C), which are keys for SAM formation and maintenance (Nagasaki et al., 2007). Thus, although waf1 affects both miRNAs and ta-siRNAs, SAM defects in waf1 are mainly due to aberrations in ta-siRNAs.
Figure 8.
Phenotypes of double mutants between waf1 and sho. A, C, and E, waf1-2 sho1-1. B, D, and F, waf1-1 sho2. A and B, Germination of double mutant seeds. The shoot does not emerge, but the seminal root is elongated. C and D, Longitudinal sections of germinating double mutant embryos. A shoot-like structure is not formed. E and F, Double mutant seeds from which the seminal root started elongation but soon arrested (arrows). Bars = 500 μm. [See online article for color version of this figure.]
DISCUSSION
WAF1 Is an Ortholog of Arabidopsis HEN1
HEN1 orthologs are present in various organisms, including plants, insects, and mammals. Although HEN1 functions in methylating miRNA and siRNA duplexes in Arabidopsis, HEN1 orthologs in animals, piwmit/dmHEN1 in Drosophila, and mHEN1 in mouse methylate single-stranded piRNAs (Horwich et al., 2007; Kirino and Mourelatos, 2007; Saito et al., 2007). One of the structural differences between Arabidopsis HEN1 and insect/mammalian HEN1 orthologs is the existence of a dsRNA-binding motif in Arabidopsis (Chen, 2007). Our database analysis revealed that WAF1 is a single-copy gene in the rice genome with strong amino acid sequence similarity to Arabidopsis HEN1. In addition, both WAF1 and HEN1 contain the dsRNA-binding motif in their N-terminal regions (Fig. 5). This suggests that WAF1 methylates dsRNA. In the waf1 mutant, both miRNA and ta-siRNA are decreased, and they are sensitive to the chemical reaction β-elimination (Fig. 6). All of these results strongly suggest that WAF1 is orthologous to Arabidopsis HEN1 and functions as a methyltransferase that stabilizes small RNA duplexes.
It was reported that Arabidopsis hen1 mutants exhibit pleiotropic phenotypes throughout their life cycle, including reduced size of aerial organs, dwarfism, an upward curling of the leaf edge, delayed flowering, and low fertility (Chen et al., 2002). Likewise, waf1 exhibits a variety of phenotypes in various organs from embryogenesis to reproductive development. In addition, cellular abnormalities were also observed in various tissues. Although it is difficult to evaluate an abnormal phenotype in rice (monocot) as equivalent to one in Arabidopsis (dicot), several phenotypes such as wavy leaf edges and dwarfism are present in both waf1 and hen1. However, some phenotypes, such as seedling lethality and abnormal root development, may be specific to waf1. Seedling lethality and the floral phenotype are possibly associated with a dysfunction of the SAM and have not been reported in hen1. One difference in the SAM phenotype is that the dependence of shoot development on the ta-siRNA pathway differs between rice (monocot) and Arabidopsis (dicot); that is, the ta-siRNA pathway is more important for SAM maintenance in rice than in Arabidopsis (Nagasaki et al., 2007). This is supported by phenotypic differences of ta-siRNA biogenesis mutants between rice and Arabidopsis. The complete loss of TAS3-derived ta-siRNAs in the rdr6 and ago7 mutants in Arabidopsis does not cause any defect in the SAM (Peragine et al., 2004; Fahlgren et al., 2006; Hunter et al., 2006), whereas the rice loss-of-function mutants SHL2 and SHL4 (RDR6 and AGO7 ortholog, respectively) completely lack an embryonic SAM (Nagasaki et al., 2007).
