Abstract
Actinorhizal symbioses are mutualistic interactions between plants and the soil bacteria Frankia that lead to the formation of nitrogen-fixing root nodules. Little is known about the signaling mechanisms controlling the different steps of the establishment of the symbiosis. The plant hormone auxin has been suggested to play a role. Here we report that auxin accumulates within Frankia-infected cells in actinorhizal nodules of Casuarina glauca. Using a combination of computational modeling and experimental approaches, we establish that this localized auxin accumulation is driven by the cell-specific expression of auxin transporters and by Frankia auxin biosynthesis in planta. Our results indicate that the plant actively restricts auxin accumulation to Frankia-infected cells during the symbiotic interaction.
Actinorhizal symbioses are mutualistic associations between plants belonging to eight angiosperm families collectively called actinorhizal plants and the soil actinomycete Frankia. These interactions culminate with the formation of a new root organ, the actinorhizal nodule, where Frankia is hosted and fixes atmospheric nitrogen (Benson and Silvester, 1993). During intracellular infection (e.g. in Casuarina glauca or Alnus glutinosa), diffusible signals are emitted by Frankia at early stages of the interaction leading to root hair deformation. The chemical nature of these signals remains unknown but biochemical and genetic studies suggest that they are different from rhizobial Nod factors (Cérémonie et al., 1998; Normand et al., 2007). Frankia then infects some of the deformed root hairs through intracellular infection threads. A limited number of cell divisions are induced in the cortex close to the infection site leading to the formation of the prenodule. Frankia infects some prenodule cells and starts fixing nitrogen while new cell divisions occur in the pericycle close to a xylem pole forming a nodule primordium (Pawlowski and Bisseling, 1996). The nodule primordium subsequently grows and become infected with intracellular Frankia hyphae coming from the prenodule. Actinorhizal nodules have a central vasculature and the cortical symbiotic tissues containing infected and uninfected cells. Unlike legume nodules, actinorhizal nodules are structurally and developmentally related to lateral roots (Pawlowski and Bisseling, 1996).
Little is known about the signals exchanged between the two partners during the establishment of actinorhizal symbioses. The phytohormone auxin controls many developmental processes and has also been involved in plant-microbe interactions (Robert-Seilaniantz et al., 2007; Mathesius, 2008; Kazan and Manners, 2009). Recently, we studied the role of auxin influx activity during actinorhizal symbioses. Inhibition of auxin influx using the competitive inhibitor naphtoxyacetic acid perturbs actinorhizal nodule formation in C. glauca (Péret et al., 2007). Two genes encoding putative auxin influx carriers from C. glauca were cloned and characterized. One of these genes named CgAUX1 was shown to encode a functional auxin influx carrier by complementation of the Arabidopsis (Arabidopsis thaliana) aux1 mutant. Interestingly, CgAUX1 is expressed in Frankia-infected cells during actinorhizal nodule formation (Péret et al., 2007). These results together with previous data showing auxin production by Frankia suggest a role for auxin in infected cells during the symbiotic interaction (Mathesius, 2008; Péret et al., 2008; Grunewald et al., 2009).
The aim of this work was to further explore the involvement of auxin in the C. glauca-Frankia actinorhizal symbiosis. Using a combination of computational modeling and molecular and cell biology approaches, we establish that the cell-specific expression of auxin transporters leads to localized auxin accumulation in Frankia-infected cells in C. glauca nodules.
RESULTS
Auxin Accumulates in Frankia-Infected Cells in Actinorhizal Nodules of C. glauca
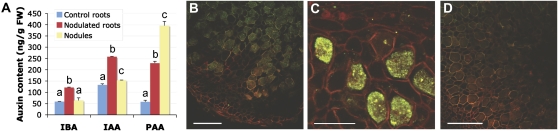
Previous reports suggested that the plant hormone auxin might be involved in the establishment of actinorhizal symbioses (Hammad et al., 2003; Péret et al., 2007). To study the involvement of auxin in the C. glauca-Frankia symbiotic interaction, we used liquid chromatography mass spectrometry (LC-MS) to quantify three major forms of auxin: indole-3-acetic acid (IAA), phenylacetic acid (PAA), and indole-3-butyric acid (IBA) in roots of plant that were not inoculated by Frankia (control), in root portions of inoculated plants that did not bear nodules, and in nodules 3 weeks after inoculation. In four independent experiments, the levels of all three auxins were significantly increased in roots of inoculated plants compared to control plants (Fig. 1A). Moreover, a dramatic increase in PAA content was observed in nodules compared to roots (Fig. 1A). These auxin quantification results show that IAA and PAA are the two major forms of auxins present in C. glauca nodules and that the symbiotic interaction with Frankia leads to increased PAA accumulation in plant tissues.
Figure 1.