Although it is largely unknown which kind(s) of small RNAs is responsible for rice-specific phenotypes, our results indicate that WAF1 is associated with multiple developmental events via the stabilization of many small RNAs. The phenotypic differences between rice waf1 and Arabidopsis hen1 suggest that during evolution rice has acquired novel small RNAs and/or target genes that are regulated by WAF1.
waf1 Phenotypes Are Shared with Those of ta-siRNA-Defective Mutants in Rice
The phenotypes of waf1, such as a malformed coleoptile and SAM, separation of leaf domains, and the elongation of an awn-like structure from the lemma, are also observed in sho mutants, which are defective in the ta-siRNA biogenesis pathway (Itoh et al., 2000, 2008). Since the amount of ta-siRNA is reduced in waf1, the shared phenotypes between waf1 and sho are likely caused by a ta-siRNA deficiency. WAF1 may affect ta-siRNA accumulation in two ways. First, WAF1 may methylate the miR390/miR390* duplex, which is required for the initiation of TAS3-derived ta-siRNA production. Second, WAF1 stabilizes 21-nt ta-siRNA duplexes processed from long dsRNA precursors. Since miR390 and ta-siRNA accumulation are reduced in waf1 (Fig. 6A), WAF1 activity is required for both the initiation and maintenance of the ta-siRNA pathway. In either case, a reduced amount of ta-siRNA should lead to up-regulation of the target genes, OsETTs. In the sho mutants, it is thought that the defects in SAM maintenance and leaf morphogenesis are caused by the ectopic expression of OsETT as a direct result of ta-siRNA deficiency and/or due to the reduced expression of HD-ZIP III genes (OSHBs) as an indirect effect of ta-siRNA deficiency (Nagasaki et al., 2007). In waf1, up-regulation of OsETT and slight down-regulation of OSHB genes were observed as in sho mutants. Thus, the overexpression of OsETTs and down-regulation of OSHBs are possible causes of the phenotypes common to waf1 and sho mutants. However, decreased miR166 accumulation would be expected to cause the overexpression of OSHB genes, targets of miR166. This inconsistency in the relationship between miR166 and the OSHBs may be explained by the transcriptional regulation of OSHBs by OsETTs. Considering the complementary expression of OSHBs in the adaxial domain of the leaf and OsETTs in the abaxial domain, it is probable that OsETTs suppress OSHB expression in the abaxial domain independently of miR166.
The waf1-2 sho1-1 and waf1-1 sho2 double mutants showed the complete loss of embryonic shoots that is exhibited by strong alleles of shl2 and shl4. Since SHO1 and SHL4/SHO2 are associated with ta-siRNA production, the phenotype of the double mutants indicates that the waf1 mutation enhances the mild ta-siRNA-defective phenotype of the sho mutants, suggesting that the defects in SAM maintenance in waf1 are caused mainly by reduced levels of ta-siRNA.
However, no abnormal root phenotypes have been reported in sho/shl mutants. This indicates that the root phenotypes observed in waf1 are not caused by a ta-siRNA deficiency but rather are associated with reduced amounts of some types of miRNAs. It has been reported that ARF and NAC family transcription factor genes targeted by miR160 and miR164, respectively, are involved in root development in Arabidopsis (Guo et al., 2005; Wang et al., 2005; Gutierrez et al., 2009). Recently, it was reported that suppression of HD-ZIP III genes through the activation of miR165/166 is required for xylem patterning in Arabidopsis (Carlsbecker et al., 2010). Thus, the altered expression of these genes could be associated with root phenotypes in waf1.
Reduced miRNA Accumulation Does Not Uniformly Affect the Expression Levels of Target Genes
Most miRNAs in plants modulate target mRNA levels by transcript cleavage, but some miRNAs repress protein synthesis by translational inhibition of target mRNAs (Aukerman and Sakai, 2003; Chen, 2004; Jiao et al., 2010). Because the accumulation of all of the miRNAs examined was greatly reduced, the expression of the target genes was expected to be enhanced. The expression profiles of putative target genes in waf1 (Fig. 7) were somewhat confusing, because several target genes showed reduced expression, and even when expression was enhanced, the extent of elevation was not great.