Auxin accumulates in Frankia-infected cells in C. glauca nodules. A, Auxin content measured by LC-MS in C. glauca roots of noninoculated plants (control), roots, and nodules harvested 3 weeks after inoculation by Frankia. Significance was tested by an ANOVA test (P < 0.01). FW, Fresh weight. B, Immunolocalization of IAA. The signal is found in cells infected by Frankia. C, Immunolocalization of PAA. A very strong signal is detected in cells infected by Frankia. D, Control immunolocalization. The section was hybridized only with the secondary antibody. No signal is detected in infected cells. Autofluorescence of the cell wall of infected cells can be detected. Scale bars: 100 μm (B and D), 50 μm (C).
To localize the sites of auxin accumulation in actinorhizal nodules, we generated transgenic C. glauca and Allocasuarina verticillata plants containing the molecular markers for auxin perception ProGH3-GUS (Hagen et al., 1991), ProDR5-GUS (Ulmasov et al., 1997), or ProIAA2-GUS (Swarup et al., 2001). GH3 is an auxin-responsive gene from soybean (Glycine max) and the ProGH3-GUS marker has been successfully used to study changes in auxin distribution during legume nodule formation (Mathesius et al., 1998). DR5 is a synthetic auxin-responsive promoter derived from GH3 (Ulmasov et al., 1997), and IAA2 is an auxin-induced Arabidopsis gene. Both ProDR5-GUS and ProIAA2-GUS are commonly used as markers to analyze changes in auxin perception in Arabidopsis. In transgenic Casuarinaceae, those markers exhibited very weak basal levels of expression, and were not or very weakly induced by exogenous auxin (Supplemental Fig. S1). No expression was found in nodules. These results indicate that these molecular markers cannot be used in Casuarinaceae trees. Therefore, an immunolocalization approach was used to investigate the distribution of IAA and PAA within C. glauca nodules. For Arabidopsis, IAA immunolocalization was shown to reflect total auxin distribution (conjugated and free IAA), while ProDR5-GUS reveals sites of free auxin perception (Aloni et al., 2003; de Reuille et al., 2006). In C. glauca nodules, a specific monoclonal IAA antibody binds strongly to Frankia-infected cells (Fig. 1B). Similarly, a polyclonal PAA antibody revealed high levels of PAA accumulation in these cells (Fig. 1C). No significant hybridization was found with the IAA or PAA antibody in uninfected cells in the nodule cortex. Some binding was detected in vascular tissues with the IAA antibody. No signal was detected under the following control conditions: hybridization with only the secondary antibody (Fig. 1D), pretreatment with ethanol to remove auxin, or auxin saturation of the antibody. These results indicate that both IAA and PAA accumulate in Frankia-infected cells of C. glauca nodules. This conclusion is supported by the recent finding that EuNOD-ARP1, a gene from the actinorhizal plant Eleagnus umbellata shown to be auxin inducible in leaves, is expressed in Frankia-infected cells in nodules (Kim et al., 2007).
Distribution of Auxin Carriers Predicts Auxin Accumulation in Frankia-Infected Cells of C. glauca Nodules
We previously found that the CgAUX1 gene that encodes an auxin influx carrier functionally equivalent to Arabidopsis AtAUX1 is expressed in the vascular tissues and in Frankia-infected cells in C. glauca nodules (Péret et al., 2007). To test whether this CgAUX1 expression might be sufficient to explain the pattern of auxin accumulation, we used a computational modeling approach similar to the ones that have been applied successfully to study developmental processes such as phyllotaxis (de Reuille et al., 2006; Chickarmane et al., 2010). Auxin fluxes and accumulation patterns in a tissue can be inferred from the cellular localization of transporters.
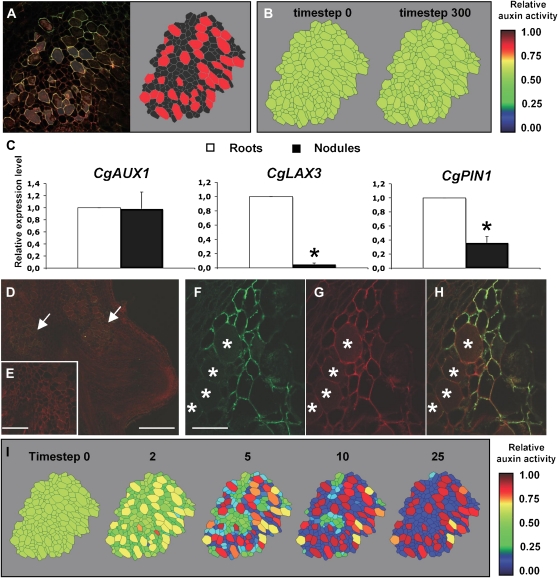
We generated cellular models for auxin transport occurring in C. glauca nodule symbiotic tissues. First, the cortical tissue of C. glauca nodules was observed to have a very specific geometry and topology (Supplemental Fig. S2). For instance, infected cells (easily detected using Frankia autofluorescence or 4′,6-diamino-phenylindole staining) were bigger (5–10 times) and more connected, i.e. were in contact with more cells, than noninfected cells (mean connectivity, i.e. number of contact with other cells, was 6.99 and 4.99, respectively). Because of the infection mechanism, infected cells tend to be organized in files. Thus, confocal images of C. glauca nodules were digitized and used to generate our virtual tissues to keep the specific geometry and topology since fluxes are known to be dependent on those properties. Three in silico models of cortical tissues were generated from confocal images of three different nodules (Fig. 2A; Supplemental Fig. S2). Two different cell types corresponding to infected and uninfected cells were defined in the virtual tissues.