Recently, small RNA target transcripts in rice have been globally identified and verified by degradome sequencing (Li et al., 2010). The analysis is informative to estimate the abundance and frequency of miRNA-directed cleavage of transcripts. The degradome analyses revealed that the miRNA accumulation level and the cleavage abundance of its target genes are not negatively correlated (Jiao et al., 2008; Li et al., 2010) and that the cleavage frequencies vary among transcripts for the same miRNA (Li et al., 2010). These findings would partially account for our results. For example, two slightly up-regulated ARFs with a miR160 target site in the waf1 have very abundant cleavage frequencies in the degradome data (Li et al., 2010), and the rest of down-regulated ARFs have low cleavage frequencies. A similar tendency was also observed in NAC genes regulated by miR164. Thus, the differential cleavage efficiencies possibly affect the change of miRNA target gene expression in waf1. However, the degradome data did not always justify our results, as in the case of SPL, HD-ZIP III, and ETT genes, although some SPL genes are repressed translationally by miR156 (Jiao et al., 2010; Li et al., 2010).
Besides cleavage efficiency and translational repression of miRNA target transcript, there are several possible explanations for the moderate change of miRNA target gene expression in waf1. First, many miRNAs show a highly specific accumulation pattern. In this case, the miRNAs could cleave the target mRNA only in a restricted region and stage, and the overall elevation of the target mRNA expression would be subtle. Supporting this, many miRNAs are known to have temporally and spatially specific expression patterns (Válóczi et al., 2006).
Second, the small residual amount of miRNAs in waf1 may be enough to cleave target mRNAs. Some miRNAs regulate miRNA biogenesis genes such as DCL1 and AGO1 (Xie et al., 2003). Although the accumulation of the corresponding miRNA was not examined in this study, a feedback mechanism might buffer the drastic up-regulation of target genes in waf1.
A third possible explanation is concerned with technical problems associated with the detection of target transcripts. It is known that 3′ cleaved products of some target mRNAs are stable and easy to detect by northern hybridization, whereas 3′ cleaved products of other mRNAs are degraded by exoribonuclease activity (Souret et al., 2004). If a microarray probe is located in the 3′ region of an mRNA downstream of the cleavage site, the expression level of the gene in the wild type would be overestimated as the sum of the intact mRNA and the cleaved 3′ fragment. As a result, moderate expression changes of the target genes may be observed in spite of the increased uncleaved mRNA in waf1. However, in the case of HD-ZIP III genes, the expression data obtained from microarray (Fig. 7) and real-time PCR amplifying downstream (Supplemental Fig. S2B, left) and upstream (Supplemental Fig. S2B, right) regions of the cleavage site were similar, although microarray probes of one of the HD-ZIP III genes (OSHB2) are located downstream of the cleavage site. This indicates that the microarray result approximately reflects the true expression level of at least HD-ZIP III genes.
Finally, an unknown mechanism may operate to keep target mRNA levels moderate in waf1. miRNAs in waf1 are unmethylated and possibly uridylated at the 3′ end of the terminal nucleotide. These unusual miRNAs may affect the efficiency of mRNA degradation. For example, unmethylated small RNAs might function as primers that recruit RDRs and promote mRNA degradation by an RNA interference-like mechanism. Recently, whole-genome and high-resolution tiling array transcriptome analysis in Arabidopsis has revealed greater amounts of antisense transcripts near the miRNA-binding sites of the target genes (Luo et al., 2009). Moreover, these antisense transcripts are elevated in the hen1 mutant background. This could result in secondary cleavages of the target transcripts by the antisense small RNAs at nonoriginal cleavage sites of the miRNA, which may contribute to homeostasis of the target gene expression in waf1.
Although the reason for this inconsistent relationship between the amounts of miRNAs and their targets, which were also indicated by previous studies (Li et al., 2010), is unclear, similar situations have been reported in Arabidopsis. The overexpression of miR166g in the jba-1D mutant causes down-regulation of its target genes, PHV, PHB, and CNA. However, as for the other target genes, REV is up-regulated and the expression of ATHB8 is unchanged (Williams et al., 2005). Our findings, together with those of previous reports, suggest the existence of a complex regulatory mechanism modifying the effects of miRNA/ta-siRNA on the expression of the target gene and provide useful information for understanding the regulatory relationship between miRNAs and target genes.