Figure 2.
Expression of auxin transporters in C. glauca nodules is sufficient to predict an auxin accumulation in Frankia-infected cells. A, Example of digitization of nodule tissue. Confocal microscopy images of nodule tissue autofluorescence (left section) were used as a base for manual digitization of the geometry in a virtual representation of the nodule (right section, see also Supplemental Fig. S1). Information regarding the infection of the cells was added in the virtual tissue (infected cells here denoted in red). B, Distribution of auxin in a virtual nodule with infected cells expressing CgAUX1, starting from a uniform auxin level. Infected cells accumulate auxin very slowly in this condition (see Supplemental Fig. S2 for other virtual nodules and starting auxin distribution). Auxin activity is the relative auxin concentration within the cells and its representation is capped at value 1. C, Expression of CgAUX1, CgLAX3, and CgPIN1 in C. glauca uninfected roots and nodules harvested 3 weeks after inoculation. RNA abundance was determined by qRT-PCR and presented are means ± sd of three independent experiments. *P < 0.05, Statistical randomization test (Rest 2009 software; Qiagen). D to H, Immunolocalization of PIN1-like proteins in C. glauca nodules. D, PIN1-like proteins are detected (arrows) close to the nodule apex, i.e. in the region close to the meristem. E, No signal was detected in control immunolocalization experiments without the PIN1. F to H, Close-up in the apex of a C. glauca nodule showing the signal corresponding to the PIN1 antibody (F), autofluorescence (G), and the merged image (H). Infected cells are marked with asterisks. I, Evolution of the distribution of auxin in a virtual nodule with infected cells expressing CgAUX1 and noninfected cells expressing CgPIN1, starting from a uniform auxin level. Auxin activity is the relative auxin concentration within the cells and its representation is capped at value 1. The generated auxin distribution was shown to be robust to CgPIN1 perturbation (see Supplemental Fig. S4). Scale bars: 250 μm (D), 100 μm (E), and 50 μm (F–H).
After the addition of AUX1-like auxin influx carrier activity to the infected cells in the in silico model, the dynamic of auxin distribution was tested in the three virtual tissues. Simulations were conducted with either auxin initially homogeneously distributed in the tissue or with auxin coming from the outside of the tissue (as would be the case if auxin was provided by the vascular stream). In all our simulations, the addition of AUX1 activity in infected cells was not sufficient to generate a significant auxin accumulation in infected cells (Fig. 2B; Supplemental Fig. S3). This result suggests that other components are needed to generate a specific auxin accumulation in Frankia-infected cells.
A search of a C. glauca EST database (Hocher et al., 2006), currently containing around 35,000 ESTs, for other auxin transport proteins that might be active in nodules identified one EST corresponding to an auxin efflux carrier of the PIN family (EST CG-R02f_005_F13 homologous to PIN1 and named CgPIN1 thereafter) and 20 ESTs corresponding to the previously described putative auxin influx carriers CgAUX1 and CgLAX3 (Péret et al., 2007). Expression analyses showed that CgPIN1 and CgLAX3 were down-regulated 2.5 and more than 16 times, respectively, in nodules compared to uninfected roots (Fig. 2C). CgAUX1 expression was not significantly different in nodules compared to uninfected roots (Fig. 2C). In conclusion, we identified genes encoding an auxin influx carrier (CgAUX1) and a putative auxin efflux carrier (CgPIN1) expressed in C. glauca nodules.
To determine the localization of PIN1-like proteins in C. glauca nodules, we performed immunolocalization experiments using polyclonal anti-AtPIN1 antibodies that have been previously shown to detect PIN1-like proteins in Arabidopsis and maize (Zea mays; Carraro et al., 2006). We found a strong signal occurring in the membrane of uninfected cortical cells surrounding Frankia-infected cells in C. glauca nodules (Fig. 2, D–G). This signal was strongest in the apical region of the nodule close to the nodule meristem (Fig. 2D) and appeared all around the periphery of cells, suggesting that auxin transport was not polarized (Fig. 2, F and H). Some signal was also found in the vasculature close to the meristem. Thus in C. glauca nodules, noninfected cortical cells expressed a gene encoding a PIN1-like auxin efflux carrier, while infected cells expressed a gene encoding an auxin influx carrier (CgAUX1). While we cannot rule out that other transporters might be involved in auxin transport in the cortical tissue, no other EST corresponding to an auxin carrier was found in our database that contains 35,000 C. glauca ESTs (including 15,000 ESTs from nodule).
The effects of these auxin influx and efflux carrier activities in infected and uninfected cells, respectively, were tested in our virtual symbiotic tissues model system. We found in our three in silico models that this cell-specific localization of auxin transporters was sufficient to cause a rapid auxin accumulation specifically in Frankia-infected cells (Fig. 2I; Supplemental Fig. S4) as was observed in our auxin immunolocalization experiments. This prediction was very robust and held for a wide range of parameter values (Supplemental Fig. S5).
Frankia Is a Potential Source of Auxin Production during the Symbiotic Association
In our models, if auxin was supplied from outside the tissue it accumulated in those infected cells that were close to the auxin source (Supplemental Fig. S4). This prediction is in disagreement with our immunolocalization results that showed strong signal in all of the infected cells irrespective of their position in the symbiotic tissues. This inconsistency suggests that auxin comes from within symbiotic tissues rather than from an external source.
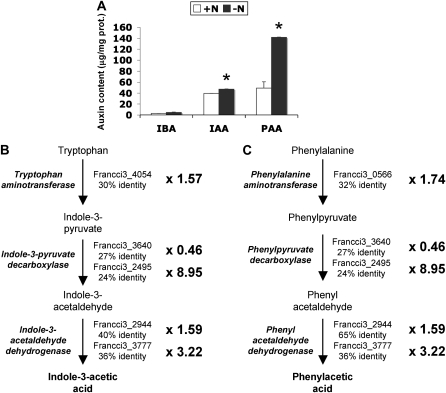
Auxin production has been demonstrated for several different Frankia strains, but not strain CcI3 (Wheeler et al., 1984; Berry et al., 1989; Hammad et al., 2003). IAA, IBA, and PAA production by the C. glauca-infective Frankia strain CcI3 was quantified by LC-MS analysis of the supernatant from in vitro cultures. Although Frankia CcI3 cultivated in BAP medium produced all three auxin types, IAA and PAA were the dominant forms of auxin and IBA was minor form (<2% of the pool; Fig. 3A). Frankia CcI3 growth in BAP medium without nitrogen source caused a significant increase in IAA and PAA production (Fig. 3A). A >3-fold increase in PAA production was detected under these conditions. These results indicate that auxin production by Frankia CcI3 in vitro is dependent on the nitrogen status and that PAA may be the major form biosynthesized.
Figure 3.
Auxin production by Frankia. A, Auxin content measured by LC-MS in the supernatant of in vitro Frankia CcI3 cultures. Bacteria were grown in BAP medium with (+N) or without (−N) nitrogen sources. Data are means ± sd. Significance was tested by an ANOVA test. * P < 0.01 compared with noninoculated. B to C, Putative IAA (B) and PAA (C) biosynthetic pathways identified in Frankia CcI3 using genome data mining. Frankia CcI3 orthologs were identified for the three key enzymes in the indole-3-pyruvate and the phenyl pyruvate pathways (see Supplemental Table S1). Expression of the corresponding genes in Frankia CcI3 bacteria grown in BAP medium with or without a nitrogen source was analyzed by qRT-PCR. Fold changes values represent the mean ratios of expression changes in BAP−N relative to BAP+N medium in two independent biological replicates.
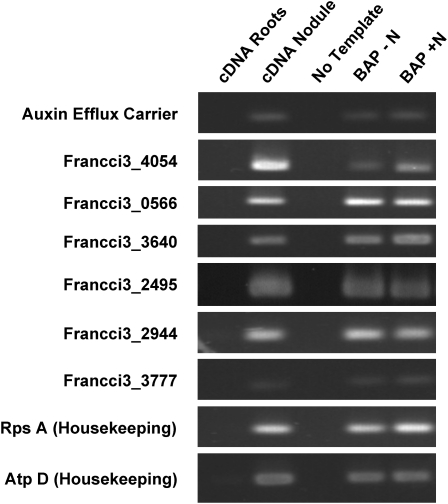
The availability of several Frankia genome databases including Frankia CcI3 (Normand et al., 2007) allowed a genome-mining approach to identify genes potentially involved in auxin biosynthesis and transport. All of the Frankia genomes contained predicted genes for complete indole-3-pyruvate and phenyl pyruvate pathways for IAA and PAA biosynthesis, respectively (Fig. 3B). Since many beneficial plant-associated bacteria preferentially use the indole-3-pyruvate pathway for IAA biosynthesis (Spaepen et al., 2007), this result is not surprising. Frankia CcI3 orthologs were identified for the three key enzymes in the indole-3-pyruvate and the phenyl pyruvate pathways. Interestingly, most of these genes were found by quantitative reverse transcription (qRT)-PCR to be overexpressed when bacteria were grown in BAP medium without nitrogen source (Fig. 3, B and C), i.e. under conditions that caused a significant increase in IAA and PAA production. A gene encoding a putative microbial auxin efflux carrier (Francci3_1249) was also identified from the automatic annotation of Frankia CcI3 and orthologs were found in all Frankia genomes. RT-PCR experiments showed that these genes were expressed in Frankia grown in vitro and in C. glauca nodules (Fig. 4), suggesting that Frankia IAA and PAA biosynthetic pathways are active in planta.
Figure 4.