CONCLUSION
We identified the WAF1 gene encoding an RNA methyltransferase that is an ortholog of Arabidopsis HEN1. The diverse phenotypic abnormalities of waf1 indicate that stabilization of miRNA and ta-siRNA via WAF1 function is required for various developmental processes. In addition, the maintenance of ta-siRNAs is more essential for shoot development in rice than in Arabidopsis, especially for SAM maintenance. Our results also suggest that there are complex mechanisms that regulate the relationships between the amount of small RNA and the target mRNA level.
MATERIALS AND METHODS
Plant Materials
Two allelic recessive mutants of rice (Oryza sativa) showing abnormal embryos and seedlings were identified from an M2 population of cv Kinmaze mutagenized with N-methyl-N-nitrosourea. We designated them waf1-1 and waf1-2, respectively, because of their most conspicuous phenotype in the vegetative phase. For observation at the early vegetative stage, mutant and wild-type seeds were sown on Murashige and Skoog medium supplemented with 3% Suc and 1% agar at pH 5.8 in a plant box at 28°C. Otherwise, plants were grown in pots or a paddy field. Transgenic plants were grown in a biohazard greenhouse at 30°C in the daytime and 25°C at night.
Histological Analysis
For paraffin sections, samples were fixed in formalin:glacial acetic acid:50% ethanol (1:1:18) for 24 h. They were then dehydrated in a graded ethanol series, substituted with xylene, and embedded in Paraplast Plus (Oxford Labware). The samples were sectioned at 8 μm thick using a rotary microtome. Sections were stained with 0.05% toluidine blue and observed using a light microscope.
In Situ Hybridization
Tissues were fixed with 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 m sodium phosphate buffer, dehydrated in a graded ethanol series, substituted with xylene, and embedded in Paraplast Plus. Microtome sections (8 μm thick) were applied to glass slides treated with Vectabond (Vector Laboratories). Digoxigenin-labeled antisense and sense RNA probes were prepared from full-length cDNAs of OSH1. As the sense probes did not give specific signals, only antisense probe data are presented. In situ hybridization and immunological detection with alkaline phosphatase were performed according to the methods of Kouchi and Hata (1993).
Mapping and Identification of the WAF1 Gene
A heterozygous WAF1-1/waf1-1 plant (subsp. japonica) was crossed with cv Kasalath (subsp. indica), and mutant plants showing the waf1 phenotype in the F2 population were used for mapping. Using cleaved-amplified polymorphic sequence and sequence-tagged site markers, the WAF1 locus was roughly mapped on the short arm of chromosome 7. Using 41 mutant plants of the F2 generation, the WAF1 locus was further limited within the region covering one bacterial artificial chromosome and two P1-derived artificial chromosome clones (OJ1714_H10, P0428D12, and P0039H02). Since we found a gene annotated as a putative HEN1 of Arabidopsis (Arabidopsis thaliana) in this region, we compared the genomic sequence of the gene between the wild type and waf1-1.
The amino acid sequences for WAF1 and HEN1 were found in GenBank (accession nos. AB583903 and AAL05056, respectively). Multiple sequence alignments were performed and manually adjusted to optimize alignments using GENETYX software (Genetyx).
WAF1 genomic DNA, including approximately 4 kb upstream and 1.5 kb downstream, was used for a complementation test. This fragment was introduced into waf1-1 homozygous plants by the Agrobacterium tumefaciens-mediated transformation method (Hiei et al., 1994).
RT-PCR
Total RNA was extracted from 100 mg of tissue from each sample (5-DAP immature embryos, roots, shoots, immature leaves, mature leaves, and inflorescence apices) using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. After RNase-free DNase (TaKaRa) treatment, 1.5 μg of RNA was reverse transcribed using an Omniscript kit (Qiagen). We performed semiquantitative RT-PCR for appropriate cycles at 94°C for 15 s, 60°C for 30 s, and 68°C for 30 s using the primers 5′-AATCATGCGTGAGTCAGCTG-3′ and 5′-CATTGGCACTTGTCTTGAG-3′ for WAF1 and 5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCAGGCATCTG-3′ for Actin.
Isolation and Analysis of Small RNAs
Total RNA of wild-type and waf1 shoots at 2 weeks after germination was extracted using TRIzol reagent, and 10 μg of RNA was used for northern hybridization analysis. Low-molecular-weight RNA was resolved on a 15% polyacrylamide gel containing 7 m urea, transferred to a nylon membrane, and incubated with 32P end-labeled oligonucleotide probes complementary to miR156, miR166, miR160, miR164, miR166, miR319, miR390, and ta-siD8. For the β-elimination experiment, miR156 RNA oligonucleotide was synthesized, and the small RNA fraction of wild-type and waf1-1 shoots at 2 weeks after germination was extracted using the High Pure miRNA Isolation Kit (Roche). Then, 0.01 μg of RNA oligonucleotide, 2 μg of small RNA in the wild type, and 20 μg in waf1-1 was incubated in 5× borate buffer (148 mm borax, 148 mm boric acid, pH 8.6), with (+β) and without (−β) 200 mm NaIO4, and was subjected to northern hybridization analysis using oligonucleotide probes for miR156.
DNA Microarray Analysis and Target Gene Search for miRNAs
Microarray analysis was performed using a GeneChip rice genome array (Affymetrix). Preparation of labeled target complementary RNA, subsequent purification, and fragmentation were carried out using One-Cycle target labeling and control reagents (Affymetrix). Double-stranded cDNA was prepared from 10 μg of total RNA of wild-type and waf1-1 shoots 2 weeks after germination. Hybridization, washing, staining, and scanning were performed as described in the supplier’s protocol. A 10-mg aliquot of fragmented complementary RNA was subjected to hybridization. Three independent replicates were used. Data analysis was performed using GeneChip Operating Software (Affymetrix) and GeneSpring 7 (Agilent Technologies).
From the array data, we extracted expression data for putative target genes of miR156, miR166, miR160, miR164, miR166, miR319, miR390, and ta-siD7-8. A putative target gene search was performed at the Web site http://bioinformatics.cau.edu.cn/PMRD/. The putative target genes obtained for each miRNA/ta-siRNA were aligned, and those constituting a gene family were selected. Potential target genes not constituting a gene family were not used.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB583903 (WAF1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of waf1-1 by the WAF1 genomic fragment.
Supplemental Figure S2. Expression levels of a few small RNAs and their target genes quantified by real-time RT-PCR.
Supplementary Material
Acknowledgments
We thank Ken-Ichiro Ichikawa (University of Tokyo) for assistance in cultivating rice plants at the Experimental Farm of the University of Tokyo.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2007) A marked end. Nat Struct Mol Biol 14: 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RN, Wiley D, Sarkar A, Springer N, Timmermans MCP, Scanlon MJ. (2010) ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-Nava M, Poethig SR. (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Sato Y, Nagato Y. (2008) The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central-marginal axis in rice. Plant Cell Physiol 49: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Itoh JI, Kitano H, Matsuoka M, Nagato Y. (2000) Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12: 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM. (2008) Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. (2007) The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Hata S. (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Luo QJ, Samanta MP, Köksal F, Janda J, Galbraith DW, Richardson CR, Ou-Yang F, Rock CD. (2009) Evidence for antisense transcription associated with microRNA target mRNAs in Arabidopsis. PLoS Genet 5: e1000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. (2009) Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. (2007) Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Tkaczuk KL, Obarska A, Bujnicki JM. (2006) Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A, Várallyay E, Kauppinen S, Burgyán J, Havelda Z. (2006) Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J 47: 140–151 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. (2006) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.