Expression of Frankia genes in the putative IAA and PAA pathways. Expression of the corresponding genes was tested in culture and in plant tissues using RT-PCR. All of the Frankia CcI3 putative IAA and PAA biosynthetic genes were expressed in culture and in planta, but were not expressed in plant root tissues. Furthermore, the two housekeeping genes and the auxin efflux carrier were expressed under these conditions.
Auxin biosynthesis by Frankia was included in our in silico model as a source of auxin. This modification led to a rapid accumulation of auxin in all infected plant cells (Fig. 5). If CgPIN1 activity is removed in this new model, auxin accumulation occurs both in infected cells and in neighboring uninfected cells (Fig. 5). On the other hand, we did not observe any effect after removing CgAUX1 activity in the model. This result suggests that CgPIN1 is necessary to restrict auxin accumulation to plant cells infected by Frankia.
Figure 5.
Simulation of auxin distribution in virtual tissues with auxin synthesis by Frankia. We considered an additional intracellular compartment in infected cell in which Frankia would release auxin. Simulation showed that it would lead to auxin accumulation specifically in infected cells provided that CgPIN1 was expressed in the noninfected cell. Auxin activity is the relative auxin concentration within the cells and its representation is capped at value 1. The equations that govern diffusive and active transport are discussed in detail in Supplemental Text S1 (model notes).
DISCUSSION
Two nitrogen-fixing root nodule symbioses between plants and bacteria have been described, actinorhizal symbioses, formed by members of the Fagales, Rosales, and Cucurbitales with Frankia bacteria, and the interaction of legumes with rhizobia. Both symbioses involve intracellular accommodation of bacteria within host cells. While symbiotic signaling mechanisms have been widely studied in the legume-rhizobia symbiosis (Oldroyd et al., 2009) very little is known about signaling mechanisms involved in plant-bacteria recognition in actinorhizal symbioses. Previous studies have suggested a role for auxin in the establishment of actinorhizal symbioses (Hammad et al., 2003; Péret et al., 2007, 2008). Here, we report that auxin accumulates in cortical cells infected by Frankia in C. glauca nodules. This result is in agreement with the expression of the auxin-responsive EuNOD-ARP1 gene in Frankia-infected cells in E. umbellata actinorhizal nodules (Kim et al., 2007) and with the expression of an auxin-responsive AUX/IAA gene in Frankia-infected cells in C. glauca nodules (V. Vaissayre, A. Crabos, V. Hocher, A. Champion, and L. Laplaze, unpublished data). These results together further indicate that auxin-responsive genes are activated in infected cells and therefore that at least part of the auxin we detected using immunolocalization is active.
Results from a combination of experimental biology and computational modeling approaches indicate that this specific accumulation of auxin in Frankia-infected cell is driven by auxin import into infected cells by expression of AUX1 in these cells, and auxin export from uninfected cells by expression of a PIN1-like gene in those cells. Our data also suggest that Frankia produces auxins (both IAA and PAA) in planta, but our data do not allow us to conclude that Frankia is the only source of auxin in nodules. Although Frankia mutant strains defective in auxin biosynthesis would be very helpful to clarify this point, techniques to generate stable transgenic or mutant Frankia strains have not yet been established (Lavire and Cournoyer, 2003).
However, it is clear that the arrangement of auxin transporters limits the auxin response to those cells that are infected by Frankia and prevents the diffusion of this very active biomolecule to other nodule tissues or parts of the plant. This localization raises the question of the role of auxin in infected cells and we propose three potential functions. First, auxin accumulation in infected cells could drive cell growth and help explain why infected cells are hypertrophied. Another potential role for auxin is cell wall remodeling processes that occur during infection. The infection threads are surrounded by the plant cell membrane and a new cell wall-like structure composed mainly of pectin derivatives (Lalonde and Knowles, 1975). Auxin is known to regulate genes involved in cell wall remodeling, pectin biosynthesis, and methylation (Lerouxel et al., 2006; Swarup et al., 2008). Thus, auxin perception in infected plant cells might be necessary for the growth of the infection threads. Finally, auxin could be involved in preventing, controlling, or limiting the plant defense mechanisms. Some bacterial pathogens such as Pseudomonas syringae modulate auxin responses and/or production to colonize their host (Chen et al., 2007; Robert-Seilaniantz et al., 2007; Kazan and Manners, 2009). A similar process might occur in C. glauca cells infected by Frankia.
A role for auxin in infected cells might not be a general feature of nitrogen-fixation endosymbioses. While the role of auxin in legume nodule development is now well documented (Mathesius, 2008), little is known about its role in the infection process. Although legume infection by rhizobia strain deficient in IAA production or overproducing auxin causes changes in nodule development and nitrogen fixation (Pii et al., 2007; Camerini et al., 2008), the molecular markers of auxin perception ProDR5-GUS and ProGH3-GUS are not expressed in infected cells of legume nodules (Mathesius et al., 1998; Pacios-Bras et al., 2003) and no data on auxin immunolocalization are available. It thus remains to be shown whether auxin accumulation is a common feature of endosymbiotic infection of plant cells by nitrogen-fixing bacteria.
MATERIALS AND METHODS
Plant and Bacterial Material
Casuarina glauca seeds were provided by Carter Seeds and grown as described in Péret et al. (2007). Transgenic C. glauca and Allocasuarina glauca plants were produced and analyzed as described (Péret et al., 2007). Frankia strain CcI3 was grown and used to inoculate C. glauca, as described (Péret et al., 2007).
Auxin Quantification
Frozen samples were ground in liquid nitrogen and auxins were extracted twice for 2 h at 4°C in the dark in 80% (v/v) methanol added with 1% butylated hydroxytoluene. For auxin quantification, 100 pmol of [2H6]IAA and 100 pmol of [13C]PAA (Euriso-Top) were added to the samples. After centrifugation (13,000g, 20 min, 4°C), the supernatants were collected and passed through a C18 cartridge (Waters). Afterward, auxins were purified using a DEAE-Sephadex A25 cartridge (formic acid conditions; Amersham Pharmacia) coupled to C18 cartridge as described in Prinsen et al. (2000). Auxins were directly eluted from the C18 column with ethyl ether. Mass analysis was conducted using a Quattro LC with an ESI Z-spray interface (MicroMass), MassLynx software, an Alliance 2695 RP-HPLC system (Waters), and a Waters 2487 UV detector set at 280 nm. An Xterra C18 column (100 × 2 mm, 5 μm, Waters) was used with a mix comprising solvent A water with 15 mm formic acid and solvent B methanol with a gradient profile (starting with 60:40, A/B, v/v, for 2 min; linear gradient up to 0:100, A/B over 15 min; a washing step 0:100, A/B for 5 min, and final equilibration at 60:40, A/B for 5 min) at a 0.3 mL/min flow rate. Chromatograms were analyzed using the Masslynx software (Waters), and the IAA, PAA, and IBA concentrations were calculated according to the principle of isotope dilution (Prinsen et al., 1998). Samples were quantified in single reaction monitoring mode based on the area of major MS signals ([M-H+]).
Auxin Immunolocalization
For IAA and PAA immunolocalization, fresh nodules sections of 50 to 60 μm were obtained with a vibratome (MicroM HM650V) and were fixed overnight, in fixative solution (4% w/v 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride [EDAC, Sigma], 4% w/v paraformaldehyde [Fluka] in phosphate-buffered saline [10 mm pH 7.4]) at 4°C. Sections were incubated overnight in 2% blocking buffer solution (Roche) at 4°C followed by overnight incubation with primary antibody, anti-IAA mouse monoclonal hybridoma antibody (Agdia) at a concentration of 0.05 mg/mL, or anti-PAA rabbit polyclonal antibody (Abcam) at 1:200 dilution. Sections were then rinsed (three times 15 min) in phosphate-buffered saline (10 mm pH 7.4). Alexa Fluor 488 goat anti-mouse IgG1 (γ1) antibody (2 mg/mL; Invitrogen) for IAA or Alexa Fluor 488 goat anti-rabbit IgG antibody (2 mg/mL; Invitrogen) for PAA was used as a secondary antibody at 1:250 dilution at room temperature during 1 h in the dark. After rinsing (five times for 10 min each), sections were mounted in Mowiol (Calbiochem) and examined using a confocal microscope 510 META Zeiss (Laser Argon 488, BP 505–530 and LP 560 nm) with LCI Neo-Neofluar 25×/0.8 1 mm Korr DIC27 and EC Plan-Neofluar 10×/0.3 objectives. For the ethanol controls, sections were pretreated with ethanol for 1 h at each concentration (70% v/v and 80% v/v in phosphate-buffered saline 10 mm pH 7.4) prior to fixation to extract IAA or PAA from the nodule sections. To verify the binding specificity of the primary anti-IAA mouse monoclonal antibody was mixed with 10 times its concentration of IAA overnight. This was to allow the binding of the free IAA to the primary antibody. The solution was applied to the fixed sections, i.e. the bound primary antibody would not be able to bind to the antigen on the sections. The same was done to test the primary anti-PAA rabbit polyclonal antibody against PAA. Another control treatment was also done whereby the secondary antibody was used alone. Nodules from 24 and 16 plants from four independent inoculation experiments were used for IAA and PAA immunolocalization, respectively.
PIN1 Immunolocalization
Fresh nodules sections of 50 to 60 μm were obtained with a vibratome (MicroM HM650V) and were fixed for 1 h in fixative solution (4% w/v paraformaldehyde [Fluka] in phosphate-buffered saline 10 mm pH 7.4) under vacuum at room temperature. Sections were incubated overnight in 2% blocking buffer solution (Roche) at 4°C followed by overnight incubation with primary antibody, anti-PIN1 goat polyclonal antibody (200 μg/mL; Santa Cruz Biotechnology) at 1:200 dilution. Sections were rinsed three times (15 min) in phosphate-buffered saline 10 mm pH 7.4. Alexa Fluor 488 F(ab’)2 fragment of rabbit anti-goat IgG (H + L) antibody (2 mg/mL; Invitrogen) was used as a secondary antibody at 1:250 dilution at room temperature during 1 h in the dark. After rinsing, sections were mounted in Mowiol (Calbiochem) and examined using a Zeiss confocal microscope 510 META (Laser Argon 488, BP 505–530 and LP 560 nm) with LCI Neo-Neofluar 25×/0.8 1 mm Korr DIC27 and EC Plan-Neofluar 10×/0.3 objectives. For the control treatment, the primary antibody was not used at all.
Gene Expression Analyses
C. glauca gene expression analyses were performed by qRT-PCR using specific primers (Supplemental Table S2) as described (Gherbi et al., 2008). Reactions were preformed in triplicates and the comparative threshold-cycle method was used to quantify gene expression. The results were standardized with CgUBI expression levels (Gherbi et al., 2008).
Total RNA was extracted from Frankia as described previously (Niemann and Tisa, 2008). To study Frankia gene expression in planta, total RNA was extracted using the RNeasy plant mini kit (Qiagen GmbH). RNA was quantified with a NanoDrop (ThermoFisher). Four-hundred nanograms of total RNA were reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and random hexamers primers. The RT-PCR was preformed using AmpliTaq Gold 360 master mix (Applied Biosystems) and 100 ng of cDNA as a template. qPCR was performed using SYBR Green PCR master mix (Applied Biosystems) and specific primer sets (Supplemental Table S2). Parameters for the Applied Biosystems 7300 were as follows: (1) one cycle 95°C 10 min, (2) 40 cycles 95°C for 15 s and 60°C for 30 s, ended by one cycle at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s run. Reactions were preformed in triplicates and the comparative threshold-cycle method was used to quantify gene expression. The results were standardized with rpsA expression levels. For RT-PCR, the thermocycler parameters were as follows: (1) initial denaturation at 95°C 5 min, (2) 35 cycles of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, and primer extension at 72°C for 30 s, and (3) a final extension step at 72°C 10 min. Amplicons were resolved by gel electrophoresis.
Modeling
Tissues were manually digitized from confocal images to standard vector graphic files (SVG) and automatically converted for use in the OpenAlea modeling software platform (Pradal et al., 2008). The models take into account cells and apoplastic compartments for computing the auxin transport (see Supplemental Text S1 for additional details). The model parameters (membrane permeability and active transports, auxin dissociation constant, cell and apoplastic pH, cell membrane potential) were defined in accordance with values from previously published studies (see Supplemental Text S1). Computational modeling was based on IAA transport parameters as they were the only auxin transport parameters that are well documented. However, our model is an abstraction of the physiology of the nodule, and as such is not tied to an auxin in particular. Similar results would be obtained for PAA assuming that IAA and PAA transport physiology are similar.
Bioinformatics Studies
The FASTA amino acid sequences of the three Frankia genomes (CcI3, National Center for Biotechnology Information [NCBI] RefSeq: NC_007777; ACN14a, NCBI RefSeq: NC_00827; EAN1pec, NCBI RefSeq: NC_009921 and from the three Frankia draft genomes [EuI1c, http://genome.ornl.gov/microbial/fran_eui1c/; EUN1f, http://genome.ornl.gov/microbial/fran_eun1f/; the uncultured Frankia symbiont of Datisca glomerata http://genome.ornl.gov/microbial/fran_sym/]) were obtained from GenBank or the Department of Energy Joint Genome Institute Genome Portal site (http://genome.jgi-psf.org/). Functionally analyzed genes for indole acetic acid and phenyl acetic acid biosynthesis pathways were identified by the use of Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto, 2000) and published literature (Patten and Glick, 1996; Spaepen et al., 2007). Proteins representing different biosynthesis pathways were used (Supplemental Table S1). BLASTP analyses were performed against each Frankia genome database using each of the above protein sequences as a query sequence for the representative IAA and PAA biosynthesis pathways.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FQ375841 (CgPIN1) and NC_007777.1 (Frankia sp. CcI3 genome).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression pattern of the ProIAA2-GUS molecular markers for auxin perception in C. glauca.
Supplemental Figure S2. Topology and digitization of symbiotic tissues.
Supplemental Figure S3. Simulation of auxin distribution in virtual tissues with infected cells expressing CgAUX1.
Supplemental Figure S4. Simulation of auxin distribution in virtual tissues with infected cells expressing CgAUX1 and noninfected cells expressing CgPIN1.
Supplemental Figure S5. Robustness of auxin distribution against CgPIN1 perturbation.
Supplemental Table S1. Genome data mining results.
Supplemental Table S2. Primers used for gene expression analyses.
Supplemental Text S1. Model description.
Supplementary Material
Acknowledgments
We thank Pr. K. Ljung (Swedish University of Agricultural Sciences, Umea, Sweden) for providing us with [2H6]IAA, Pr. T. Guilfoyle (University of Missouri) for the ProDR5-GUS and ProGH3-GUS constructs, and Dr. R. Swarup (University of Nottingham, UK) for the ProIAA2-GUS construct. We are grateful to Dr. D. Bogusz (Institut de Recherche pour le Développement, France), Dr. A. Champion (Institut de Recherche pour le Développement, France), Dr. S. Guyomarc’h (Université Montpellier 2, France), and Dr. C. Godin (Institut National de Recherche en Informatique et Automatique, France) for helpful discussions and critical reading of this manuscript.
References
- Aloni R, Schwalm K, Langhans M, Ullrich CI. (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216: 841–853 [DOI] [PubMed] [Google Scholar]
- Benson DR, Silvester WB. (1993) Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev 57: 293–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry AM, Kahn RKS, Booth MC. (1989) Identification of indole compounds secreted by Frankia HFPAri3 in defined culture-medium. Plant Soil 118: 205–209 [Google Scholar]
- Camerini S, Senatore B, Lonardo E, Imperlini E, Bianco C, Moschetti G, Rotino GL, Campion B, Defez R. (2008) Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch Microbiol 190: 67–77 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S. (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cérémonie H, Cournoyer B, Maillet F, Normand P, Fernandez MP. (1998) Genetic complementation of rhizobial nod mutants with Frankia DNA: artifact or reality? Mol Gen Genet 260: 115–119 [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V, Roeder AH, Tarr PT, Cunha A, Tobin C, Meyerowitz EM. (2010) Computational morphodynamics: a modeling framework to understand plant growth. Annu Rev Plant Biol 61: 65–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuille PB, Bohn-Courseau I, Ljung K, Morin H, Carraro N, Godin C, Traas J. (2006) Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci USA 103: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Péret B, Laplaze L, Franche C, et al. (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105: 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Van Isterdael G, Beeckman T, Gheysen G, Mathesius U. (2009) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Martin G, Li Y, Guilfoyle TJ. (1991) Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol 17: 567–579 [DOI] [PubMed] [Google Scholar]
- Hammad Y, Nalin R, Marechal J, Fiasson K, Pepin R, Berry AM, Normand P, Domenach A-M. (2003) A possible role for phenyl acetic acid (PAA) on Alnus glutinosa nodulation by Frankia. Plant Soil 254: 193–205 [Google Scholar]
- Hocher V, Auguy F, Argout X, Laplaze L, Franche C, Bogusz D. (2006) Expressed sequence-tag analysis in Casuarina glauca actinorhizal nodule and root. New Phytol 169: 681–688 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Kim HB, Lee H, Oh CJ, Lee NH, An CS. (2007) Expression of EuNOD-ARP1 encoding auxin-repressed protein homolog is upregulated by auxin and localized to the fixation zone in root nodules of Elaeagnus umbellata. Mol Cells 23: 115–121 [PubMed] [Google Scholar]
- Lalonde M, Knowles R. (1975) Ultrastructure, composition, and biogenesis of encapsulation material surrounding endophyte in Alnus crispa var Mollis root nodules. Can J Bot 53: 1951–1971 [Google Scholar]
- Lavire C, Cournoyer B. (2003) Progress on the genetics of the N-2-fixing actinorhizal symbiont Frankia. Plant Soil 254: 125–137 [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. (2006) Biosynthesis of plant cell wall polysaccharides—a complex process. Curr Opin Plant Biol 9: 621–630 [DOI] [PubMed] [Google Scholar]
- Mathesius U. (2008) Auxin: at the root of nodule development? Funct Plant Biol 35: 651–668 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA. (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Niemann J, Tisa LS. (2008) Nitric oxide and oxygen regulate truncated hemoglobin gene expression in Frankia strain CcI3. J Bacteriol 190: 7864–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, et al. (2007) Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res 17: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Harrison MJ, Paszkowski U. (2009) Reprogramming plant cells for endosymbiosis. Science 324: 753–754 [DOI] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HR, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP. (2003) Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR. (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42: 207–220 [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T. (1996) Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell 8: 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Svistoonoff S, Lahouze B, Auguy F, Santi C, Doumas P, Laplaze L. (2008) A role for auxin during actinorhizal symbioses formation? Plant Signal Behav 3: 34–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup R, Jansen L, Devos G, Auguy F, Collin M, Santi C, Hocher V, Franche C, Bogusz D, et al. (2007) Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii Y, Crimi M, Cremonese G, Spena A, Pandolfini T. (2007) Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C. (2008) OpenAlea: a visual programming and component-based software platform for plant modelling. Funct Plant Biol 35: 751–760 [DOI] [PubMed] [Google Scholar]
- Prinsen E, Van Dongen W, Esmans EL, Van Onckelen HA. (1998) Micro and capillary liquid chromatography tandem mass spectrometry: a new dimension in phytohormone research. J Chromatogr A 826: 25–37 [Google Scholar]
- Prinsen E, Van Laer S, Oden S, Van Onckelen H. (2000) Auxin analysis. Methods Mol Biol 141: 49–65 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler CT, Crozier A, Sandberg G. (1984) The biosynthesis of indole-3-acetic-acid by Frankia. Plant Soil 78: 99–104 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.